Wrangling process: Difference between revisions
(Stub of this page. More sections to come.) |
(Adding a few more sections to page.) |
||
| Line 3: | Line 3: | ||
For a collection, most steps will apply to each dataset in that collection, however, a few (e.g. ‘Respond to submitters’) only apply to the collection as a whole. | For a collection, most steps will apply to each dataset in that collection, however, a few (e.g. ‘Respond to submitters’) only apply to the collection as a whole. | ||
=Respond to submitters= | ==Respond to submitters== | ||
This step only applies to ‘live wrangled’ datasets. Researchers will email us at cells@ucsc.edu requesting that we host their data. When you respond to them, do these NN things in your response (unless they’ve already mentioned them): | This step only applies to ‘live wrangled’ datasets. Researchers will email us at cells@ucsc.edu requesting that we host their data. When you respond to them, do these NN things in your response (unless they’ve already mentioned them): | ||
| Line 47: | Line 47: | ||
::Thanks! | ::Thanks! | ||
=Make a directory= | ==Make a directory== | ||
Make a directory with the dataset short name in /hive/data/inside/cells/datasets. The submitters should have supplied you with one since it’s mentioned on the submission guidelines page. If not, you can ask them if they had a short name in mind and share the short name requirements with them. You will most likely have to adjust their suggested name. | Make a directory with the dataset short name in /hive/data/inside/cells/datasets. The submitters should have supplied you with one since it’s mentioned on the submission guidelines page. If not, you can ask them if they had a short name in mind and share the short name requirements with them. You will most likely have to adjust their suggested name. | ||
| Line 73: | Line 73: | ||
(You may notice that there are quite a few datasets that don’t seem to follow these guidelines. These were created before we established these rules and are ‘grandfathered’ in. '''You can’t change a short name once it’s been published to the main site.''') | (You may notice that there are quite a few datasets that don’t seem to follow these guidelines. These were created before we established these rules and are ‘grandfathered’ in. '''You can’t change a short name once it’s been published to the main site.''') | ||
=Download files= | ==Make entry in spreadsheet== | ||
==Download files== | |||
Within the directory made in the last step, make an ‘orig’ directory - place ‘original’ files there. The files downloaded to orig should remain (mostly) unchanged from those you downloaded. | Within the directory made in the last step, make an ‘orig’ directory - place ‘original’ files there. The files downloaded to orig should remain (mostly) unchanged from those you downloaded. | ||
| Line 94: | Line 96: | ||
(If you do need to go this route, it’s probably best to do this while on the UCSC network to save your own bandwidth.) | (If you do need to go this route, it’s probably best to do this while on the UCSC network to save your own bandwidth.) | ||
== Import data == | |||
You will use different utilities depending on your starting files: | |||
* [https://cellbrowser.readthedocs.io/en/master/howto.html#how-to-create-a-cell-browser-using-a-scanpy-h5ad-file cbImportScanpy] for h5ad or loom | |||
** Use h5adMetaInfo to find an input field for the -c/–clusterField option | |||
** cbImportScanpy has some default fields hardcoded (so you can skip -c in these cases): | |||
::["CellType", "cell_type", "Celltypes", "Cell_type", "celltype", "annotated_cell_identity.text", "BroadCellType", "Class"] | |||
* cbImportSeurat for RDS, Rdata, or Robj | |||
** Use rdsMetaInfo to find an input field for the -c/–clusterField option | |||
** [https://cellbrowser.readthedocs.io/en/master/howto.html#how-to-create-a-cell-browser-using-a-seurat-rds-file cbImportSeurat] defaults to active.ident, so if that looks sufficient, it may not be necessary to use -c. | |||
* For tsv/csv files, you will be starting with a matrix file, metadata, and layout coordinates. | |||
** Use tabInfo on meta.tsv to find a field to use as the default color/label fields | |||
** Create a default cellbrowser.conf with cbBuild --init then adjust the default file names as needed | |||
** If the submitter provided cluster markers, use those. If not, generate them using cbScanpy [link to other section] | |||
If you need to generate the UMAP/tSNE coordinates, use [cbScanpy]. | |||
==Commit cellbrowser/desc.conf files== | |||
This is only for public datasets (i.e. those without visibility=”hide” in their cellbrowser.conf). | |||
The [https://github.com/ucscGenomeBrowser/cellbrowser-confs cellbrowser-confs] repo houses the configuration files for all of the public datasets in the Cell Browser. Add your cellbrowser.conf and desc.conf files to this repo early so that you can track the changes that you and others make throughout the submission process. | |||
git add cellbrowser.conf desc.conf | |||
git commit -m “Initial commit of cellbrowser.conf and desc.conf files for BLAH dataset” | |||
For a collection, you will need to commit the desc.conf and cellbrowser.conf for each dataset in that collection, either individually or all at once, such as: | |||
git add cellbrowser.conf desc.conf all-tissues/desc.conf all-tissues/cellbrowser.conf immune/cellbrowser.conf immune/desc.conf | |||
==Annotate marker genes (human-only)== | |||
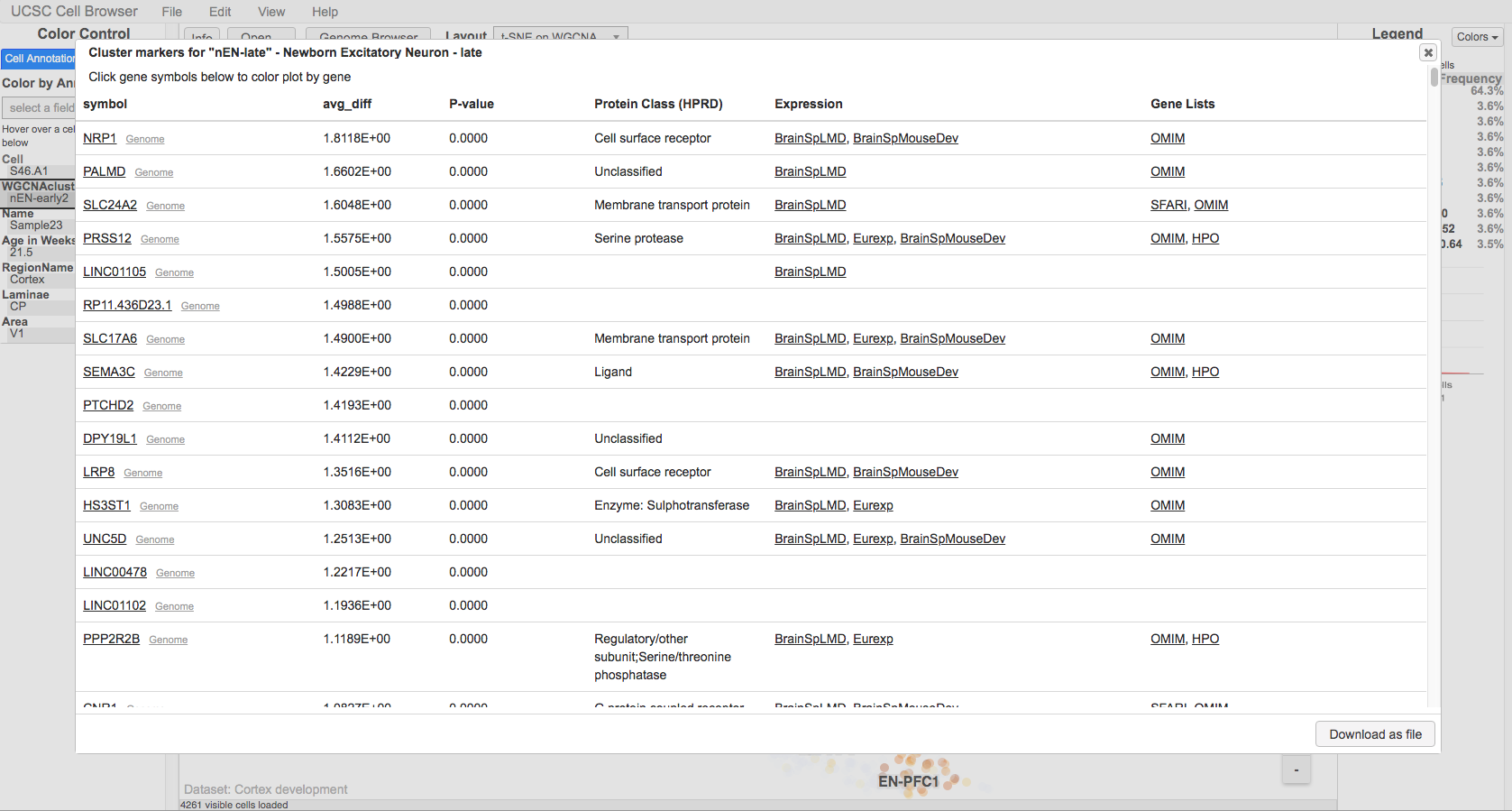
Annotating the marker genes file will add linkouts to the marker gene pop-up to a number of different resources, such as OMIM: | |||
[[File:cellbrowserAnnotatedMarkerGenes.png]] | |||
To annotate the marker genes run: | |||
cbMarkerAnnotate markers.tsv markers.annotated.tsv | |||
This places the annotated marker genes into a new file called markers.annotated.tsv. Be sure to update the ‘markers’ line in the cellbrowser.conf to point to this new file. | |||
[[Category:Cell Browser]] | [[Category:Cell Browser]] | ||
Revision as of 15:57, 23 May 2022
This page is intended to serve as a set of guidelines for wrangling a dataset into the Cell Browser, both those from archives (e.g. GEO) or those submitted to us by an external collaborator (aka live wrangling). It should be noted that this list is not comprehensive; there may be things that need to be done that aren’t covered here, or you might skip steps that aren’t relevant to your current dataset.
For a collection, most steps will apply to each dataset in that collection, however, a few (e.g. ‘Respond to submitters’) only apply to the collection as a whole.
Respond to submitters
This step only applies to ‘live wrangled’ datasets. Researchers will email us at cells@ucsc.edu requesting that we host their data. When you respond to them, do these NN things in your response (unless they’ve already mentioned them):
- Let them know we can host it
- Point them to the submission guidelines
- Ask if this is for a publication (so you can gauge their timeline)
- Ask if they want the dataset hidden
It’s best to respond to these emails within 24-48 hrs of receiving them.
Example emails/responses:
- Zhiwei Li, NN dataset
- Their email:
- Dear Sir or Madam,
- I have a single cell dataset of mouse lung in allergic asthma, and the relevant paper is accepted to be published in Allergy,
- I have set a UCSC cell browser in my local computer, and I want share the single cell data to the the website http://cells.ucsc.edu for public access, please tell me how to do it. Thank you.
- Best wishes,
- Dr. Zhiwei Li
- Our response:
- Hello, Zhiwei.
- We would be happy to host your data on the UCSC Cell Browser. Please take a look at our submission guidelines: https://cellbrowser.readthedocs.io/en/master/submission.html. Let us know if you have any questions.
- Thank you!
- Angela Ting, NN dataset
- Their email:
- To whom this may concern,
- We are preparing to resubmit our manuscript containing normal human ureter single-cell data to Developmental Cell (https://www.biorxiv.org/content/10.1101/2021.12.22.473889v1). The raw data and expression matrix have already been accepted by GEO, but we would like to deposit this data with UCSC cell browser to enable convenient access/utilization by the broader scientific community.
- Please advise.
- Our response
- Hi, Angela.
- We'd be happy to host your data on the Cell Browser. Please review this page for more information about submitting data: https://cellbrowser.readthedocs.io/en/master/submission.html. After you've prepared everything for submission, feel free to share the required files and we can get started on the import. Let us know if you have any questions about the process!
- Thanks!
Make a directory
Make a directory with the dataset short name in /hive/data/inside/cells/datasets. The submitters should have supplied you with one since it’s mentioned on the submission guidelines page. If not, you can ask them if they had a short name in mind and share the short name requirements with them. You will most likely have to adjust their suggested name.
If it’s a dataset you’re wrangling from the archives, you will have to think of a short name that captures the main idea of the dataset while adhering to our requirements.
Short name requirements:
- 4 words or less
- All lowercase
- Separate words with “-”
- Aim for 20 characters or less
Some common shortenings/contractions we use:
- dev for developing
- org for organoids
- vasc for vascular
Some examples of good short names:
- tabula-sapiens
- mouse-dev-brain
- mouse-gastrulation
- hgap
- covid19-brain
(You may notice that there are quite a few datasets that don’t seem to follow these guidelines. These were created before we established these rules and are ‘grandfathered’ in. You can’t change a short name once it’s been published to the main site.)
Make entry in spreadsheet
Download files
Within the directory made in the last step, make an ‘orig’ directory - place ‘original’ files there. The files downloaded to orig should remain (mostly) unchanged from those you downloaded.
The easiest way of downloading a file is via aria2c:
aria2c https://ftp.ncbi.nlm.nih.gov/geo/series/GSE133nnn/GSE133345/suppl/GSE133345_Annotations_of_all_1231_embryonic_cells_updated_0620.txt.gz aria2c -o TS_germ_line.h5ad.gz 'https://figshare.com/ndownloader/files/34702051'
In this second example, the -o option allows us to specify a name for the final file, rather than wget’s default of assigning the name based on the last part of the URL ('34702051' in this case).
If you have multiple files, place all of the URLs into a single file and use the ‘-i’ option:
aria2c -i my_files.lst
The utility rclone is another option for downloading files, though it does take some effort to set up. See our internal instructions. Once you have it set up, it is fairly easy to use (quite similar to .
If all else fails, you may need to download files to your computer and then upload those to hgwdev using scp:
scp <files> <uname>@hgwdev.gi.ucsc.edu:/hive/data/inside/cells/datasets/<dname>/orig
(If you do need to go this route, it’s probably best to do this while on the UCSC network to save your own bandwidth.)
Import data
You will use different utilities depending on your starting files:
- cbImportScanpy for h5ad or loom
- Use h5adMetaInfo to find an input field for the -c/–clusterField option
- cbImportScanpy has some default fields hardcoded (so you can skip -c in these cases):
- ["CellType", "cell_type", "Celltypes", "Cell_type", "celltype", "annotated_cell_identity.text", "BroadCellType", "Class"]
- cbImportSeurat for RDS, Rdata, or Robj
- Use rdsMetaInfo to find an input field for the -c/–clusterField option
- cbImportSeurat defaults to active.ident, so if that looks sufficient, it may not be necessary to use -c.
- For tsv/csv files, you will be starting with a matrix file, metadata, and layout coordinates.
- Use tabInfo on meta.tsv to find a field to use as the default color/label fields
- Create a default cellbrowser.conf with cbBuild --init then adjust the default file names as needed
- If the submitter provided cluster markers, use those. If not, generate them using cbScanpy [link to other section]
If you need to generate the UMAP/tSNE coordinates, use [cbScanpy].
Commit cellbrowser/desc.conf files
This is only for public datasets (i.e. those without visibility=”hide” in their cellbrowser.conf).
The cellbrowser-confs repo houses the configuration files for all of the public datasets in the Cell Browser. Add your cellbrowser.conf and desc.conf files to this repo early so that you can track the changes that you and others make throughout the submission process.
git add cellbrowser.conf desc.conf
git commit -m “Initial commit of cellbrowser.conf and desc.conf files for BLAH dataset”
For a collection, you will need to commit the desc.conf and cellbrowser.conf for each dataset in that collection, either individually or all at once, such as:
git add cellbrowser.conf desc.conf all-tissues/desc.conf all-tissues/cellbrowser.conf immune/cellbrowser.conf immune/desc.conf
Annotate marker genes (human-only)
Annotating the marker genes file will add linkouts to the marker gene pop-up to a number of different resources, such as OMIM:
To annotate the marker genes run:
cbMarkerAnnotate markers.tsv markers.annotated.tsv
This places the annotated marker genes into a new file called markers.annotated.tsv. Be sure to update the ‘markers’ line in the cellbrowser.conf to point to this new file.