Cell Browser best practices: Difference between revisions
(→Providing the Unit for datasets: adding a bit more detail about Seurat units) |
(→Providing the Unit for datasets: even more seurat unit details.) |
||
| Line 68: | Line 68: | ||
Provide the unit of the values used in the expression matrix. Typical values: "read count/UMI", "log of read count/UMI", "TPM", "log of TPM", "CPM", "FPKM", "RPKM". | Provide the unit of the values used in the expression matrix. Typical values: "read count/UMI", "log of read count/UMI", "TPM", "log of TPM", "CPM", "FPKM", "RPKM". | ||
For Seurat objects, the 'counts' slot is typically 'UMI count' for 10x data or 'read count' for Smart-seq2 or similar assays. The 'data' slot is the log-normalized version of the counts slot. This Github issue has some details: https://github.com/satijalab/seurat/issues/3711. | For Seurat objects, the 'counts' slot is typically 'UMI count' for 10x data or 'read count' for Smart-seq2 or similar assays. The 'data' slot is the log-normalized version of the counts slot. This Github issue has some details: https://github.com/satijalab/seurat/issues/3711. For SCT assay datasets, it's slightly different: https://satijalab.org/seurat/reference/sctransform; in short though, the units are: counts -> (corrected) counts, data -> log1p(counts), scale.data -> pearson residuals. | ||
It's probably easiest to ask the authors if you're unsure. | It's probably easiest to ask the authors if you're unsure. | ||
Revision as of 20:13, 26 August 2022
Best Practices
Formatting configuration files
- Typically you should keep a maximum of 80-120 characters per line; you can use
gqgqin VIM in visual mode to auto format a paragraph into multiple ~80 character lines - For special characters, please refer to HTML character encoding: https://ascii.cl/htmlcodes.htm
cellbrowser.conf
Putting things into cellbrowser-confs repo
From inside a dataset directory:
git add desc.conf cellbrowser.conf git commit -m “message” git push
Only do this for public datasets. For additional help you can refer to Commit cellbrowser/desc.conf files.
Naming datasets
Dataset names should be:
- all lowercase
- 4 words or less
- less than 20 characters and separated by hyphens
The names need to be lowercase because the Cell Browser website code converts all names lowercase. There are only a few exceptions for early datasets (e.g. adultPancreas).
Layout Coordinates
- Capitalize
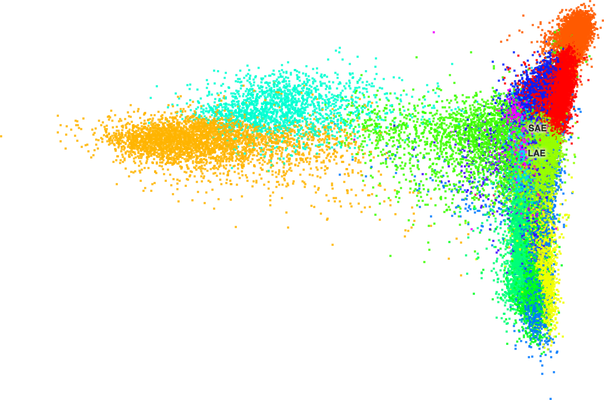
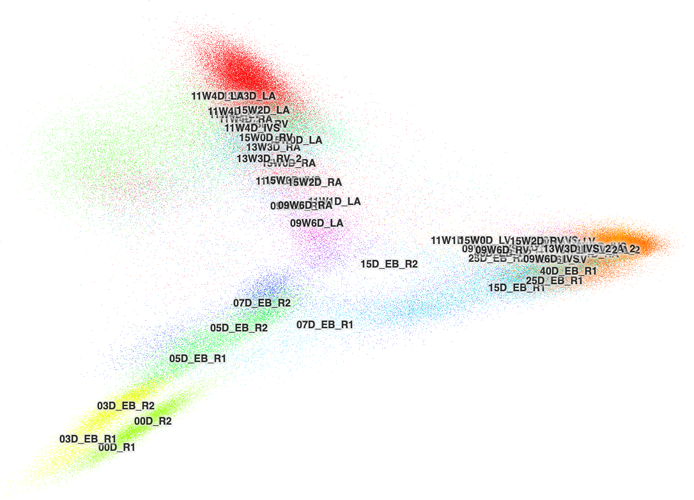
"UMAP"and"tSNE". - Remove extra layout coordinates (e.g. PCA or Harmony) because the cbImportTools export all of the possible layouts and they export only the only the first two coordinates. The CB can only handle two coordinates and so these layouts often look like a clump of cells.
The following two images are examples of PCA plots. For reference, the first image is from the "lung-airway" dataset and the second image is from the "hoc" dataset.
Finding a paper associated with a bioRxiv pub
Sometimes you will have to go back and edit the paper citation for a dataset.
In /hive/data/inside/cells/datasets run find . -name desc.conf | xargs grep "biorxiv" |grep -v "Strange\|\#" Should get results like: ./cbl-dev/desc.conf:biorxiv_url = "https://www.biorxiv.org/content/10.1101/2020.06.30.174391v1 Aldinger et al. 2020. bioRxiv." Copy this bit of the URL: 10.1101/2020.06.30.174391 And feed it to the bioRxiv API: curl https://api.biorxiv.org/details/biorxiv/10.1101/2020.06.30.174391 In the response, you should see the word "published" and if it's published, it'll have a doi otherwise it'll just say NA.
This is referenced from the Cells Redmine To Do #27316.
You could also paste the DOI into doi.org.
Providing the Unit for datasets
unit=""
Provide the unit of the values used in the expression matrix. Typical values: "read count/UMI", "log of read count/UMI", "TPM", "log of TPM", "CPM", "FPKM", "RPKM".
For Seurat objects, the 'counts' slot is typically 'UMI count' for 10x data or 'read count' for Smart-seq2 or similar assays. The 'data' slot is the log-normalized version of the counts slot. This Github issue has some details: https://github.com/satijalab/seurat/issues/3711. For SCT assay datasets, it's slightly different: https://satijalab.org/seurat/reference/sctransform; in short though, the units are: counts -> (corrected) counts, data -> log1p(counts), scale.data -> pearson residuals.
It's probably easiest to ask the authors if you're unsure.
desc.conf
Most commonly used desc.conf settings to keep consistent:
title = "First word is capitalized and the rest is all lowercased"
paper_url = “http://www.paper_url.com/xxx Last name et al. Journal. Year.”
biorxiv_url = "https://www.biorxiv.org/content/123/123.full Last name et al. bioRxiv Year."
The additional database links (GSE, Bioproject, SRA accessions, PMID, etc.) can be set to just the number, no author info:
pmid = "12343234" geo_series = "GSE25097" dbgap = "phs000424.v7.p2" arrayexpress = "xxx" sra_study = "xxxx"