Cell Browser wrangling guided examples: Difference between revisions
(→Part 1: Directory setup and data export: fixing image formatting) |
No edit summary |
||
| Line 282: | Line 282: | ||
====What next?==== | ====What next?==== | ||
Make other changes to the cellbrowser.conf and desc.conf files to see how they affect the display. What does changing radius or alpha in cellbrowser.conf do? What about adding information for the lab or institution to the desc.conf? (Don't forget to rebuild the dataset between those changes!) | Make other changes to the cellbrowser.conf and desc.conf files to see how they affect the display. What does changing radius or alpha in cellbrowser.conf do? What about adding information for the lab or institution to the desc.conf? (Don't forget to rebuild the dataset between those changes!) | ||
[[Category:Cell Browser]] | [[Category:Cell Browser]] | ||
[[Category:Cell Browser Onboarding]] | |||
Latest revision as of 16:36, 20 July 2022
This page will walk you through the basics of wrangling two cell browsers using the two main tools: cbImportSeurat and cbImportScanpy. The examples will be divided into three parts, each roughly corresponding to the stages of wrangling a dataset for the Cell Browser. All command-line steps are done on hgwdev.
Using cbImportScanpy
This section is intended to teach you the basics of using cbImportScanpy and how it fits into the wrangling process in general. To do so, we will be importing data from the h5ad file for the liver segment of Tabula Sapiens. We will use an h5ad file, which is written out using the python package AnnData and (almost) always compatible with cbImportScanpy.
Part 1: Directory setup and data export
In this first section, we will go through the process of setting up a directory in which you will download and then import the data for a cell browser.
Ensure that you are in the proper conda environment:
conda activate scanpyenv
Change into a good working directory:
cd /hive/users/${hgwdev_username}/cb
Create a directory for this dataset:
mkdir -p tabula-sapiens-liver/orig/
This command also makes an ‘orig’ directory. In the Cell Browser, we use this to store the unchanged files obtained from the submitter or downloaded from GEO/etc.
Change into that directory:
cd tabula-sapiens-liver/orig/
Copy over the h5ad file we’ll be working with:
cp /hive/data/inside/cells/exampleDatasets/TS_Liver.h5ad .
Determine what field to use as the input for the cluster field option:
h5adMetaInfo TS_Liver.h5ad
The cell_ontology_class field seems like it contains cell names that are derived from an ontology, a standardized, controlled set of names.
Go up a directory and export the data from that file:
cd ../ cbImportScanpy -i orig/TS_Liver.h5ad -o . --clusterField=cell_ontology_class
The options we've specified for cbImportScanpy are:
-i: the name of the input h5ad file-o: the output directory (with '.' indicating the current directory)--clusterField: the name we want to use as the default cluster labels (and calculate markers for)
(You can run cbImportScanpy with no arguments to see the full usage message.)
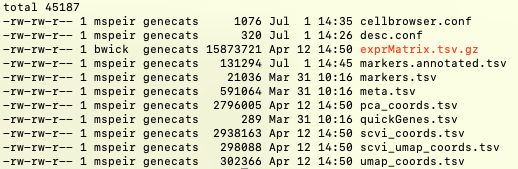
This export should not take more than 3 or 4 minutes. After it completes, you can do an ls and you should see files like meta.tsv or markers.tsv:
These and other files will be used as input to cbBuild in the next section of this guide.
Part 2: cellbrowser.conf and cbBuild
Now, we will go through the process of modifying the cellbrowser.conf and building a cell browser for this dataset into your public_html directory.
Open the cellbrowser.conf file using vim:
vim cellbrowser.conf
Edit the name and shortLabel fields of your cellbrowser.conf so that it matches the following:
name='tabula-sapiens-liver' shortLabel='Liver - Tabula Sapiens'
Build the cell browser into your public_html directory
cbBuild -o ~/public_html/cb
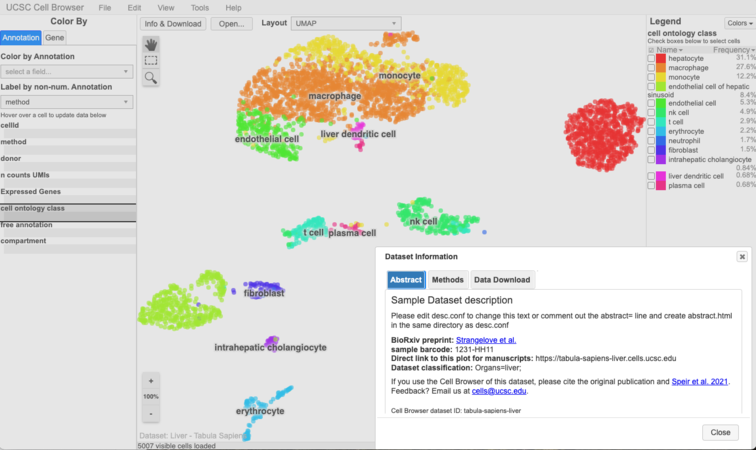
Look at your cell browser! It should be at https://hgwdev.gi.ucsc.edu/~${hgwdev_username}/cb/. It should look something like this:
When looking at the cell browser for this dataset, do you notice any changes that should be made to make the data more understandable for the average user? Maybe ‘layouts’ that need to be removed because they're uninformative? Or sample text that needs to be changed? We’ll talk more about polishing up the dataset in the next part. {image here?}
Part 3: desc.conf and final polish
Finally, we’ll cover filling out a desc.conf with some basic information about this dataset as well as polishing up any last visual details for this dataset.
Open the desc.conf file using vim:
vim desc.conf
Edit the following lines in your desc.conf to read:
title = "Liver Subset - Tabula Sapiens" abstract = """ Liver subset of the Tabula Sapiens dataset covering over 5000 cells. """ paper_url="https://www.science.org/doi/10.1126/science.abl4896 The Tabula Sapiens Consortium. Science. 2022." other_url="https://tabula-sapiens-portal.ds.czbiohub.org/ Tabula Sapiens Website"
Let's check if there are any 'colors' in the 'uns' slot of the h5ad:
h5ad TS_Liver.h5ad
AnnData object with n_obs × n_vars = 5007 × 58870
obs: 'organ_tissue', 'method', 'donor', 'anatomical_information', 'n_counts_UMIs', 'n_genes', 'cell_ontology_class', 'free_annotation', 'manually_annotated', 'compartment', 'gender'
var: 'gene_symbol', 'feature_type', 'ensemblid', 'highly_variable', 'means', 'dispersions', 'dispersions_norm', 'mean', 'std'
uns: '_scvi', '_training_mode', 'cell_ontology_class_colors', 'dendrogram_cell_type_tissue', 'dendrogram_computational_compartment_assignment', 'dendrogram_consensus_prediction', 'dendrogram_tissue_cell_type', 'donor_colors', 'donor_method_colors', 'hvg', 'method_colors', 'neighbors', 'sex_colors', 'tissue_colors', 'umap'
obsm: 'X_pca', 'X_scvi', 'X_scvi_umap', 'X_umap'
layers: 'decontXcounts', 'raw_counts'
obsp: 'connectivities', 'distances'
It looks like there are, and their names match metadata field names in obs (e.g. cell_ontology_class & cell_ontology_class_colors). Now export these colors to a file:
colorExporter -i orig/TS_Liver.h5ad -o colors.tsv
Annotate marker genes with link outs to other resources:
cbMarkerAnnotate markers.tsv markers.annotated.tsv
Make these changes to the cellbrowser.conf:
# {
# "file": "scvi_umap_coords.tsv",
# "shortLabel": "scvi_umap"
# },
# {
# "file": "scvi_coords.tsv",
# "shortLabel": "scvi"
# },
# {
# "file": "pca_coords.tsv",
# "shortLabel": "pca"
# }
markers = [{"file": "markers.annotated.tsv", "shortLabel":"Cluster Markers"}]
colors="colors.tsv"
Rebuild the dataset
cbBuild -o ~/public_html/cb
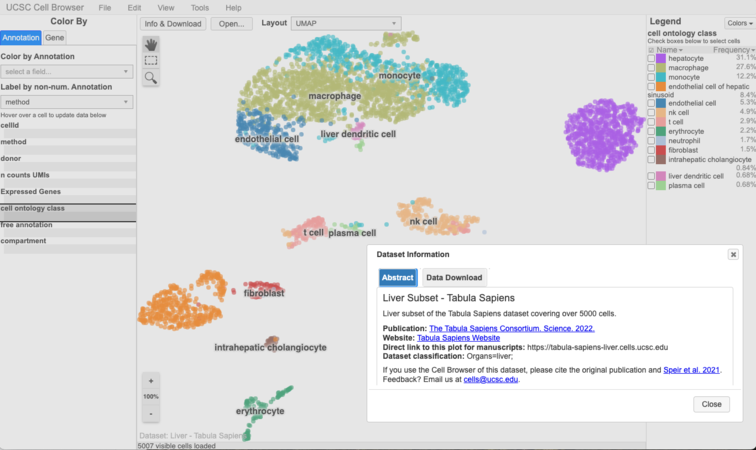
Check it out: https://hgwdev.gi.ucsc.edu/~${hgwdev_username}/cb/. It should look something like this:
What next?
Make other changes to the cellbrowser.conf and desc.conf files to see how they affect the display. (Don't forget to rebuild the dataset between those changes!)
Using cbImportSeurat
In this section we'll walk through how to create a cell browser starting with a Seurat RDS file, which is quite similar to using cbImportScanpy.
Part 1: Directory setup and data export
In this section, we'll set up the required directory structure for this new dataset and export the data from the RDS file.
Ensure that you are in the proper conda environment:
conda activate seuratenv
Change into a good working directory:
cd /hive/users/${hgwdev_username}/cb
Create a directory for this dataset:
mkdir -p mouse-dev-neocortex/orig/
This command also makes an ‘orig’ directory which we use to store the unchanged files obtained from the submitter or downloaded from GEO/etc.
Change into that directory:
cd mouse-dev-neocortex/orig/
Copy over the RDS file we’ll be working with:
cp /hive/data/inside/cells/exampleDatasets/Li_et_al_2020_UCSC_seurat_object.rds .
Go up a directory so that you are now just in the mouse-dev-neocortex directory. Now export the data from the RDS file:
cd ../ cbImportSeurat -i orig/Li_et_al_2020_UCSC_seurat_object.rds -o . --clusterField=clusters
The options we've specified for cbImportSeurat are:
-i: the name of the input RDS file-o: the output directory (with '.' indicating the current directory)--clusterField: the name we want to use as the default cluster labels (and calculate markers for)
(You can run cbImportSeurat with no arguments to see the full usage message.)
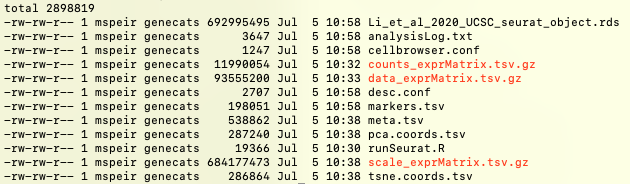
This export may take up to 30 minutes. After it completes, you can do an ls and you should see files like meta.tsv or markers.tsv:
These and other files will be used as input to cbBuild in the next section of this guide.
Part 2: cellbrowser.conf and cbBuild
Next up is modifying the cellbrowser.conf and building a cell browser for this dataset.
First, open the cellbrowser.conf file using vim:
vim cellbrowser.conf
Edit the name and shortLabel fields of your cellbrowser.conf so that it matches the following:
name='mouse-dev-neocortex' shortLabel='Developing Mouse Neocortex'
Build the cell browser into your public_html directory
cbBuild -o ~/public_html/cb
Look at your cell browser! It should be at https://hgwdev.gi.ucsc.edu/~${hgwdev_username}/cb/. It should look something like this:
Similar to the dataset we imported as part of the cbImportScanpy example above, do you notice any changes that could be made that might make the data understandable to a user? Maybe ‘layouts’ that need to be removed because they're uninformative? Or sample text that needs to be changed? We’ll go through some of those changes in the next part.
Part 3: desc.conf and final polish
Finally, we’ll cover filling out a desc.conf with some basic information about this dataset as well as polishing up any last visual details for this dataset.
Open the desc.conf file using vim:
vim desc.conf
For this dataset, we can pull from the paper Li et al itself to fill out the title, abstract, and other information. Edit the following lines in your desc.conf to read:
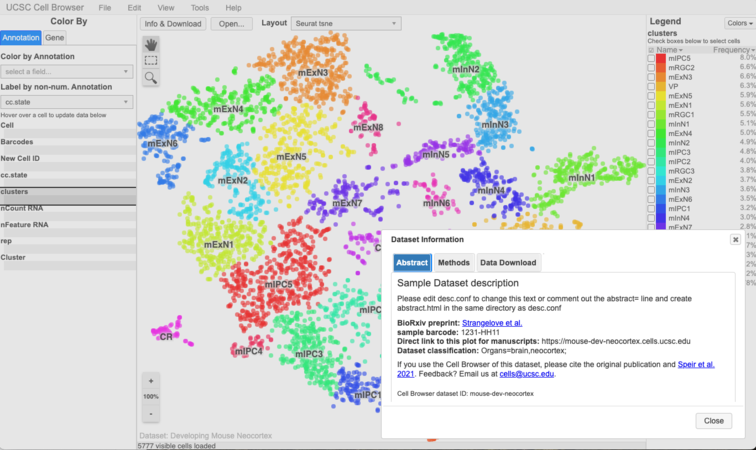
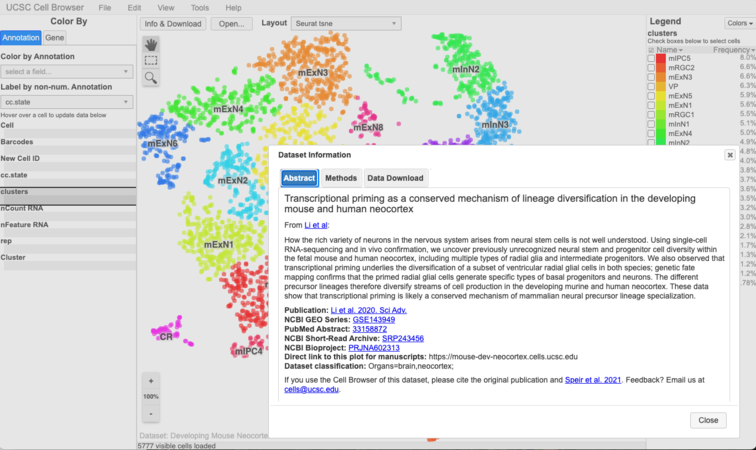
title = "Transcriptional priming as a conserved mechanism of lineage diversification in the developing mouse and human neocortex" abstract = """ <p> From <a href="https://www.science.org/doi/10.1126/sciadv.abd2068" target="_blank">Li et al</a>: <p> How the rich variety of neurons in the nervous system arises from neural stem cells is not well understood. Using single-cell RNA-sequencing and in vivo confirmation, we uncover previously unrecognized neural stem and progenitor cell diversity within the fetal mouse and human neocortex, including multiple types of radial glia and intermediate progenitors. We also observed that transcriptional priming underlies the diversification of a subset of ventricular radial glial cells in both species; genetic fate mapping confirms that the primed radial glial cells generate specific types of basal progenitors and neurons. The different precursor lineages therefore diversify streams of cell production in the developing murine and human neocortex. These data show that transcriptional priming is likely a conserved mechanism of mammalian neural precursor lineage specialization. """ methods=""" <section>Dimension reduction and clustering</section> <p> We used PCA and t-distributed SNE as our main dimension reduction approaches. PCA was performed with RunPCA function (Seurat) using HVGs. Following PCA, we conducted JACKSTRAW analysis with 100 iterations to identify statistically significant (P < 0.01) PCs that were driving systematic variation. We used t-SNE to present data in 2D coordinates, generated by RunTSNE function in Seurat. Significant PCs identified by JACKSTRAW analysis were used as input. Perplexity was set to 30. t-SNE plots were generated using R package ggplot2. Clustering was done with the Luvain-Jaccard algorithm using t-SNE coordinates by FindClusters function from Seurat with default setting. """ paper_url="https://advances.sciencemag.org/content/6/45/eabd2068 Li et al. 2020. Sci Adv." pmid = "33158872" geo_series = "GSE143949" sra_study = "SRP243456" bioproject = "PRJNA602313"
Make these changes to the cellbrowser.conf:
coords=[{"file": "tsne.coords.tsv", "shortLabel": "Seurat tsne"}]
body_parts=["brain","neocortex"]
Rebuild the dataset
cbBuild -o ~/public_html/cb
Check it out: https://hgwdev.gi.ucsc.edu/~${hgwdev_username}/cb/. It should look something like this:
What next?
Make other changes to the cellbrowser.conf and desc.conf files to see how they affect the display. What does changing radius or alpha in cellbrowser.conf do? What about adding information for the lab or institution to the desc.conf? (Don't forget to rebuild the dataset between those changes!)