Iron sulfur clusters: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 75: | Line 75: | ||
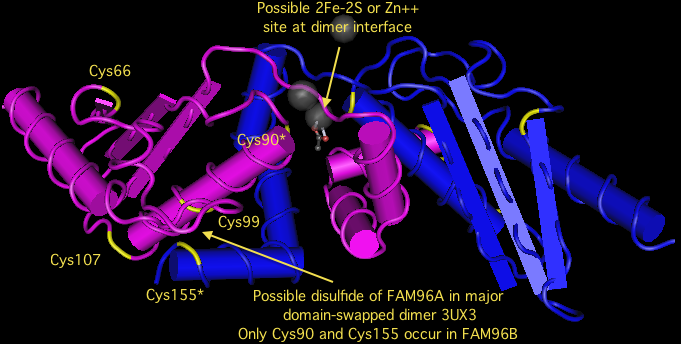
FAM96B is remarkably conserved throughout eukaryotes. It duplicated in the earliest metazoa, giving rise to FAM96A after the divergence of choanflagellates but before those of sponge, trichoplax, ctenophore or cnidarian. Both FAM96B and FAM96A were retained in all metazoan lineages. In vertebrates but not earlier diverging deuterostomes, FAM96A acquired an umistakable signal peptide, meaning it was no longer targeted to the cytoplasm. The species with the signal peptide are exactly those with an extra pair of invariant cysteines, suggesting a disulfide suitable for an oxidizing subcellular compartment such as endoplasmic reticulum. | FAM96B is remarkably conserved throughout eukaryotes. It duplicated in the earliest metazoa, giving rise to FAM96A after the divergence of choanflagellates but before those of sponge, trichoplax, ctenophore or cnidarian. Both FAM96B and FAM96A were retained in all metazoan lineages. In vertebrates but not earlier diverging deuterostomes, FAM96A acquired an umistakable signal peptide, meaning it was no longer targeted to the cytoplasm. The species with the signal peptide are exactly those with an extra pair of invariant cysteines, suggesting a disulfide suitable for an oxidizing subcellular compartment such as endoplasmic reticulum. | ||
However, these new cysteines are not in proper crystallographic position to form a disulfide | However, these new cysteines are not in proper crystallographic position to form a disulfide, though the new Cys99 and the long conserved near-terminal Cys155 are within range and may form a disulfide under certain conditions. The positions of two Cys90, one from each monomer, may also be in position to form a disulfide, 2Fe-2S, or zinc ligand provided the protein is purified anaerobically or reconstituted using some sort of activity assay. The phylogenetic conservation of cysteines is explored in the alignment below. | ||
The two encoded proteins both bind CIA01. However they must have distinct functions in vivo to account for retention of both proteins in so many lineages for so long. The usual explanations -- specialized time of expression during development or in differentiated populations of cells -- are not applicable to single-celled organisms. Since the signal peptide in FAM96A arose fairly late, it too cannot explain retention in earlier diverging species. (Note however tools that recognize signal peptides are not adequately trained on these species.) Since the duplication is restricted to metazoans (ie animals), it could possibly be associated with dietary, rather than diffusive, acquisition of iron. | The two encoded proteins both bind CIA01. However they must have distinct functions in vivo to account for retention of both proteins in so many lineages for so long. The usual explanations -- specialized time of expression during development or in differentiated populations of cells -- are not applicable to single-celled organisms. Since the signal peptide in FAM96A arose fairly late, it too cannot explain retention in earlier diverging species. (Note however tools that recognize signal peptides are not adequately trained on these species.) Since the duplication is restricted to metazoans (ie animals), it could possibly be associated with dietary, rather than diffusive, acquisition of iron. | ||
| Line 119: | Line 119: | ||
[[Image:SufT.png|left]] | [[Image:SufT.png|left]] | ||
<br clear=all> | <br clear=all> | ||
[[Image:Phenyl.png|left]][[Image:DUF59s.gif| | [[Image:Phenyl.png|left]][[Image:DUF59s.gif|430px|thumb|left|Click to enlarge]] | ||
Another important clue to FAM98B/SufT function may come from additional insights into the steps of the [http://www.ncbi.nlm.nih.gov/ | Another important clue to FAM98B/SufT function may come from additional insights into the steps of the phenylacetic acid degradation pathway] -- an esoteric topic but [http://www.ncbi.nlm.nih.gov/pubmed/22398448,21296885,20660314,12846838,9748275,17259607,9600981,19208251 exceedingly well-studied]. | ||
An epic annotation error has propagated to thousands of GenBank and UniProt entries: the DUF59 domain of the phenylacetate catabolic pathway lies in PaaD, not PaaJ (which has only thiolase domains). Some entries state -- nonsensically -- that the PaaD protein is the product of the PaaJ gene; overall 85% of PaaD homologs are incorrectly named PaaJ (with the E.coli [http://www.uniprot.org/uniprot/P76080 accession P76080 for ydbQ] as correct starting point for PaaD): | |||
>PaaD_escCol Escherichia coli <font color=red>8 conserved cysteines</font> P76080 PaaJ ydbQ DUF59 unacceptable synonym: PaaJ | |||
MQRLATIAPPQVHEIWALLSQIPDPEIPVLTITDLGMVRNVTQMGEGWVIGFTPTYSG<font color=red>C</font>PATEHLIGAIREAMTTNGFTPVQV | |||
VLQLDPAWTTDWMTPDARERLREYGISPPAGHS<font color=red>C</font>HAHLPPEVR<font color=red>C</font>PR<font color=red>C</font>ASVHTTLISEFGSTA<font color=red>C</font>KALYR<font color=red>C</font>DS<font color=red>C</font>REPFDYFK<font color=red>C</font>I* | |||
The first alternative implies that each enzyme closely associated to a DUF59 contains a | The initial aromatic ring oxygenation (using O2) utilizes a multisubunit complex requiring PaaA, PaaB, PaaC, and PaaE for in vitro reconstitution of catalytic activity. PaaD has no place in the complex but an [http://www.ncbi.nlm.nih.gov/pubmed/16997993 essential role in vivo] as established by mutation and supported by position within the Paa operon in many species. | ||
Together, this suggests the DUF59 domain of PaaD establishes and/or repairs the [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3060524/?tool=pubmed 2Fe-2S cluster of PaaE reductase] (which might be especially vulnerable to reactive oxygen species in the oligomer). Such a role for PaaD would not be a great departure from what its homolog does in eukaryotes. Note PaaA itself has a di-iron center though its regulatory paralog PaaC does not. | |||
The DUF59 domain appears in multiple contexts in bacteria -- SufT, PaaD, YITW, MIP18 and rhamnose reductase-related protein. Some of this is attributable to poor quality computer annotation and nomenclatural confusion across species, but DUF59 does reside in at least two distinct operons, the iron sulfur cluster system Suf and the phenylacetic acid catabolism pathway Paa, the difference being that the DUF59 acts on a broader class of iron-sulfur apoproteins in bacterial species with a SufT-containing operon. | |||
The question here is whether all remaining DUF59 homologs assist in iron sulfur complex assembly, either specialized to a one apoenzyme or as part of a general system maturating numerous unrelated apoenzymes (like eukaryotic FAM96B). Alternatively -- consistent with poor percentage identity -- the DUF59 domain duplicated and diverged into dimly related functions with little in common apart from a shared fold. However this would allow multiple DUF59 domains within a single prokaryotic genome, so far not observed. | |||
The first alternative implies that each enzyme closely associated to a DUF59 contains a Fe-S cluster. However these have proven notoriously difficult to detect in the case of DNA helicases, polymerases and primases. The problem arises from loss during aerobic protein purification (perhaps with replacement by Zn) and lack of reliable bioinformatic signature for iron-sulfur clusters, notably an 'insufficient' number of conserved liganding cysteines (due to composite dimeric sites or glutathione contributing unsuspected cysteines), a lack of consistent pattern spacing in the cysteines, and ambiguity relative dedicated zinc binding sites. | |||
<br clear=all> | <br clear=all> | ||
Alignment of FAM96A and FAM96B in phylogenetic order: | Alignment of FAM96A and FAM96B in phylogenetic order: | ||
| Line 222: | Line 232: | ||
FAM96B_tryCru DPIDSLEVFHHIRSIRDPEHPNTLEELKVVEPELIRV---DEVKQTVRVQFTPTVPH<font color = red>C</font>SMTTLIGL<font color = red>C</font>ISLKLQRSLPRGTKVDVYVTPGSHEQEEQVNKQLNDKERVAAALENKNLLNVVES<font color = red>C</font>LNEFE | FAM96B_tryCru DPIDSLEVFHHIRSIRDPEHPNTLEELKVVEPELIRV---DEVKQTVRVQFTPTVPH<font color = red>C</font>SMTTLIGL<font color = red>C</font>ISLKLQRSLPRGTKVDVYVTPGSHEQEEQVNKQLNDKERVAAALENKNLLNVVES<font color = red>C</font>LNEFE | ||
FAM96B_leiInf DPIDAWEVFEIIRRIRDPEHPNSLEQLKVVEPSLITV---DWKKRHIRVLFTPTVPH<font color = red>C</font>SLTTLIGLSIRLQLERSLPEYTKVDIYVTPGTHEQEAQVNKQLNDKERVAAALEN<font color = red>C</font>NLLNVVES<font color = red>C</font>INEFD | FAM96B_leiInf DPIDAWEVFEIIRRIRDPEHPNSLEQLKVVEPSLITV---DWKKRHIRVLFTPTVPH<font color = red>C</font>SLTTLIGLSIRLQLERSLPEYTKVDIYVTPGTHEQEAQVNKQLNDKERVAAALEN<font color = red>C</font>NLLNVVES<font color = red>C</font>INEFD | ||
=== NARFL (IOP1) === | === NARFL (IOP1) === | ||
Revision as of 03:46, 24 June 2012
Introduction
The surprisingly numerous nuclear proteins containing 4Fe-4S clusters are made from their respective apoproteins in the cytoplasm during the final stages of an assembly process that begins within mitochondria and ends with an embedded cluster in polymerases, helicases, primases, telomerases, and photolyases with no explained need for a cofactor otherwise associated with oxidation and reduction.
These 4Fe-4S clusters do not spontaneously associate with their target protein because they do not occur in free solution, being quite unstable to unwanted oxidation. Instead, nascent clusters are attached to a series of mediating proteins, carrier scaffolds and conformational chaperones throughout a complex process of maturation. That process and the gene products involved -- which are conserved from yeast to human -- have been recently reviewed in depth and new results (1,2) have clarified the roles of the four main protein components that collaborate on the final stage of cytoplasmic assembly.
Not all extra-mitochondrial 4Fe-4S cluster proteins are assembled in this pathway, but the molecular basis for specificity has not yet been determined. Indeed, a surprising number of proteins -- some studied for decades -- have only been recognized as iron sulfur proteins in 2011-12.
Indeed, the list of proteins is still incomplete because many unrelated homology classes have Fe-S clusters, meaning no single diagnostic pattern can be used to scan the entire proteome. Often four conserved cysteines coordinate the cubane complex but their spacing within the primary sequence is not uniform and difficult to distinguish from cysteine patterns that bind intrinsic zinc. Further confusing matters, seemingly artifactual zinc can replace bona fide 4Fe-4S clusters in proteins purified for crystallography in the presence of oxygen (1,2,3).
The early and middle stages of intra-mitochondrial iron sulfur cluster assembly are carried out by gene products of bacterial origin, relics of alphaproteobacterial endosymbiosis transferred long ago to the nuclear genome. However, not all components of final cytoplasmic assembly have such a clear origin whereas most targeted apoproteins (such as primase large subunit PRIM2) are clearly those of the archaeal parent. Thus the final stage of assembly presents two worlds in collision: bacterial proteins assembly iron sulfur clusters in unfamiliar archaeal proteins.
Bioinformatics, while a poor substitute for experimentation, is fast and easy, so it is best to exhaust the possibilities there first. Nothing is proven but sometimes it can suggest interesting directions.
MMS19: a large all-scaffold protein
MMS19 is a large protein involved in cytoplasmic iron sulfur assembly first studied with bioinformatic tools 12 years ago (1,2). Revisiting that with modern comparative genomics methods, MMS19 emerges as a modular scaffolding protein over its entire length, conserved in its features -- though not particularly in amino acid sequence -- from the earliest diverging eukaryotes to human.
The C-terminus of MMS19 was initially classified as HEAT repeats. Today we know these are not found as individual units but instead work together to form a long twisted spiral of consecutive modules called an ARM domain. An individual HEAT unit consists of a small 3-helix bundle, a generic super-secondary structure analogous to a beta-alpha-beta Rossmann fold unit, meaning most occurrences of HEAT in the eukaryotic proteome are not truly homologous despite structural similarity but instead represent convergent evolution analogous to Rossmann-like fold units forming many unrelated beta propellers or TIM barrels.
Since these domains are catalytically inert and lack conserved cysteins or other conserved motifs, MMS19 can contribute as organizing principle to the cytoplasmic iron assembly complex (and other nuclear complexes) but not to the actual business of forming 4Fe-4S on target apoproteins.
The size of MMS19 -- over a thousand residues -- makes it a difficult target for structure determination. As of June 2012, no deposited structure at PDB provides a template upon which the MMS19 can be threaded. In the interim, MMS19 might be structurally modeled using the known beta-catenin structure -- despite its lack of authentic homology, it too is comprised almost entirely of HEAT units.
The number of HEAT repeats in an ARM domain is subject to expansion and contraction over evolutionary time. The individual units often align poorly with each other and generally lack conserved residue signatures despite initial reports, yet this level of variation does not necessarily affect the overall fold. However, this lack of diagnostic features makes it difficult to reliably identify remote homologs of HEAT repeats because the primary sequences can be diverged beyond recognition and homological alignments go out of register when the number of repeats differs.
An accurate count of the number of individual HEAT domains in MMS19 from a given species is also difficult because some domains are more accurately represented in the HMMer profile than others, here giving different counts between mouse and human at UniProt despite 90% sequence identity over their entire length. Indeed the five species with manually reviewed UniProt entries are all in conflict, with the nominal number of HEAT units range from 7 in human to 18 in slime mold, despite similar lengths and overall alignment implying the same actual domain structure.
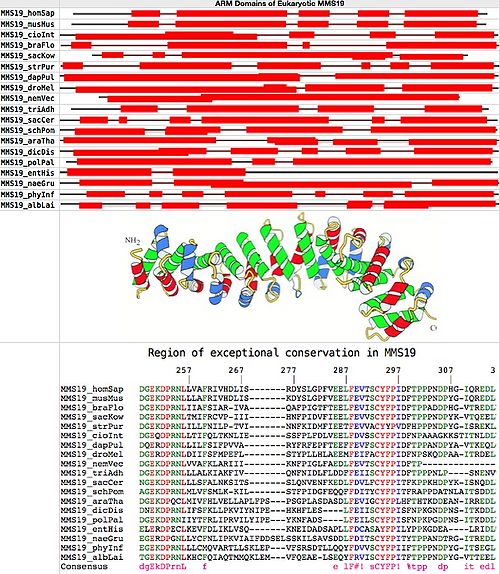
MMS19 is a single-copy gene without paralogs in all eukaryotes, implying simple orthology (no retained duplications or losses). Below, 20 full length fasta sequences from GenBank were chosen for uniform distribution over the eukaryotic phylogenetic tree. Superfamily proved to be the most consistent, sensitive and selective online tool for ARM domain detection. The figure at bottom establishes that MMS19 consists entirely of HEAT units and spacers, which in effect form a single ARM.
It emerges upon alignment with MultAlin that conservation is mediocre overall except for one previously notedspecial region of exceptional conservation containing two blocks of invariant residues from human to yeast to amoeba. This region must have already been established in the last common ancestor of all eukaryotes and play a very special role in MMS19 even today to account its conservation over trillions of years of cumulative branch length. However that role remains a complete mystery. Sequence conservation in MMS19 is otherwise not exceptional: typically 27-34% identity relative to human, of which some portion is accidental.
Ultra-conserved region in MMS19: human: 182 DGEKDPRNLLVAFRIVHDLISRDYSLGPFVEELFEVTSCYFPIDFTPPPNDPHGIQREDL 241 yeast: 184 NGEKDPRNLLLSFALNKSITSSLQNVENFKEDLFDVLFCYFPITFKPPKHDPYKISNQDL 24
Regardless of Blast query -- full-length MMS19, this ultra-conserved region, or reconstructed ancestral sequence -- no counterpart to MMS19 occurs among 2,500 complete bacteria and archaea genomes, even though unambiguous orthologs to human MMS19 are readily found in the earliest diverging eukaryotes. MMS19 may thus represent a eukaryotic innovation needed to organize more complex cytoplasmic iron assembly, or be too simplified and diverged (or just lost) in prokaryotes. As method of last resort, prokaryotic operons containing other cytoplasmic iron sulfur assembly proteins could be scanned for adjacent HEAT-like domains or comparable scaffolding proteins.
There are no matches to MMS19 at PDB using Blastp. Since the fold is widespread and generic, structural matches in DALI do not imply homology. On the other hand, this allows the crystallographic structure of a non-homologous ARM protein (beta-catennin pdb: 1LUJ) to serve as provisional structural template. Bound E-cadherin, ICAT, XTCF3 complexes have been also been determined which may suggest a binding mode for cytoplasmic iron sulfur and helicase-type proteins on HEAT repeats of MMS19.
MMS19 could determine selectivity among the overall set of iron sulfur apoproteins if only those interacting with DNA (or comparable nucleotide) have binding propensity for HEAT units. This specificity would then vary by organism, as would the effects of knock-in replacement. Only those apoproteins that align along the linear scaffolding structure in close enough proximity to CIA effector proteins receive an iron sulfur complex. Not every protein arising in this context need directly bind a HEAT domain -- they could bind another protein with that capacity.
This scaffolding scenario requires multiple non-homologous proteins to have HEAT binding sites, which seemingly requires convergent evolution on a significant scale since a shared mobile binding domain can be ruled out. If MMM19 is truly a eukaryotic innovation, then cytoplasmic iron assembly complex initially functioned without the scaffolding until MMS19 and these binding sites evolved.
That seems implausible, so some other common ground must account for HEAT binding. One option, based on the super-helical configuration and major groove of the overall ARM domain, supposes that that MMM19 spoofs a DNA helix or nucleotide base in shape and charge (along the lines of W536 in CRY1B photolyase). This would explain most of the specificity -- each archaeal apoproteins of DNA metabolism needing an iron sulfur cluster already has a DNA binding site, so already an appropriate MMM19 HEAT binding site.
In early endosymbiosis, retained bacterial cluster assembly machinery collides with nuclear-encoded archaeal iron-sulfur protein motifs previously maturated by a different system, a conflict that had to be seamlessly resolved without ever a gap in continued functionality.
CIAO1: a WD40 multi-protein scaffold
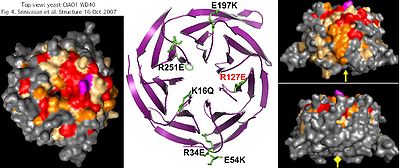
The bioinformatic analysis of CIAO1 is straightforward: it consists of a WD40 domain in its entirety. There is no CIAO2 -- this is a single-copy gene in all eukaryotes. These ubiquitous 7-propeller blade domains can arise in non-homologous proteins as a common supersecondary structure rather than from spread of a mobile domain -- the human genome encodes some 257 proteins with a WD40 repeat. However a degree of coincidental sequence alignment can arise from common constraints (such as a conserved glycine/histidine and the tryptophan/aspartate pairs).
WD40 domains are not catalytic and so, like MMS19, not involved here mechanistically in Fe-S formation, transfer or repair. Thus CIAO1 is likely a structural scaffolding protein coordinating larger multi-protein complexes, so its acronym (Chinese for bridge) is appropriate. Crystallographic structures have been determined both in yeast PDB: 2HES and human PDB: 3FM0.
Strongly conserved surface residues -- which likely mediate oligomeric interactions -- mostly lie on the top and one side of CIAO1. Despite this, of four substitutions tested, only R127E (purple) affected in vivo functionality, as assayed bu plasmid rescue of CIAO1 depleted cells and levels of the assembled Fe-S protein isopropylmalate isomerase. This does not explain observed conservation of other surface residues such as K16, R34, E54, E197 and R251 which are unlikely to play a role in internal structure or stability.
No journal article ever accompanied the human structural determination but differences from yeast are likely minor in view of demonstrated replacement capability. Since human CIAO1 forms a hetero-oligomer with FAM96A and likely FAM96B as well, this suggests yeast CIAO1 forms a similar oligomer with its counterpart yeast FAM96B consistent with the late role of CIAO1 in cytosolic FeS assembly.
It is thus feasible to separately determine conserved residues in FAM96A and FAM96B using ConSurf. Their common ground would then include the binding site for CIAO1, presumably either its previously established conserved top or side residues. Since FAM96B forms a domain-swapped dimer and CIAO1 binds stoichiometrically, a symmetric heterotetramer can be expected.
However nothing is accomplished in terms of coordinated docking unless CIAO1 has a second binding site for another component of cytosolic FeS assembly. CFD1 is a possibility here in view of the CFD1-CIAO1 fusion protein in S. pombe, as are NBP35, ERCC2 and ANT2.
CIAO1 has weak blast matches in both bacteria and archaea but these are not associated with any of the three iron sulfur cluster assembly system operons (ISC, SUF, NIR) and may simply represent convergence in WD40 proteins. Matches to early-diverging eukaryotes -- a half dozen are provided below -- are much more persuasive because back-blastp to human uniquely recovers CIAO1. These exhibit extensive conserved regions considering the immense phylogenetic span and rapid evolution of some clades. Narrowly ineage-specific indels, presumably in loop regions, can be removed to create an idealized alignment that better reflects conserved residues.
FAM98B is homologous to bacterial SufT
FAM96B is remarkably conserved throughout eukaryotes. It duplicated in the earliest metazoa, giving rise to FAM96A after the divergence of choanflagellates but before those of sponge, trichoplax, ctenophore or cnidarian. Both FAM96B and FAM96A were retained in all metazoan lineages. In vertebrates but not earlier diverging deuterostomes, FAM96A acquired an umistakable signal peptide, meaning it was no longer targeted to the cytoplasm. The species with the signal peptide are exactly those with an extra pair of invariant cysteines, suggesting a disulfide suitable for an oxidizing subcellular compartment such as endoplasmic reticulum.
However, these new cysteines are not in proper crystallographic position to form a disulfide, though the new Cys99 and the long conserved near-terminal Cys155 are within range and may form a disulfide under certain conditions. The positions of two Cys90, one from each monomer, may also be in position to form a disulfide, 2Fe-2S, or zinc ligand provided the protein is purified anaerobically or reconstituted using some sort of activity assay. The phylogenetic conservation of cysteines is explored in the alignment below.
The two encoded proteins both bind CIA01. However they must have distinct functions in vivo to account for retention of both proteins in so many lineages for so long. The usual explanations -- specialized time of expression during development or in differentiated populations of cells -- are not applicable to single-celled organisms. Since the signal peptide in FAM96A arose fairly late, it too cannot explain retention in earlier diverging species. (Note however tools that recognize signal peptides are not adequately trained on these species.) Since the duplication is restricted to metazoans (ie animals), it could possibly be associated with dietary, rather than diffusive, acquisition of iron.
The placement and phase of introns in FAM96B and FAM96A -- largely conserved -- implies that FAM96B was largely intronated prior to gene duplication. Although the two genes did go their separate ways to a certain extent during the subsequent 600 million years (both gains and losses of introns occurred), the patterns remain very closely related today even comparing human to sponge. Most remarkably, the first intron was already in place in early diverging stramenopiles (eg Phytophthora infestans) and the last exon already present in amoebozoa (eg Dictyostelium discoideum). Below phase numbers such as 00 and 12 show codon bp overhangs and ^^ indicates absence of an exon break.
FAM96A_homSap MQRVSGLLSWTLSRVLWLSGLSEPGAARQPRIMEEKALEVY 12 DLIRTIR ^^ DPEKPNTLEELEVVSESCVEVQEINEEEYLVIIRFTPTVPHCSLATLI 12 GLCLRVKLQRCLPFKHK 00 LEIYISEGTHSTEED 12 INKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_sacKow MNKDETLRIKGAEDDQEKELAEEIY 12 DIIRTIR ^^ DPEKPQTLEDLDVVYEDGVLVNHRGTDEFLVNVEFTPTVPHCTLATLI 12 GLCIRVKLQRTLPHSYK 00 LDIFIKKGTHSTEDE 12 INKQINDKERIAAAMENPNLKDLVDNCVVDLE FAM96A_triCas MPDLLELVEFEDSPRKEVKEVSEDDSELKYTVY 12 DLIRTIK ^^ DPEKPNTLEELNVVYEEGEVKERTSGNVSVVRVEFNPTVPHCSLATLI ^^ GLCIRIKLERCIPYRIK ^^ LDIYIKAGAHTTEHE 12 INKQINDKERIAAAMENPNLREMVENCIVEED FAM96A_nemVec MSNTNDPSFGASRIDNDVNSQSNRNLALDVY 12 DLIKDIK 00 DPEKPQTLEDLKVVYESCVEVQKVAGQDHITIT FTPTVPHCSLATLI 12 GLCIRVKLEKSLPEKFK 00 LDIYLKKGTHSTENE 12 INKQINDKERIAAAMENPNLRKIVENCIDEDNGYD FAM96A_acrPal MSENKILSTAADSSFDNLVLVQEVF 12 DIVKDIR 00 DPELPQTLEELHVIEEEFIKIDKIENDEYIIKIEFTPTVPHCSLATLI 12 GLCLRVKLERSLPYKFK 00 LDIFLSRGTHSTENE 12 INKQINDKERIAAAMENPNLKKIVEECILDAN FAM96A_triAdh 12 ELIRDIK 00 DPELPQTLEELNVVTEDEIFVRNMKQGEACIRINFTPTVPHCSLATLI 12 GLCIRVKLQRCLDQDYK 00 LDIYVTKGSHDTEDG 12 VNKQINDKERVAAAIENPNVKKLVEECLQEVQY FAM96B_homSap MVGGGGVGGGLLENANPLIYQRSGERPVTAGEEDEQVPDSIDAREIF 12 DLIRSINDPEHPLTLEELNVVEQVRVQ 00 VSDPESTVAVAFTPTIPHCSMATLI ^^ GLSIKVKLLRSLPQRFK 00 MDVHITPGTHASEHA 12 VNKQLADKERVAAALENTHLLEVVNQCLSARS FAM96B_braFlo MSSPDRLQNANPQLYGRTSEREVTPEELNEDVEDAIDAREIF ^^ DILSSINDPEHPLTLEELNVIEQSRIT 00 VDEDNNHVSVEFTPTIPHCSMATLI ^^ GLSIRVKLLRALPTRFK 00 VDVHITPGTHQSEHA 12 VNKQLADKERVAAALENQHLLEVVNQCLSTRN FAM96B_droMel MPTEIENINPNVYDRIKERVLTANEEDENVPDPFDKREIF 12 DLIRNINDPEHPLTLEELHVVQEDLIR ^^ INDSQNSVHISFTPTIPHCSMATLI ^^ GLSIRVKLLRSLPPRFK ^^ VTVEITPGTHASELA ^^ VNKQLADKERVAAALENNHLAEVINQCIAKG FAM96B_nemVec MAAVTDRLENVNPTVFQRLKERVVLAEEEDDNIVDKIDDREIF 12 DMIRSINDPEHPLTLEELNVVEQALID ^^ VSDDESYVKVQFTPTIPHCSMATLI ^^ GLAIRVRLLRSLPDRFK 00 VDVKITPGTHQSEIA 12 VNKQLADKERVAAALENNHLLDVIDQCLVSKK FAM96B_triAdh MSKFSSAIGLENINPTVFSKSKEREILPEELDDNIVDKIDEREIF 12 DIIRSINDPEHPLTLEELNVVEECKID ^^ VDDDNNFVKVHFTPTIPHCSMATLI ^^ GLCIRVRLIRSLPERFK 00 VDITVTPGSHSSEIA 12 VNKQLADKERVAAAMENSNLLKVVNQCLAMD FAM96B_ampQue MMAADNANPVVFEVAKQRPVTVEEEKDDVYDEIDAREVF 12 DLIRHINDPEHPLTLEELNVVQEDLIC ^^ INNKENFVSVHFTPTIPHCSMATLI ^^ GLSIRVCLLRSLPNRFK 00 IDVIITPGSHMSEQA 12 VNKQLADKERIAAAIENSHLLNVVHQCLNTKRKQ FAM96B_monBre MALENAAPRVYGKVQERVTLDNDFDDDVIDPFDSREIF 12 DLVRHINDPEHPLTLEELNVVRL 12 DQILVDDAQNYVRVQFTPTIPHCSMASLI ^^ GLCLRVRLLRALPPRFK 00 VDVEIFPGTHATEAS 12 INKQLADKERVAAALENPNLKTVVNECLQLDDKYLD
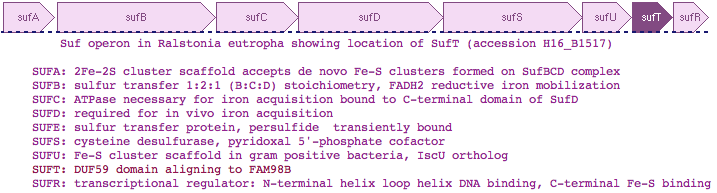
FAM96B and FAM96A are comprised of a single domain of unknown function, DUF59, a fall-out of automated primary sequence clustering. DUF59 is an uncommon domain -- FAM96B/FAM96A are the only human proteins (out of an 18,500 member proteome) to contain it. Thus it is remarkable that the top Blastp matches to prokaryotes are a DUF59 domain in the bacterial SufT gene. SufT is part of a large operon of an alternative system for formation of 4Fe-4S proteins. Further, a SufT-related domain occurs in Q6STH5 of Arabidopsis thaliana (in chloroplasts, cyanobacterial endosympbiont proteins assemble Fe-S complexes) fused N-terminally to a homolog of NUBP2/CFD1 which participate in cytosolic Fe-S cluster assembly. Recall CFD1 is fused to CIAO1 in S. pombe, consistent with the three proteins forming a heteroligomer.
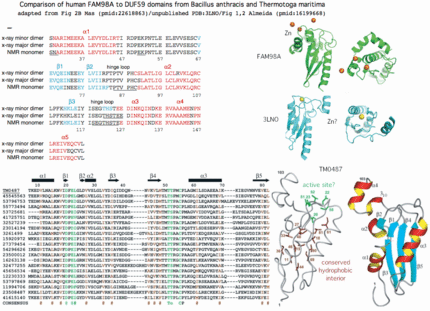
The underlying premise of DUF59 as valid domain defined by sequence is validated by good structural alignments across the five PDB structures available for it. While the diversified roles do not always have a clear connection to iron sulfur cluster metabolism, features such as active site and reaction catalyzed (if any) might transfer to a direct function for SufT and human FAM96B and FAM96A.
Gene PDB UniProt PubMed Species Comment FAM96A 3UX2 Q9H5X1 22683786,2261886 Homo sapiens cytosolic 4Fe-4S cluster formation FAM96B '3UX2' Q9Y3D0 22678362,22678361 Homo sapiens from FAM96A utilizing >50% sequence identity DUF59 1WCJ TM0487 16199668,15213465 Thermotogo maritima implications for 216 homologous DUF59 protein. 1UWD DUF59 3LNO Q81XF6 -------- Bacillus anthracis article never published DUF59 3CQ1 Q53W28 -------- Thermus thermophilus assigned to dDTP-4-Keto-L-Rhamnose Reductase TTHB138, aka 2CU6 HCF101 ---- Q6STH5 14690502,19817716 Arabidopsis thaliana chloroplast 4Fe-4S cluster formation PaaD ---- G8RCQ5 16199668 Staphylococcus aureus aromatic ring hydroxylating enzyme PaaJ ---- O84984 9748275,9600981 Pseudomonas putida thiolase in phenylacetic acid degradation aka PhaH SufT ---- Q0K120 -------- Ralstonia eutropha iron sulfur cluster assembly protein
Thus although human FAM98B and bacterial SufT are very diverged in primary sequence, multiple lines of evidence support bona fide homology, a surprising result that mixes -- within human -- a component of the 'backup' bacterial SUF system of iron sulfur complex formation together with the main bacterial ISC system. SufT has been located within the SUF operon in at least three genera of bacteria as SufABCDSUTR in Ralstonia, Cupriavidus and Pseudogulbenkiania -- this can't be coincidence given the rarity of the DUF59 domain and involvement of both in iron sulfur cluster formation.
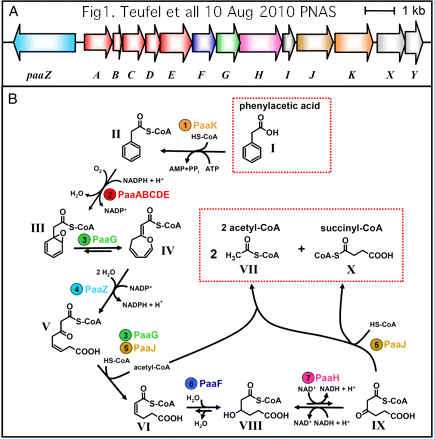
Another important clue to FAM98B/SufT function may come from additional insights into the steps of the phenylacetic acid degradation pathway] -- an esoteric topic but exceedingly well-studied.
An epic annotation error has propagated to thousands of GenBank and UniProt entries: the DUF59 domain of the phenylacetate catabolic pathway lies in PaaD, not PaaJ (which has only thiolase domains). Some entries state -- nonsensically -- that the PaaD protein is the product of the PaaJ gene; overall 85% of PaaD homologs are incorrectly named PaaJ (with the E.coli accession P76080 for ydbQ as correct starting point for PaaD):
>PaaD_escCol Escherichia coli 8 conserved cysteines P76080 PaaJ ydbQ DUF59 unacceptable synonym: PaaJ MQRLATIAPPQVHEIWALLSQIPDPEIPVLTITDLGMVRNVTQMGEGWVIGFTPTYSGCPATEHLIGAIREAMTTNGFTPVQV VLQLDPAWTTDWMTPDARERLREYGISPPAGHSCHAHLPPEVRCPRCASVHTTLISEFGSTACKALYRCDSCREPFDYFKCI*
The initial aromatic ring oxygenation (using O2) utilizes a multisubunit complex requiring PaaA, PaaB, PaaC, and PaaE for in vitro reconstitution of catalytic activity. PaaD has no place in the complex but an essential role in vivo as established by mutation and supported by position within the Paa operon in many species.
Together, this suggests the DUF59 domain of PaaD establishes and/or repairs the 2Fe-2S cluster of PaaE reductase (which might be especially vulnerable to reactive oxygen species in the oligomer). Such a role for PaaD would not be a great departure from what its homolog does in eukaryotes. Note PaaA itself has a di-iron center though its regulatory paralog PaaC does not.
The DUF59 domain appears in multiple contexts in bacteria -- SufT, PaaD, YITW, MIP18 and rhamnose reductase-related protein. Some of this is attributable to poor quality computer annotation and nomenclatural confusion across species, but DUF59 does reside in at least two distinct operons, the iron sulfur cluster system Suf and the phenylacetic acid catabolism pathway Paa, the difference being that the DUF59 acts on a broader class of iron-sulfur apoproteins in bacterial species with a SufT-containing operon.
The question here is whether all remaining DUF59 homologs assist in iron sulfur complex assembly, either specialized to a one apoenzyme or as part of a general system maturating numerous unrelated apoenzymes (like eukaryotic FAM96B). Alternatively -- consistent with poor percentage identity -- the DUF59 domain duplicated and diverged into dimly related functions with little in common apart from a shared fold. However this would allow multiple DUF59 domains within a single prokaryotic genome, so far not observed.
The first alternative implies that each enzyme closely associated to a DUF59 contains a Fe-S cluster. However these have proven notoriously difficult to detect in the case of DNA helicases, polymerases and primases. The problem arises from loss during aerobic protein purification (perhaps with replacement by Zn) and lack of reliable bioinformatic signature for iron-sulfur clusters, notably an 'insufficient' number of conserved liganding cysteines (due to composite dimeric sites or glutathione contributing unsuspected cysteines), a lack of consistent pattern spacing in the cysteines, and ambiguity relative dedicated zinc binding sites.
Alignment of FAM96A and FAM96B in phylogenetic order: FAM96A_homSap --EKALEVYDLIRTIRDPEKPNTLEELEVVSESCVEVQEINEEEYLVIIRFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_calJac --EKALEVYDLIRTIRDPEKPSTLEELEVVSESCVEVQEINEEEYLVIIRFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_otoGar --EKALEIYDLIRTIRDPEKPNTLEELEVVTESCVEVQEINEEDYLVIIKFTPTVPHCSLATLIGLCLRVKLQRCFPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_tupBel --EKALEVYDLIRTIRDPEKPNTLEELDVVTESCVEVQEINEDDYLVIIRFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_musMus --EKALEVYDLIRTIRDPEKPNTLEELEVVTESCVEVQEINEDDYLVIIKFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_oryCun --EKALEVYDLIRTIRDPEKPNTLEELEVVTESCVEVQEINEDDYLVVIRFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_bosTau --EKALEVYDLIRTIRDPEKPNTLEELEVVTESCVEVQEINEDDYLVIIRFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_canFam --EKALEVYDLIRTIRDPEKPNTLEELEVVTESSVEVQEINEEDYLVIIRFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_loxAfr --EKALEVYDLIRNIRDPEKPNTLEELEVVTESCVEVQEINEDDYLVIIRFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_choHof --EKALEVYDLIKIIQDPEKPNTLEEPEVATESCVEVQEINEEDYLVII-FTPTVPHCCLATLIGLCLRVKLQRCLPFKHNLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_monDom --EKALEVYDIIRTIRDPEKPNTLEELEVVTESCVEVKEIHEEDYLVIIRFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVIEPD FAM96A_ornAna --DKALEVYDLIRTIRDPEKPNTLEELEVVTESCVKVKEVDEDDYLVIIRFTPTVPHCSLATLIGLCLRVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_taeGut --DRAIEVYDIIRTIRDPEKPNTLEELEVVTENCVQVQEIGEDEYLVIIRFTPTVPHCSLATLIGLCLRIKLQRCLPFRHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVTEPD FAM96A_galgal --DKALEVYDIIRTIRDPEKPNTLEELDVVTESCVQVDEIGEEEYLVVIRFTPTVPHCSLATLIGLCLRIKLQRCLPFRHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVTEPD FAM96A_chrPic --DRALEVYDIIRTIRDPEKPNTLEELEVVTESCVEVHEIGEDEYLVIIRFTPTVPHCSLATLIGLCLRIKLQRCLPFKHKLEIYISEGAHSTEEDVNKQINDKERVAAAMENPNLREIVEQCVTEPD FAM96A_anoCar --ERALEVYDIIRTIRDPEKPNTLEELDVVTESCVEVHETSEDEYLVTIRFTPTVPHCSLATLIGLCLRIKLQRCLPFKHKLEIFISEGAHSIEEDINKQINDKERVAAAMENPNLREIVEQCVLEPD FAM96A_xenTro --ERALEVYDIIRNIRDPEKPNTLEDLDVVSESCVSVQELDEECYLVVIRFTPTVPHCSLATLIGLCLRVKLQRCLSFKHKLEIYISEGTHSTEEDINKQINDKERVSAAMENPNLREIVEQCVTEPD FAM96A_danRef --EKALEVYDVIRTIRDPEKPNTLEELDVVTEKCVEVQELGDDEYLIVIKFSPTVPHCSLATLIGLCLQVKLQRCLPFKHKLEIYITEGTHSIEEDINKQINDKERVAAAMENPNLREIVEQCVTEPD FAM96A_oreNil --EKALEVYDVIKSIRDPEKPNTLEELEVVTEKCVEVQELGEDEYLIIIRFSPTVPHCSLATLIGLCLQVKLQRCLPFKHKLEIYISEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVTEPD FAM96A_oryLat --EKALEVYDVIRSIRDPEKPNTLEELEVVTEKCVEVQDLGEDEYLIIIKFSPTVPHCSLATLIGLCLQVKLQRCLPFKHKLEIYLSEGTHSTEEDINKQINDKERVAAAMENPNLREIVEQCVTEPD FAM96A_cioInt MEDYEGTIYDIIRTIKDPEKPGSLEDLDVVYEEGVSVKTSENHRCNVEVKFRPTIKHCSLATLIGLCLHVKLQRTLPTTHKIRVFVKEGSHNTEDEVNKQINDKERIAAAMENPNIRKMVENCIKEPD FAM96A_braFlo LDDLSDIVYDLIRDIRDPEKDNTLEELDVVYESGVHVEPWGEDKFHISIEFTPTVPHCSLATLIGLCLRVKLENNLPQHYKLDITVKEGTHSTGPEINKQINDKERIAAAMENPDLRAVVNKCVQDPE FAM96A_sacKow EKELAEEIYDIIRTIRDPEKPQTLEDLDVVYEDGVLVNHRGTDEFLVNVEFTPTVPHCTLATLIGLCIRVKLQRTLPHSYKLDIFIKKGTHSTEDEINKQINDKERIAAAMENPNLKDLVDNCVVDLE FAM96A_strPur LNGMAGDIYDIIRDIQDPEKPNTLEDLEVVYEEGVTVAALETEEQLINIEFTPTVPHCSLATLIGLCLRVRLERSLPNKHKLDIIVKKGTHATEDDINKQINDKERIAAAMENPNLRKLVEHCVSIED FAM96A_triCas DSELKYTVYDLIRTIKDPEKPNTLEELNVVYEEGVEVKERTSGNVSVVVEFNPTVPHCSLATLIGLCIRIKLERCIPYRIKLDIYIKAGAHTTEHEINKQINDKERIAAAMENPNLREMVENCIVEED FAM96A_nemVec NRNLALDVYDLIKDIKDPEKPQTLEDLKVVYESCVEVQKVAGQDHIT-ITFTPTVPHCSLATLIGLCIRVKLEKSLPEKFKLDIYLKKGTHSTENEINKQINDKERIAAAMENPNLRKIVENCIDEDN FAM96A_acrPal NLVLVQEVFDIVKDIRDPELPQTLEELHVIEEEFIKIDKIENDEYIIKIEFTPTVPHCSLATLIGLCLRVKLERSLPYKFKLDIFLSRGTHSTENEINKQINDKERIAAAMENPNLKKIVEECILDAN FAM96A_triAdh NQKLCSQIFELIRDIKDPELPQTLEELNVVTEDEIFVRNMKQGEACIRINFTPTVPHCSLATLIGLCIRVKLQRCLDQDYKLDIYVTKGSHDTEDGVNKQINDKERVAAAIENPNVKKLVEECLQEVQ FAM96B_homSap DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRVQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAVNKQLADKERVAAALENTHLLEVVNQCLSARS FAM96B_papHam DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRVQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAVNKQLADKERVAAALENTHLLEVVNQCLSARP FAM96B_micMur DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRVQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAVNKQLADKERVAAALENTHLLEVVNQCLSARS FAM96B_tupBel DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRVQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAVNKQLADKERVAAALENTHLLEVVNQCLSARS FAM96B_musMus DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRIQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAVNKQLADKERVAAALENTHLLEVVNQCLSars FAM96B_oryCun DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRVQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAVNKQLADKERVAAALENTHLLEVVNQCLSARS FAM96B_bosTau DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRVQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAVNKQLADKERVAAALENTHLLEVVNQCLSARS FAM96B_canFam DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRVQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAGKKQLADKERVAPPLENTHLLEVVNQCLSARS FAM96B_loxAfr DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRVQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAVNKQLADKERVAAALENTHLLEVVNQCLSARS FAM96B_choHof DSIDAREIFDLIRSINDPEHPLTLEELNVVEQVRVQV---SDPESTVAVAFTPTIPHCSMATLIGLSIKVKLLRSLPQRFKMDVHITPGTHASEHAVNKQLADKERVAAALENTHLLEVVNQCLSARS FAM96B_macEug DSIDDREIFGLIRSINDPEHPLTLEELNVVEQVRVKV---NDRESTVAVEFTPTIPHCSMATLIGLSIKVKLIRSLPERFKMDVHITPGTHASEHAVNKQLADKERVAAALENSHLLEVVNQCLSARS FAM96B_monDom DSIDDREIFVLIRSINDPEHPLTLEELNVVEQVRVKV---NDRESTVAVEFTPTIPHCSMATLIGLSIKVKLIRSLPERFKMDVHITPGTHASEHAVNKQLADKERVAAALENSHLLEVVNQCLSARS FAM96B_galGal DSIDDREIFDLIRSINDPEHPLTLEELNVVEQVRVKV---NDAESTVAVEFTPTIPHCSMATLIGLSIKVKLIRSLPERFKMDVHITPGTHASEHAVNKQLADKERVAAALENSHLLEVVNQCLSARS FAM96B_xenTro DRIDDREIFDLIRCINDPEHPLTLEELNVVEEIRVKV---SDEESTVSVEFTPTIPHCSMATLIGLSIKVKLLRSLPERFKVDVHITPGTHASEHAVNKQLADKERVAAALENSHLLEVVNQCLSGRS FAM96B_tetNig DPIDDREIFDLIRTINDPEHPLSLEELNVVEQVRVKV---NDAESTVDVEFTPTIPHCSMATLIGLSIKVKLLRCLPNRFKIDVHITPGTHASEEAVNKQLADKERVAAALENSSLLEVVNQCLSSRG FAM96B_gasAcu DPIDDREIFDLIRAINDPEHPLSLEELNVVEQVRVQV---NDEESIVGIEFTPTIPHCSMATLIGLSIKVKLLRSLPDRFKIDVHITPGTHASEEAVNKQLADKERVAAALENSSLLEVVNQLTPTRG FAM96B_danRer DPIDVREIFDLIRSINDPEHPLSLEELNVVEQVRVNV---NDEESTVSVEFTPTIPHCSMATLIGLSIKVKLLRSLPDRFKIDVHITPGTHASEDAVNKQLADKERVAAALENSQLLEVVNQCLSSRG FAM96B_petMar DEIDSREVFDLIRGINDPEHPLTLEELKVVEEAYVSV---TDAESMVVVAFTPTIPHCSMATLIGLAIRVQLLRCLPDRFKVDVHIAPGMHASEHAVNKQLADKERVAAALENSHLLGVVNQCLGGRK FAM96B_cioInt DPFDRREIFDLIRDINDPEHPLTLEDLRVVSENDIEV---DDEKSFIKVSFTPTIPHCSMATLIGLAIRVRLLRSLPPRFKVEVEISPGSHQSEKAVNKQLGDKERVAAALENNHLLNVVNQCLTgrk FAM96B_braFlo DAIDAREIFDILSSINDPEHPLTLEELNVIEQSRITV---DEDNNHVSVEFTPTIPHCSMATLIGLSIRVKLLRALPTRFKVDVHITPGTHQSEHAVNKQLADKERVAAALENQHLLEVVNQCLSTRN FAM96B_strPur DAIDTREVFDLIRNINDPEHPLTLEELNVVQQAEVEV---DDPGNVVKVTFTPTIPHCSMATLIGLAIRVKLIRSLPSRFKVDINIKPGTHVSENAVNKQLADKERVAAALENNHLLEVVNQCLTQRD FAM96B_monFav DKIDEREVFDLIRSINDPEHPLTLEQLNVVEQSLVEV---DDTNNYVKIQFTPTIPHCSMATLIGLAIRVQLLRSLPDRFKVDISITPGTHASEDAVNKQLADKERVAAALENTHLLEVVNQCLAVRH FAM96B_nemVec DKIDDREIFDMIRSINDPEHPLTLEELNVVEQALIDV---SDDESYVKVQFTPTIPHCSMATLIGLAIRVRLLRSLPDRFKVDVKITPGTHQSEIAVNKQLADKERVAAALENNHLLDVIDQCLVSKK FAM96B_plePil dkiddreivdmirsINDPEHPNSLEELSVVQLDLITC---NDTDNYVDVKFTPTIPHCSMATLIGLSLKVKLLRSLASRFKVDVRITPGSHSTEEAINKQLADKERVAAALENPQLVNMVNQCIYGkk FAM96B_ampQue DEIDAREVFDLIRHINDPEHPLTLEELNVVQEDLICI---NNKENFVSVHFTPTIPHCSMATLIGLSIRVCLLRSLPNRFKIDVIITPGSHMSEQAINKQLADKERIAAAIENSHLLNVVHQCLNTKR FAM96B_subDom DEIDAREVFDLVKNINDPEHPLTLEQLNVVQLGHIDV---SDVDSSVTVYFTPTIPHCSMATLIGLSIRVRLLRALPARFKVDVMISPGTHASEVAVNKQLADKERIAAALENNHLLDVVNSCLTGVr FAM96B_triAdh DKIDEREIFDIIRSINDPEHPLTLEELNVVEECKIDV---DDDNNFVKVHFTPTIPHCSMATLIGLCIRVRLIRSLPERFKVDITVTPGSHSSEIAVNKQLADKERVAAAMENSNLLKVVNQCLAMDr FAM96B_monBre DPFDSREIFDLVRHINDPEHPLTLEELNVVRLDQILV---DDAQNYVRVQFTPTIPHCSMASLIGLCLRVRLLRALPPRFKVDVEIFPGTHATEASINKQLADKERVAAALENPNLKTVVNECLQLDD FAM96B_sacCer DLIDAQEIYDLIAHISDPEHPLSLGQLSVVNLEDIDVHDSGNQNAEVVIKITPTITHCSLATLIGLGIRVRLERSLPPRFRITILLKKGTHDSENQVNKQLNDKERVAAACENEQLLGVVSKMLVTCK FAM96B_ajeDer EPIDEQEIYDLIATIADPEHPISLGALAVVSLPDISIKPPDSPLRTVSVLITPTITHCSLATVIGLGVRVRLEQSLPPRFRVDVRIKEGTHSTADEVNKQLADKERVAAALENGTLMGVIGKMLETCQ FAM96B_ashGos DPVDPQEIYDLIAHISDPEHPLTLGQLAVVNLPDIEVRDSGDPHAEVVVRITPTITHCSLATLIGLGIRVRLERSLTPRFRITVLLKKGSHQSENQVNKQLNDKERVAAACENEQLVEVVSKMLSTCK FAM96B_canAlb DPIDEQEIFDLIATISDPEHPLTLAQLAVVNLSDIKITNDGGGGSEVLIKITPTITHCSLATLIGLGIRVRLDRSLPSRYRIKILIKEGTHQSENQVNKQLNDKERVAAACENDQLLNVISQMLSTCK FAM96B_dekBru EPIDAQEIYDLTASISDPEHPLTLGQLAVXNLNDIEVKNASDKSGEILLRITPTISQCSLATLIGLGIRVRLDRCLPKRFRITILLKEGTHQTEKQVNKQLNDKERVSAAAENPQLLKVISNMLSSCE FAM96B_kluLac DPIDAQEIYDLIAHISDPEHPLTLGQLAVVNLADIEVHDTNGKDAEVIVRITPTITHCSLATLIGLGIRVRLERSLSPRFRITILLKKGTHQSENQVNKQLNDKERVAAACENDQLLGVVSKMLSTCK FAM96B_komPas ESVDALEIYDLISSISDPEHPLTLGQLAVVNLEDIQLDDSGNPNAEVIIKITPTITHCSLATLIGLGIRVRLERCLPPRYRIIIKVKEKTHQSENQVNKQLNDKERVSAACENDQLLKVISQMLSSCK FAM96B_parBra EPIDEQEIFDLISTIADPEHPISLGSLAVVSLPDISIRPPDSPLRTVTVLITPTITHCSLATVIGLGVRVRLEQSLPHRFRVDVRIKEGTHSTADEVNKQLADKERVAAALENGTLMGVIGRMLETCQ FAM96B_schPom DPIDPQEIYDLLAKINDPEHPLTLAQLSVVKLEDIEVVDNVEGDSYITVHITPTIPHCSMCTLIGLCIRVRLERCLPPRFHVDVKVKKGTHASESQVNKQLNDKERVAAACENEQLLSVLNGMMATCV FAM96B_triAtr EAIDEQEIYDLISNITDPEHPVSLGQLSVINLPDIHITPVPSPNVQVTVELTPTVTHCSLATVLGLGVRVRLEQVLPPNYRVEVICKENSHSQDDQVNKQLSDKERVAAALENDSLKSVLDKMLESCI FAM96B_yarLip EPIDSQEIYDLIATISDPEHPLTLGQLAVVKLEDIWVHDTGDKNAEIVVKITPTITHCSLATLIGLGIRVRLERALPPRFRFTITVKEGTHQSENQVNKQLNDKERVAAACENEQLLGVISGMLATCQ FAM96B_micSpp DPVDAIEVFYHIKNINDPEHPYSLEQLDIVSVENIRV---HSEAQFIQVYFTPTVPHCSMATLIGLAIRRKLQESLAGRFKTEVLVFPGSHSSESAVNKQLNDKERVAAALENTNLLEKVNLCLRGNL FAM96B_ostLuc DAVDALEIFDHVRDINDPEHPYSLERLNVVGASAIEC---DDARNRVRVEFTPTVPHCSMATLIGLSIRVKLLRTLPRRFKVDVVIAPGTHASERAVNKQLNDKERVAAALENGNLLEKVDLCLSGKT FAM96B_araTha EPIDQLEIFDHIRDIKDPEHPNTLEDLRVVTEDSVEV---DDENSYVRVTFTPTVEHCSMATVIGLCVRVKLLRSLPSRYKIDIRVAPGSHATEDALNKQLNDKERVAAALENPNLVEMVDECLPSEE FAM96B_vitVin EPVDQQEIFDHIRDIKDPEHPYSLEELKVITEDAIEV---DDKRSYVRVTFTPTVEHCSMATVIGLCLRVKLLRSLPSRYKVDIKVAPGTHATEAAVNKQLNDKERVAAALENPNLLDMVDECLAPSY FAM96B_popTri EPIDQLEVFDHIRDIKDPEHPYSLEELKVITEDAIEV---DDNHSYVRVTFTPTVEHCSMATVIGLCLRVKLMRSLPQRYKVDIRVAPGTHATESAVNKQLNDKERVAAALENPNLVDMVDECLAPSY FAM96B_zeaMay EPIDQLEIFDHIRDIKDPEHPYSLEQLNVVTEDSIEL---NDESNYVRVTFTPTVEHCSMATIIGLCIRVKLVRSLPPRYKVDIRVAPGSHATEAAVNKQLNDKERVAAALENPNLLDMVEECLSPTF FAM96B_braDis EPIDQLEIFDHIRDIKDPEHPYSLEELNVVTEESVEI---NDKLSHVRVTFTPTVEHCSMATVIGLCVRVKLIRSLPPRYKVDIRVAPGSHATETAVNKQLNDKERVAAALENPNLLDIVEECLAPTF FAM96B_dicDis DEFDEQEIFDLVRSITDPEHPLTLEQLNVVRIENVNI---NLENSYILLYFTPTVPHCSMANLIGLSIKEKLARSLPKRFKVDVIVTPGSHSSESSVNKQLNDKERVSAALDSSSILTIVNECIKQN- FAM96B_polPal DDFDVYEIFDLVRDINDPEHPLTLEQLNVVRHENIKI---DISNNIIRLYFTPTVPHCSMANIIGLSIKEKLSRSLPQRFKVDVKVTPGSHSSEQSVNKQLNDKERVSAALDSSSILNVVNECIKLPI FAM96B_entDis EDIDQLEIYEHIRRIKDPEHPVTLEQLKVISPDLINV---DDKGNHIIVKFTPTVDNCTMATLIGLTIRTKLMRILPPRIKLDIYLTKGTHQTEEDVNKQLNDKERIAAALEKQTLLQLVNKCLIlpi FAM96B_phyInf DPFEPDEVFEILRHINDPEHPLTLEQLKVMSLENVHV---DDVNSRVKIFFTPTIPHCSMATLIGLCLRVKLLRSLPSRFKVDILITPGTHSSEAAVNKQLNDKERVAAALENSHLLTVVNKCIAHTD FAM96B_thaPse DAITVNEIFDIVRNIQDPEHPLTLEQLNVVRLELIKV---VDSFSTVHVQFTPTIPHCSMATLIGLSLRVKLLRSLPPRFKVVVEIESGTHASEHAVNKQLADKERVRAALENEHLLGVVNKCIAGVA FAM96B_phaTri DMVDADEVFEIIRNIQDPEHPLTLEQLGVVSKRQIDV---HDSYSTLDVRFTPTIPHCSMATHIGLCLRVKLDRSLPPRFKVKVRIEPGSHSSETAINKQLADKERVCAALENKHLLGIVNRCIIDGM FAM96B_naeGru DEFDALEVYDLIRNINDPEHPLSLEQLKVTQHDLITV---DNKNNLIVIYFTPTITHCSMATLIGLSIRVKLLRSLPKRFKVDIFITPGTHQSEDQVNKQLNDKERVAAALENERLLSVVNRCIAQS- FAM96B_triVag EAIDSLELYNYIRLIKDPEHPFSLEQLHIVSPDDIKV---DDKEGRVNLVFTPTVPNCSLPAVLGLCIRERLLQVLPQRFKIFITVARGKHIQEDSINRQLRDKERCLAALERRNIRTMIDNCIACDD FAM96B_cryMur SEITPMDIFEIIRRIKDPEYPLTLEQLNVVELKNISV---DNNANRVIVYFTPTITSCSQASLIGLSILFKLTFTLPSRFKVIIKVTPGSYDSEEALNKQMRDKERVRAALENMQIFKAITRGIVNSD FAM96B_babBov DEFEVTEIFNIIRNIKDPEYSYTLESLKIVEPENIDI---DQENAIVTVKFTPTVPHCSQATIIGLMIYVKLQQSLPLHFKIDVQITEGTHNTEDAINKQLLDKERVAAALENPVLLDMINDGIYNTV FAM96B_tetThe DEIDQLEIFDLIRHIDDPEHPLTLEQLNVLQPENIKV---NIDHKLVTVLFTPTIPHCSLAQIIGLMIKVKLIRSLPRDYKVDVYITPGTHVQELSVNKQINDKERVMAAIENPSILRVVNKGVSNSD FAM96B_theAnn ESFDEEEIFDIIRTIKDPEYSYSLEDLNVVSKDNIFI---DEDTSTISVFFTPTVPHCTQASIIGLMIFVKLYQSLPPYFKIDVQISKGTHNTEEMINKQLLDKERISAALEYPPILKMINKGILFLQ FAM96B_tryCru DPIDSLEVFHHIRSIRDPEHPNTLEELKVVEPELIRV---DEVKQTVRVQFTPTVPHCSMTTLIGLCISLKLQRSLPRGTKVDVYVTPGSHEQEEQVNKQLNDKERVAAALENKNLLNVVESCLNEFE FAM96B_leiInf DPIDAWEVFEIIRRIRDPEHPNSLEQLKVVEPSLITV---DWKKRHIRVLFTPTVPHCSLTTLIGLSIRLQLERSLPEYTKVDIYVTPGTHEQEAQVNKQLNDKERVAAALENCNLLNVVESCINEFD
NARFL (IOP1)
(coming shortly)
ERCC2 (XPD)
(coming shortly)
Curated reference sequences
It serves no current purpose to collect all possible full length MMS19 sequences from GenBank, so only a sample of 20 uniformly distributed over the eukaryotic phylogenetic tree is provided here. MMS19 presents no real homology complications, being present as a single-copy gene. Genes in early diverging eukaryotes are assumed single-exon, ie taken as the largest open reading frame enveloping the match to the ultra-conserved region. MMS19 is studied experimentally only in yeast and human.
The apparent absence in Giardia and various obligate parasites could be attributable to a reduced genome, extreme sequence divergence relative to available probes, or incomplete assembly -- it is inconceivable that these species lack core iron sulfur proteins of DNA metabolism. Indeed, the conserved cysteine pattern of primase large subunit are readily located in these species. It remains conceivable however that the very earliest diverging eukaryotes retain components of the archaeal iron sulfur cluster formation system.
The yeast gene, sometimes called MET18 in that literature, is unsurprisingly single-exon (only 283 of 6000 yeast proteins have them) and not located in a yeast-type operon. While some immediate neighbors are involved in DNA processes, none are homologous to iron sulfur cluster assembly components or have recognized 4Fe-4S cofactors themselves.
Gene Position Description MET18 chrIX:113806 DNA repair, TFIIH regulator, nucleotide excision repair, RNA polymerase II, telomere maintenance RRT14 chrIX:117024 rDNA transcription, localizes to nucleolus, involved in ribosome biogenesis STH1 chrIX:117992 ATPase component in chromatin remodeling, expression of early meiotic genes, helicase-related protein homologous to Snf2p KGD1 chrIX:122689 mitochondrial alpha-ketoglutarate dehydrogenase ASG1 chrIX:102782 zinc cluster transcriptional regulator stress response CSM2 chrIX:99860 homologous recombination repair, accurate chromosome segregation during meiosis SIM1 chrIX:128151 may participate in DNA replication
The human gene has 31 coding exons. These do not correspond to natural structural breaks in the tertiary structure (eg HEAT units) and the ultra-conserved regions is spread across parts of 3 exons. Thus despite its modular structure, MMS19 had already completed its internal expansion of domain units prior to the main era of exon formation and could not today expand further by exon duplication because these would present issues of compatible [phasing] as well as not corresponding cleanly to structural units.
Exon structure of human MMS19: columns show exon number, amino acid size, intron phasing (donor bp overhang), primary sequence, and ultra-conserved region. 1 37 1 MAAAAAVEAAAPMGALWGLVHDFVVGQQEGPADQVAA 2 17 2 DVKSGNYTVLQVVEALG 3 33 1 SSLENPEPRTRARAIQLLSQVLLHCHTLLLEKE 4 29 0 VVHLILFYENRLKDHHLVIPSVLQGLKAL 5 25 0 SLCVALPPGLAVSVLKAIFQEVHVQ 6 23 1 SLPQVDRHTVYNIITNFMRTREE 7 43 1 ELKSLGADFTFGFIQVMDGEKDPRNLLVAFRIVHDLISRDYSL 8 21 0 GPFVEELFEVTSCYFPIDFTP 9 29 0 PPNDPHGIQREDLILSLRAVLASTPRFAE 10 25 0 FLLPLLIEKVDSEVLSAKLDSLQTL 11 26 0 NACCAVYGQKELKDFLPSLWASIRRE 12 46 1 VFQTASERVEAEGLAALHSLTACLSRSVLRADAEDLLDSFLSNILQ 13 52 0 DCRHHLCEPDMKLVWPSAKLLQAAAGASARACDSVTSNVLPLLLEQFHKHSQ 14 26 1 SSQRRTILEMLLGFLKLQQKWSYEDK 15 42 1 DQRPLNGFKDQLCSLVFMALTDPSTQLQLVGIRTLTVLGAQP 16 28 2 DLLSYEDLELAVGHLYRLSFLKEDSQSC 17 33 1 RVAALEASGTLAALYPVAFSSHLVPKLAEELRV 18 50 1 GESNLTNGDEPTQCSRHLCCLQALSAVSTHPSIVKETLPLLLQHLWQVNR 19 52 1 GNMVAQSSDVIAVCQSLRQMAEKCQQDPESCWYFHQTAIPCLLALAVQASMP 20 34 2 EKEPSVLRKVLLEDEVLAAMVSVIGTATTHLSPE 21 34 0 LAAQSVTHIVPLFLDGNVSFLPENSFPSRFQPFQ 22 23 0 DGSSGQRRLIALLMAFVCSLPRN 23 42 1 VEIPQLNQLMRELLELSCCHSCPFSSTAAAKCFAGLLNKHPA 24 34 0 GQQLDEFLQLAVDKVEAGLGSGPCRSQAFTLLLW 25 19 0 VTKALVLRYHPLSSCLTAR 26 62 1 LMGLLSDPELGPAAADGFSLLMSDCTDVLTRAGHAEVRIMFRQRFFTDNVPALVQGFHAAPQ 27 28 0 DVKPNYLKGLSHVLNRLPKPVLLPELPT 28 55 0 LLSLLLEALSCPDCVVQLSTLSCLQPLLLEAPQVMSLHVDTLVTKFLNLSSSPSM 29 20 0 AVRIAALQCMHALTRLPTPV 30 34 2 LLPYKPQVIRALAKPLDDKKRLVRKEAVSARGEW 31 9 0 FLLGSPGS*
Alignment of ultra-conserved region: MMS19_homSap FTFGFIQVMDGEKDPRNLLVAFRIVHDLI-------SRDYSLGPFVEELFEVTSCYFPIDFTPPPNDPHG-IQREDLILSLR MMS19_musMus FTFGFIQVMDGEKDPRNLLLAFRIVHDLI-------SKDYSLGPFVEELFEVTSCYFPIDFTPPPNDPYG-IQREDLILSLR MMS19_cioInt FLYQYIQVIDGEQDPRNLLTIFQLTKNLI-------ESSFPLFDLVEELFDVSSCYFPIDFNPAAAGKKSTITNLDLVSSLR MMS19_braFlo FVWGFIQAMDGEKDPRNLIIAFSIAR-IV-------AQAFPIGTFTEELFEVISCYFPIDFTPPADDPHG-VTREDLVLGLR MMS19_strPur FVLGLLHAMDGEKDPRNLILLFNILP-TV-------INNFKIDMFIEETFEVVACYYPVDFHPPPNDPYG-ISREKLALSLK MMS19_sacKow FVFGYIQCMDGEKDPRNLTMIFRCVP-II-------IHNFPIDVFIEELFEVVSCYFPIDFTPPPNDPYK-VTQEELVLGLR MMS19_dapPul FVFGLIQLADQERDPRNLLILFSIFPVVA--------RYFRFEPFTEEFFEVFSCYFPIDFTPPANDPYA-VTKEQLCDGLR MMS19_droMel FVYGLINSIDGERDPRNLDIIFSFMPEFL--------STYPLLHLAEEMFEIFACYFPIDFNPSKQDPAA-ITRDELSKKLT MMS19_nemVec LVFGFLQAMDGEKDPRNLVVAFKLAR-II-------IKNFPIGLFAEDLFEVTSCYFPIDFTP------------------- MMS19_triAdh FVFGYIQVMDGEKDPRNLLLALKIAKFIV--------QNFNIDLFLDDFFEIISCYFPIDFTPPPNLP----SNENVTK--- MMS19_sacCer FIETFLHVANGEKDPRNLLLSFALNKSIT-------SSLQNVENFKEDLFDVLFCYFPITFKPPKHDPYK-ISNQDLKTALR MMS19_schPom FFSGICSTFAGEKDPRNLMLVFSMLK-KI-------LSTFPIDGFEQQFFDITYCYFPITFRAPPDATNLAITSDDLKIALR MMS19_araTha LVYAMCEAIDGEKDPQCLMIVFHLVELLAPLFP---SPSGPLASDASDLFEVIGCYFPLHFTHTKDDEAN-IRREDLSRGLL MMS19_dicDis FMVGYLQFIDNEKDPRNLIFSFKLLPKVIYNIP---EHKHFLES----LFEIISCYFPISFNPKGNDPNS-ITKDDLSNSLL MMS19_pytUlt FAQAFLNAMEGEKDPRNLLLCLQIARELLAKLE---VVFDRHDAVLQQYFDVVSCYFPITFTPPPNDPYG-ITSEELILSLR MMS19_sapPar LMDGFLRAMSGEKDPRNLLFCLRFAAELLTTYA---NVVDAD--VAKGFFDATSCYFPITFRPPPNDPYG-ITSEDLVLALR MMS19_polPal FMAGFLQFIDGEKDPRNIIYTFRLIPRVILYIP---EYKNFADS----LFEILSCYFPISFNPKPGDPNS-ITKDDLVSSLL MMS19_entHis -IDTCVQLIELERDPECLKEVFDLIKLVS-------QKNEIDADSAPLLFDCASAYFPILYPPKGDEA----LRIDLTNKIL MMS19_naeGru FLNGFIQSLEGERDPGNLLYCFNLIPKVIAIFDDSELSSKILSAVSDDLFDITSCYFPITYTPPANDTRG-ITREDLSRSLK MMS19_phyInf LAQTFLSAMEGEKDPRNLLLCMQVARTLLSKLE---PVFSRSDTLLQQYFDVVSCYFPIIFTPPPNDPYG-ITSEGLILSLR MMS19_albLai FIRSFLNAMTGEKDPRNLKHCFQIAQTMMQKLE---MVFQEAE-LSEQYFRVISCYFPITFTPPPNDPYG-VTTEELIRSLR Consensus f g q dgEkDPrnL f lF#v sCYFPI ftpp ndp it edl lr
Summary of MMS19 reference sequences: MMS19_homSap Homo sapiens mammal Q96T76 1030 aa 7 HEAT 100% MMS19_musMus Mus musculus mammal Q9D071 1031 aa 9 HEAT 89% MMS19_cioInt Ciona intestinalis urochordate XP_002128657 1026 aa x HEAT 32% MMS19_braFlo Branchiostoma floridae cephalochordate XP_002588594 1027 aa x HEAT 42% MMS19_strPur Strongylocentrotus purpuratus echinoderm XP_001194909 975 aa x HEAT 36% MMS19_sacKow Saccoglossus kowalevskii hemichordate XP_002735310 1007 aa x HEAT 40% MMS19_dapPul Daphnia pulex crustacean EFX86854 961 aa x HEAT 38% MMS19_droMel Drosophila melanogaster insect NP_649519 959 aa x HEAT 30% MMS19_nemVec Nematostella vectensis cnidarian XM_001629116 897 aa x HEAT 32% MMS19_triAdh Trichoplax adhaerens single-celled metazoan XP_002114595 959 aa x HEAT 39% MMS19_sacCer Saccharomyces cerevisiae budding yeast P40469 MET18 1032 aa 13 HEAT 29% MMS19_schPom Schizosaccharomyces pombe fission yeast Q9UTR1 1018 aa 14 HEAT 25% MMS19_araTha Arabidopsis thaliana plant NM_124186 1134 aa x HEAT 28% armadillo/beta-catenin-like MMS19_dicDis Dictyostelium discoideum slime mold Q54J88 1115 aa 18 HEAT 31% MMS19_pytUlt Pythium ultimum stramenopiles ADOS01001616 957 aa 31% MMS19_sapPar Saprolegnia parasitica stramenopiles ADCG01000470 804 aa 31% MMS19_polPal Polysphondylium pallidum amoeba EFA86574 994 aa x HEAT 28% MMS19_entHis Entamoeba histolytica amoeba XP_651925 868 aa x HEAT 25% MMS19_naeGru Naegleria gruberi early eukaryote: heterolobosea XP_002678884 1070 aa x HEAT 27% MMS19_phyInf Phytophthora infestans early eukaryote: stramenopiles 1114 aa x HEAT 33% MMS19_albLai Albugo laibachii early eukaryote: stramenopiles 1077 aa x HEAT 27%
>MMS19_homSap Homo sapiens mammal Q96T76 1030 aa 7 HEAT 100% MAAAAAVEAAAPMGALWGLVHDFVVGQQEGPADQVAADVKSGNYTVLQVVEALGSSLENPEPRTRARAIQLLSQVLLHCHTLLLEKEVVHLILFYENRLK DHHLVIPSVLQGLKALSLCVALPPGLAVSVLKAIFQEVHVQSLPQVDRHTVYNIITNFMRTREEELKSLGADFTFGFIQVMDGEKDPRNLLVAFRIVHDL ISRDYSLGPFVEELFEVTSCYFPIDFTPPPNDPHGIQREDLILSLRAVLASTPRFAEFLLPLLIEKVDSEVLSAKLDSLQTLNACCAVYGQKELKDFLPS LWASIRREVFQTASERVEAEGLAALHSLTACLSRSVLRADAEDLLDSFLSNILQDCRHHLCEPDMKLVWPSAKLLQAAAGASARACDSVTSNVLPLLLEQ FHKHSQSSQRRTILEMLLGFLKLQQKWSYEDKDQRPLNGFKDQLCSLVFMALTDPSTQLQLVGIRTLTVLGAQPDLLSYEDLELAVGHLYRLSFLKEDSQ SCRVAALEASGTLAALYPVAFSSHLVPKLAEELRVGESNLTNGDEPTQCSRHLCCLQALSAVSTHPSIVKETLPLLLQHLWQVNRGNMVAQSSDVIAVCQ SLRQMAEKCQQDPESCWYFHQTAIPCLLALAVQASMPEKEPSVLRKVLLEDEVLAAMVSVIGTATTHLSPELAAQSVTHIVPLFLDGNVSFLPENSFPSR FQPFQDGSSGQRRLIALLMAFVCSLPRNVEIPQLNQLMRELLELSCCHSCPFSSTAAAKCFAGLLNKHPAGQQLDEFLQLAVDKVEAGLGSGPCRSQAFT LLLWVTKALVLRYHPLSSCLTARLMGLLSDPELGPAAADGFSLLMSDCTDVLTRAGHAEVRIMFRQRFFTDNVPALVQGFHAAPQDVKPNYLKGLSHVLN RLPKPVLLPELPTLLSLLLEALSCPDCVVQLSTLSCLQPLLLEAPQVMSLHVDTLVTKFLNLSSSPSMAVRIAALQCMHALTRLPTPVLLPYKPQVIRAL AKPLDDKKRLVRKEAVSARGEWFLLGSPGS* >MMS19_musMus Mus musculus mammal Q9D071 1031 aa 9 HEAT 89% MAAATGLEEAVAPMGALCGLVQDFVMGQQEGPADQVAADVKSGGYTVLQVVEALGSSLENAEPRTRARGAQLLSQVLLQCHSLLSEKEVVHLILFYENRL KDHHLVVPSVLQGLRALSMSVALPPGLAVSVLKAIFQEVHVQSLLQVDRHTVFSIITNFMRSREEELKGLGADFTFGFIQVMDGEKDPRNLLLAFRIVHD LISKDYSLGPFVEELFEVTSCYFPIDFTPPPNDPYGIQREDLILSLRAVLASTPRFAEFLLPLLIEKVDSEILSAKLDSLQTLNACCAVYGQKELKDFLP SLWASIRREVFQTASERVEAEGLAALHSLTACLSCSVLRADAEDLLGSFLSNILQDCRHHLCEPDMKLVWPSAKLLQAAAGASARACEHLTSNVLPLLLE QFHKHSQSNQRRTILEMILGFLKLQQKWSYEDRDERPLSSFKDQLCSLVFMALTDPSTQLQLVGIRTLTVLGAQPGLLSAEDLELAVGHLYRLTFLEEDS QSCRVAALEASGTLATLYPGAFSRHLLPKLAEELHKGESDVARADGPTKCSRHFRCLQALSAVSTHPSIVKETLPLLLQHLCQANKGNMVTESSEVVAVC QSLQQVAEKCQQDPESYWYFHKTAVPCLFALAVQASMPEKESSVLRKVLLEDEVLAALASVIGTATTHLSPELAAQSVTCIVPLFLDGNTSFLPENSFPD QFQPFQDGSSGQRRLVALLTAFVCSLPRNVEIPQLNRLMRELLKQSCGHSCPFSSTAATKCFAGLLNKQPPGQQLEEFLQLAVGTVEAGLASESSRDQAF TLLLWVTKALVLRYHPLSACLTTRLMGLLSDPELGCAAADGFSLLMSDCTDVLTRAGHADVRIMFRQRFFTDNVPALVQGFHAAPQDVKPNYLKGLSHVL NRLPKPVLLPELPTLLSLLLEALSCPDSVVQLSTLSCLQPLLLEAPQIMSLHVDTLVTKFLNLSSSYSMAVRIAALQCMHALTRLPTSVLLPYKSQVIRA LAKPLDDKKRLVRKEAVSARGEWFLLGSPGS* >MMS19_cioInt Ciona intestinalis urochordate XP_002128657 1026 aa x HEAT 32% MEKVKFEMEEMIQLWLRDKNDDHKILKCAQQIENREQTIGDLVTALGPHLTNKDTKIRIDACTLLSNVIHKLPKDCLNQGELESLVQFLCSRLEDHYTLQ PVALSLLLQLSSADNLTGENACSIITSVFKEVHIQTCMQHDRLKIFQILGTLLDIHTKDVITMGRDFLYQYIQVIDGEQDPRNLLTIFQLTKNLIESSFP LFDLVEELFDVSSCYFPIDFNPAAAGKKSTITNLDLVSSLRGVLASTKQFAQYCIPLMLEKLESDVESAKIDSLETLTACLGCYGKQELEKYLSSLWSDV KREINQSSSEQIEKCCLTFLTSLLSNLSSWPVDQKSEKATDLKSFLDDVLEDCVPRLQAQSDDRSKWMAGHVVLACAKSSKKACSQIVTTVLPILLQNAQ SKSASTTLAGQSVQQSALDNLVKLTAVCGQFNFENHPVLKKKEEFFTILNELALKSEIEEQLKCIAVAGFASLLKLEILSNVELTEIASLLLKMIKLKPE SHLRGEVLSVAGYLSSQHPDVAKSHLIPCVMRRMEEGDDSCFDVLASVCTHFDVLKLVLGFIMERIVNTQVDETSEPLLHACLESLQKMTSSSWVGNTEI EYMALNLVLPLLKRCIEVTLELSVPEQCCANCHIFEDVSKECASLPILKSAAIVIRNVCQKLKPGKSTDLVIQLIASLYNNSKLSSLDIKSDVHFTPFHP KASPLQTRTLCFLPATICALHPNIEIPELAELETKLLNTCLHCTDQPSYVFAAKALSGLVNKYKKPSIPILEKLKSHFDTDPNWSLKSEEEKMMILTLLI WICKALVLSNHPDSLIFIKNLLYWMGDDSVGEVAAAGFDIILRESNEVLSPSSHSTIRLMHKQRFFLLIIPEIVSSFKTSENKTQQTNILTALSHLIGHL PKQVLMQHFTELLPLLTQALHTDNTQLLKSVLSTLFCFIQDTTEAMTAHLENLMKHFLRLSKFKQDIDVRVKAVQCIGVVTLLPPIVILPFKNDIVRHLV SVLDDRKRDVRTEASKARSEWFLVGT* >MMS19_braFlo Branchiostoma floridae cephalochordate XP_002588594 1027 aa x HEAT 42% MAALSGNVQENVLEFVQGQQDSALQSVAKAVFDGETSLLQLVESLGSSLTSTEVTTRARATQLLAEVLHRSPSNRLTEKEAEVLSAFFCDRLLDHHSVQP HVLHGLLALSAAPQLPQGEEVKIVQVIFKEVYVQSLVQTDRRAIYNILANFLDTRLEALQALGADFVWGFIQAMDGEKDPRNLIIAFSIARIVAQAFPIG TFTEELFEVISCYFPIDFTPPADDPHGVTREDLVLGLRQVLAATSKFAQFCVPMLLEKLSSDVTSAKLDSLHTLAACAEVYGADSMKSFLDLLLSAISKE VYSSIHQDVENAALAGLTAVVATLSHAVTETRSVFSLHHFLDSLLKGCKHHLCEPELKLMWPSAKLLQAAARASDPACVHVLDTAVPLLVEQFQVHPYPQ HRHTILEVTIAFIHVAHASTSGTDAPNPVVPHSDNFLTLFYSVLEDADAGLRSSGVGGMAAMIGITDVVKGKHLDLCANHLGRLVLHDADPTVQRRSTEA LAAMATAHPDVVREEVLSKLLQVLENNNPNAMDTNQSEQVCAKHVTNQYVLNTLAAVSTHPTIVRCTIPKLLSHLQALIESCPDQATQEAIATLDCVYKV VEKTVINDANVEYFVDTIVLNLMSMALSAAVNTSDENLLHDTSLLEIVAKVLRAVARSLPNSTGKGIVNSTVQAFLQGNLAAISLNTSASFEPLDVSSPW QQTQTVQLLSAIVCSVARNVDIPSISELAQKLLTLSCASDHEPTSLAAAKSLSGVVNKWDQGEQLQTFLQETRDCLEQILSKTEDEKARCRAVAVLVWLT KALVIRGHPSGSQFTKTLMALFEDEAIGRRAAEGFYVILSDSPDVLSKESHANIRLMYKQRFFMENLPALVDGFNQADDGRKQSFLCAVSHLLTFIPRQV LLGALPPLVPLLVQSLLGEDPSLQVSTLEMFSSLVQEAPQVISKNIDALIPQLLELSKNGPTMKVRMAALKSVGSMTSLPHAVVYPYRNRVVRELAVAVG DKKRMVRKEAVAARGEWFLLGSPGGK* >MMS19_strPur Strongylocentrotus purpuratus echinoderm XP_001194909 975 aa x HEAT 36% MGTYLMSTETRIRAKGVELMSEVLTSLPRAFLNQQQIQVLIEFLCARLLDHHSITQHTLKGLLAMSSQSSFPPSSSVQVMTAIFKEVQVQTMLQVDRRTV YNIVVNLLRISLTELQGMGSEFVLGLLHAMDGEKDPRNLILLFNILPTVINNFKIDMFIEETFEVVACYYPVDFHPPPNDPYGISREKLALSLKTCLSST PKFAQFCLPMVMEKLSSDLQTARLDSYQLLQACAPVYSQGDMMSYIEAIWSYCRKELMVGASVELDQEAVKTLGAVVKAVSTGIQSTSGGGGGDGDLNSF LRNILTECRQHLCKPDHRMIHPCSKLLVAVATASYPACIAILKYSVPILLDQFHIFDQTRERMVLLDIIQRLLHSGHDHIKEADDWRAIYAHHLDTVVTT VLSTLAKDQGVPDLRMAGLGTLGELVQVPVLMDQSRLELVGQELTRILLEEANEDVRCECIETVSCFASRHTEFVKSTILTTLWTTVQKGESGYQRIVVD MATIVTDTSDDLSRSLTSELMVEIAKTELNSEQHLVYLATLNTLTAHLSSAPSNLESLLSSVVHPLMKMVVSATLQSSVEAGNNPHCCGEVLVAMAEVFR TVIPKLDSSMGSKLCQCAVDVFLHGNLTSLELTNPNTSVPFSPLDPRAPVHQTQLVTVLQPIVCSLRRDIHIPSSKQLMSSLLHIAAHSRAWLASTWAAK GLAGGVNKHPAGSDLDEVLVEAESLLGQAMSSGQEGSVKQQALMAWVLLTKALVMRSHPKATAFLTTLLRLLEDAELGAQVPQTLGMLLEDMRDVLSEGL HADVKIMYKQRVFLQALPFVMALFNKDDLRTKAITALCHLLPSIPRPVLLAELPPIIPRLVQSLRVTDPRPTLPILDILESLLEETLPSLVDQADTLLPT LLELSAYQASMKVRIASLKCVGAITSFPHHLVYPHQETVVRSLAPRLDDKKRLVRQEAGKARTKWILLQQDTKG* >MMS19_sacKow Saccoglossus kowalevskii hemichordate XP_002735310 1007 aa x HEAT 40% MATSMCIEIVENYVRGEDESAIHAAEIKILELVENLGTYLTSTEKNIRCRGTRLLSEVLNRLPKNFLSSDEVRALVIFYCDRLSDHYSVTPHTLLGMLAL STYDNLPKGCEVQLVQAIYKEVHVQSMIQVDRRSVYAILSNLLDTRIKDLQSLGRDFVFGYIQCMDGEKDPRNLTMIFRCVPIIIHNFPIDVFIEELFEV VSCYFPIDFTPPPNDPYKVTQEELVLGLRKCLAATPKFAEHCLPLLMEKLSSDVQRAKIDSFLTLAECCEVYGEDDLMEFLPAMWSTIRREVFQAFSHEV EKSALTCLCSIVKTLSNAVSNANKAAGGLDEFLDLVLKDCSKHLRDPGLRLMLPTSKLLQSAASASDPACYKIISAVVPILLEQFHKCKQVNERVSLLHA ALDFIKVCKSFTFGDDTPSPVIPFKDSLASLFLSLLSDHSSQLRCIGITGLVGLMSLNAIMNINEKKLAAMHFTNIVLTDQDNKVCSEAVTALAFMSMEF SLLVKEEVLPQLIKELDSRATGTRHRFIVNTLAGISMHSDIVLTTIPVMLQHLGTLSEDNTAESLETAVNTIQSIDIVVNSNISDEQCLDFFHSKLLPQL LRITVDQALQVNNYILCKEDVVSSIATVCRNIAKVLDDRVASNLVSNTISLFLDGNLENIGLKQSSQHFRPLEISSPWQHTQLVSLLTSIICSMKTFELS SQCLELMEKLLKLSLSSEHHLTCVSAAKCYAGLVNKHKQGTDLDSSLETVVESTCRMLQDEISDQNIYNRQKALTLWLWVTKALVLRAHFKSTQFTTKLI SLFEDHQLSQMAADGFYIILSESQDVLNKDMHCDIKLMYRQRLFMQTLPRILAGFEKANEDKKQYYLSALSHLLQFIPKQVLLSELPPLMPMLVQSLYCQ DVGLYVSTSDTLSMLIQDAPTVISLYVDTLLPQLLTLSTYQQSMKVRIAALKCIGLFVTLPTHVIYPRQKEVVRRLASVLDDRKRLVRQQAVTARGLWFL LEAPKK* >MMS19_dapPul Daphnia pulex crustacean EFX86854 961 aa x HEAT 38% MAISTAIQKLRDSFNSEESANESIRCISQSIASKELTILKLVEDLQPDLLNQQNTHRCKAVSTIGTILEQLGPELKGLNEKEVELVTEFFCSKLKDHHSI LPAALQGLHALSTAPKLSPGLARLISQSIFQDVHCQSQLQHDRRAIYKTLKNLLAFHLKELQDLGQDFVFGLIQLADQERDPRNLLILFSIFPVVARYFR FEPFTEEFFEVFSCYFPIDFTPPANDPYAVTKEQLCDGLRQCLAGSPHFAEYCLPLLQEKLESDLVSAKVEALKTLELCCQTYQAGQLEKWVDSFWTGIR REVLINVNTDDLEHASLDALAALSRAFTTDGEFNSPAFTKLLKNVLTECQGHLCEPERRLMTPSSYILLAICSGSAPACALIVSQVIPLLMDQYRIRPQS NPRQFILNSLNKMVHAGLYGFTEENVAQSGLASLIPKLLELYLEVLKEDDAVLRNLSLQGLSHLIGTCLNHQDLEKVNGTLLDLLQKSTATDSVIAEIGH FFCKSAEKNENLFLEQVLVKLLDIAVSGSIPTDGCARTIRPGITTGSTQSFDSFRTKGRTRNIPAIAPLGIENIGRRRSSGVTPSHLCLIEKSGFQFIFR VILLDQNVGRNVFVTFSALYRKATEFINEQTEQYVSQHLARSPWTLSIMEATLGSLDATPSGHSLERLVNTLEPLTVCHPKADVRLSACRLMAALVNKLP EGHELEAILDSLRRKWQDPSTDRCNSVCLFVWITKALLMRSYSKLNQYIQELVDSLNDPTHGYQVAEGFKTILCDTEECLNFNCHANIRLMYRQRFFQEV VPRLLKLYRESESCNKAACFAAIANQLAFIPEGVLIAHITTLIPLLIQCLSTDQPAQLIISTINAFMGLMSDNVSAIEEYISSLVPRLLTLAKDGITMDV RRLALQCLSELRKAQSIVLLPLRSEVILRLVPCLSDKKRLVRREAALARQKWIMLGQPGCN* >MMS19_droMel Drosophila melanogaster insect NP_649519 959 aa x HEAT 30% MTTPTRATLEKALKSDQKLVNSATQIAKDLTAKAYDISALAEALGFALSSPDMEERVAGTNLLSAVLIALPQDLLQERQLEFLSTFYMDRLRDHHNVMPA IIDGIDALVHMKALPRAQIPQILQSFFEHTTCQSQTRSDRTKLFHIFQYLTENFQDELQAMAGDFVYGLINSIDGERDPRNLDIIFSFMPEFLSTYPLLH LAEEMFEIFACYFPIDFNPSKQDPAAITRDELSKKLTNCLVANNEFAEGTVVLAIEKLESELLVAKLDSIELLHQAAVKFPPSVLEPHFDQIWQALKTET FPGNDNEEILKASLKALSALLERAAHIPDISHSYQSSILGVILPHLSDVNQRLFHPATGIALVCVSGDAPYAADKILNSFLLKLQAADASSEQRIKIYYI VSQVYKLSALRGSLQKLDTTIRESVQDDVIASLRLIEQEEFDAKKEDLELQKAALSVLNESAPLLNEKQRALIYKALVQLVSHPSIDIDFTTLTVSLGAL QPVEVQSNFIDVCVRNFEIFSTFVKRKIYTNLLPLMPQIAFTQRILDLVMTQTFNDTTAEPVRLLALEALNKLLLLADQRFIVDVQQESNLLHKLIELGQ KTEGLSMQSLEQIAGALSRITQQLPLSEQSAIVSEYLPGLNLSQSADLYITKGLLGYLHKDITLDDHFERLLTDLTQLSLNSDNEQLRVIAHHLLCSMVN KMESNPANLRKVKKITEQLKVAIKKGDVRAVEILAWVGKGLVVAGFDEAADVVGDLSDLLKHPSLSTAAALGFDIIAAEYPELDLPVVKFLYKQKLFHTI MGKMGSKLANYCVHHLKAFVYVLKATPQAVIKLNIEQLGPLLFKSLEEHNEAQSLCIALGICEKFVAQQDTYFQGHLAHLIPSCLELSKYKAQHTMQVRI AALQLLYDVTKYPTFVLLPHKVDVTLALAAALDDPKRLVRNTAVKARNAWYLVGAPSPN* >MMS19_nemVec Nematostella vectensis cnidarian XM_001629116 897 aa x HEAT 32% MAALGQEEYPSLATLLQDVYQRRKNLLQVVELLGPSLTSTDTDKRCSAVQLLSSLLQKLVNYKLTDREDLKPVGSDLVFGFLQAMDGEKDPRNLVVAFKL ARIIIKNFPIGLFAEDLFEVTSCYFPIDFTPFCLPLLMEKLSSDVINAKIDSLLTLVFQTVSSELEDAAFKALSSIIKNLESSSPGQEPFLSRIFINFYT ISCYVTQCHPDVVEFKTPFLDCVIKECCANIEGADLRKVKPSGQLLQAAFVTDTTYNEITSTAVPLILSKYNDEATQGLVKKLLLDVLLGLLTASKPYYK RKGSVLASHTSALVDVLFSALVSDSPSLCRAAIAGLVSMVTLPGLLLEQKVGMFVEHLTSFVLNTKDLTVRQESNAALAFLAMEFPELIKTKLVSVLAEQ LQKEDGSAMDEENISHLQSDKSHPQYDQMLNTLSAVCTEEGVVRHVVPIILDHGEYLVTGKDLERGVLHGKISETLKCLNSIVKGTLQSSTVEPNYYTEV VIYRIIDLCTQSALQESPDCPMATPEALALVCSIVRQVISHLAVNEAEDVLHIIVSNFIEGKTPLSARAEQKFAPLEPSSPWQQSQLVTVLMAAVCSARR EVRIPRQKELVPRLQVLASGCNHRKTTVAASKCLGGIINKMAQGDDLTADLHSLKGQLQNHMDGNEEQRWRAVITWLWLTRALVTRSHPMAQEFVQKVLH LLDDVSVGRVAADGFYVIVSDCDDVMNQAMHADIKMMYKQRFFMETLPLLLKGFHDTRPECKYLYLCALSHLLQWIPKQVLLTEIPTLMPMLIQALSRDE PSLLLSTLQTLYSLVFDAPEVISRQVTSLIPNFLELAKCKASMKVRMEAIKCLGAMTTLEHHVVYVYKARVIKELACTLDDPKRLVRAEAVKCRNEW* >MMS19_triAdh Trichoplax adhaerens single-celled metazoan XP_002114595 959 aa x HEAT 39% MEKDSSAKSLQQLMDEFILGNSSAINEIIKGIYDGHIKLSTIVELLGPYLTSVEHEKRLQGMKLLSEVLQMLSMYKMQATEVQLLVAFYSDRLQDHFSIL PETLRGILALVQHQIISEEDAVTIVKGIFKEVQNQALLQADRNKVYAILAGLLDKHYEGIKIMDADFVFGYIQVMDGEKDPRNLLLALKIAKFIVQNFNI DLFLDDFFEIISCYFPIDFTPPPNLPSNENVTKEDLIIVLRESLTSTRKFAGISCAKIYTATDFQEYLQPIWTAIRQEVFLSMDDQVQELSLEALKHVVV TISSNSLQQPDQDPLNDFINMIVTETQQYLQDPELKLANPCGNVLNAVASASDRSCYSILTPIIPRLVNLYSTDKTVIFRCKVLDILIKLLNAAANCQLS EQFIAPMDWHEIVKLLQLAMDTSEEDIRLRVTASFSILIQIKDALPADEIERISNDILKRALEDPSSIVRHGSISTLATIASVLPDVIITTVIPYIRTSV TNLQLLLQCLANVKNRIENCLYLYHYLFDDILWLCVYNSLEESINSFEFKTIKIIASIGQLIYLNLDESSQKKFIDNLLELFMNGQVSVLKPMTVIDELP LKQFYPLNVASSQRQVQLIEILCKILGAIKFRDGILSPNDMITNLLDISCKSVHQPSATSAAQLLSSIINKMEEGDQLENYIKSITNTICNVLYSKNVET EMKNAVNTWIWMFAILCRYSCSLFHYSNFDIKTSFDASFQLMKALIMRSHPYSNEALIQVLKFFKLPNVGHVASAGFKIIIGDEENILCESTNAIVKFMY KNRFFMMASEKLMENYRIASKGIKHHYLTALSHLLNGVPKQMLLNHLQMLMPLLVESVSCDEESLRLSSLQTLRPLITEAPDIISNYVASMLPELLKLCN FPSSMKIRISALQSVNDLASLPIHLVVPYKSKVINELGNTVNDKKRLVFTVINPKKQQ* >MMS19_sacCer Saccharomyces cerevisiae budding yeast P40469 MET18 1032 aa 13 HEAT 29% MTPDELNSAVVTFMANLNIDDSKANETASTVTDSIVHRSIKLLEVVVALKDYFLSENEVERKKALTCLTTILAKTPKDHLSKNECSVIFQFYQSKLDDQA LAKEVLEGFAALAPMKYVSINEIAQLLRLLLDNYQQGQHLASTRLWPFKILRKIFDRFFVNGSSTEQVKRINDLFIETFLHVANGEKDPRNLLLSFALNK SITSSLQNVENFKEDLFDVLFCYFPITFKPPKHDPYKISNQDLKTALRSAITATPLFAEDAYSNLLDKLTASSPVVKNDTLLTLLECVRKFGGSSILENW TLLWNALKFEIMQNSEGNENTLLNPYNKDQQSDDVGQYTNYDACLKIINLMALQLYNFDKVSFEKFFTHVLDELKPNFKYEKDLKQTCQILSAIGSGNVE IFNKVISSTFPLFLINTSEVAKLKLLIMNFSFFVDSYIDLFGRTSKESLGTPVPNNKMAEYKDEIIMILSMALTRSSKAEVTIRTLSVIQFTKMIKMKGF LTPEEVSLIIQYFTEEILTDNNKNIYYACLEGLKTISEIYEDLVFEISLKKLLDLLPDCFEEKIRVNDEENIHIETILKIILDFTTSRHILVKESITFLA TKLNRVAKISKSREYCFLLISTIYSLFNNNNQNENVLNEEDALALKNAIEPKLFEIITQESAIVSDNYNLTLLSNVLFFTNLKIPQAAHQEELDRYNELF ISEGKIRILDTPNVLAISYAKILSALNKNCQFPQKFTVLFGTVQLLKKHAPRMTETEKLGYLELLLVLSNKFVSEKDVIGLFDWKDLSVINLEVMVWLTK GLIMQNSLESSEIAKKFIDLLSNEEIGSLVSKLFEVFVMDISSLKKFKGISWNNNVKILYKQKFFGDIFQTLVSNYKNTVDMTIKCNYLTALSLVLKHTP SQSVGPFINDLFPLLLQALDMPDPEVRVSALETLKDTTDKHHTLITEHVSTIVPLLLSLSLPHKYNSVSVRLIALQLLEMITTVVPLNYCLSYQDDVLSA LIPVLSDKKRIIRKQCVDTRQVYYELGQIPFE* >MMS19_schPom Schizosaccharomyces pombe fission yeast Q9UTR1 1018 aa 14 HEAT 25% MSSNLVALYLFSIDRSQDEANDVVDRIVEEIVTDRMGIVDLVTSIGEYLTDNNISVRAKAVLLLSQTLGELPKDRLPAKHVSVLLQFYLSRLDDEVTMKE NALGIGALLNMQNFPAQKIVDVCKALFSSTDMPKYAQATRLNILKVFETIIDNYLFFISSQTRDAFFSGICSTFAGEKDPRNLMLVFSMLKKILSTFPID GFEQQFFDITYCYFPITFRAPPDATNLAITSDDLKIALRETLVANDAFSKLLLPALFERLKASTVRIKIDALNIYIEACKTWRVGAYLWSAKDFWESIKQ EILNSTDAELQNLALGALNTLASKFYKEEGFSSSFTEFVDMILIQLSQRLLEDVNVKSCGSCAAVFASLASISVETFNYCSCNFLPSVLDLPMVNEPLEK QKGMLVFLEYVYKCLVLLYGKWRSKNQADIDNPLLVYKDKQLSFVSGSLMGTAKDETEIRMLALKVIFLMASIKNFLTESELTMVLQFLDDIAFDFSDPI KKKATECLKDLGLLKPDFLLLTSFPFAFSKLTDDVTAKSSSEETFKQYLSVLVSISEERSLFKALVIRLVEMLKDQFKSKEMSVDLVESIVQSLSVAFKE RNDRNEQEIPFFFEELLKQLFTLCFANCESMNVRCLIYVSQTINEIVRVNHFEFQEKFVGQLWKLYMENSNSDLIETEGCEKAAERFTLAASLSDQKFLN LVVLLQGGLNGLSKKLHFIEKLNIELLNLLINVVFVTESPGVKISALRLISSLINKCEKDEDISSFISSKGVTSLWDKVYTGTPKESEAALDVLAWVDKA LVSRKHSEGIPLAFKLLDTLNLQNVGDSSVKALSIIIKDDPALSKENSYVEKLLYKQRFYASVSPKILEHISTATGGEKSLYLMLLSNVIGNVPKEIVIP DMPSILPLLLQCLSLSDISVKLSTLNVIHTSVKELTSLLTEYLDTLIPSLLAIPKDMNNPTVVRLLALKCLGSLPEFTPTTNLQLFRDKVIRGLIPCLDD PKRVVRTEASRTRHKWYI* >MMS19_araTha Arabidopsis thaliana plant NM_124186 1134 aa x HEAT 28% armadillo/beta-catenin-like MMVEPNQLVQHLETFVDTNRSSSQQDDSLKAIASSLENDSLSITQLVREMEMYLTTTDNLVRARGILLLAEILDCLKAKPLNDTIVHTLVGFFSEKLADW RAMCGALVGCLALLKRKDVAGVVTDIDVQAMAKSMIQNVQVQALALHERKLAFELLECLLQQHSEAILTMGDLLVYAMCEAIDGEKDPQCLMIVFHLVEL LAPLFPSPSGPLASDASDLFEVIGCYFPLHFTHTKDDEANIRREDLSRGLLLAISSTPFFEPYAIPLLLEKLSSSLPVAKVDSLKCLKDCALKYGVDRMK KHYGALWSALKDTFYSSTGTHLSFAIESLTSPGFEMNEIHRDAVSLLQRLVKQDISFLGFVVDDTRINTVFDTIYRYPQYKEMPDPSKLEVLVISQILSV SAKASVQSCNIIFEAIFFRLMNTLGIVEKTSTGDVVQNGNSTVSTRLYHGGLHLCIELLAASKDLILGFEECSPTSGCANSGCSMVKSFSVPLIQVFTSA VCRSNDDSVVDVYLGVKGLLTMGMFRGGSSPVSRTEFENILVTLTSIITAKSGKTVVWELALKALVCIGSFIDRYHESDKAMSYMSIVVDNLVSLACSSH CGLPYQMILEATSEVCSTGPKYVEKMVQGLEEAFCSSLSDFYVNGNFESIDNCSQLLKCLTNKLLPRVAEIDGLEQLLVHFAISMWKQIEFCGVFSCDFN GREFVEAAMTTMRQVVGIALVDSQNSIIQKAYSVVSSCTLPAMESIPLTFVALEGLQRDLSSRDELILSLFASVIIAASPSASIPDAKSLIHLLLVTLLK GYIPAAQALGSMVNKLGSGSGGTNTSRDCSLEEACAIIFHADFASGKKISSNGSAKIIVGSETTMSKICLGYCGSLDLQTRAITGLAWIGKGLLMRGNER VNEIALVLVECLKSNNCSGHALHPSAMKHAADAFSIIMSDSEVCLNRKFHAVIRPLYKQRCFSTIVPILESLIMNSQTSLSRTMLHVALAHVISNVPVTV ILDNTKKLQPLILEGLSVLSLDSVEKETLFSLLLVLSGTLTDTKGQQSASDNAHIIIECLIKLTSYPHLMVVRETSIQCLVALLELPHRRIYPFRREVLQ AIEKSLDDPKRKVREEAIRCRQAWASITSGSNIF* >MMS19_dicDis Dictyostelium discoideum slime mold Q54J88 1115 aa 18 HEAT 31% MTSNITELNKWIEGYVNPQSEESVKTNAINMVLLYMKSNKIDLQDVVQGLGDYLKSNDSILRARGTLLLSEVLCRLPDLPLNQDQVHFLAMFYCDRLQDY ACSSEVVKGITGLITNHTPDYPDNQKLLRNIFSEVHPTSLTQAHRKMVLQVIDIMFNKCLSEIQELKNDFMVGYLQFIDNEKDPRNLIFSFKLLPKVIYN IPEHKHFLESLFEIISCYFPISFNPKGNDPNSITKDDLSNSLLNCFSCTPLLAEHSIPFLIDKICSNLIETKIEALQTLVYCCDRYGGFAVQPFLEEIWS TLRTLILTHKNTTVIEESKKTIFYLTRSFTKERKVLESFLSIMIKECLHHIKSSQDSKIAIYCASILYQSVSASLLSSKIILIHIFPNLFNFLSELQKQD TVQKVNEQNSVIALFNDLLKANSIAFEMYSNENKEPNPLEPFVDQLFKLFSDLLLLNSSSSIRSNSIECLSNLYISKKVHTTEQDDDDSEQITNEFLLDL EKRQFIIKSLVSLLNSSDNTLRHKSLDSLFTIASNEDPSVLNLYVIPTLLQMINHSSCNINTTNNKINNNNNNNNIVIKNNKCQDEHCNEDHSNKNENNN NSNENSNGNSTSGSDDDLKHYLEAFTKLCTHQPLLESVIPQIQVLLQHNIKETYQSNEDFEKSILILQSISFILEKSTNIKSMTICSKSILFPLIKGLYK QELISSSNDNNNNNNNNSNRFNQILTPTLKMIHSIFENISIESQKPLLEKLIKLFLNGDTLVINYQLPTTTTTIIKPFEKSSPYKYLIPIFTTIISQSKL DLSENNELKQSLYQMSLDVNVDDSIAISCSKAYSSIINKQQQQQQQDQINFNFFNDNLLKVINDTTTPLPLKIRHLDLFTWCTKALLTNGNSINIKLGSC LADIISNENVELSYHASKSFGILLSETDVLNEKSGSIIKILFEQKFFTLMFPILLESFKVSKNKELQTISSHYLIAISNLLKHVPKEILLAELNEILPIV MQSLKSSDNNDQVQLLDSSLQTLTMLINETPSSFISYLDSLIPSLIKISTKSTKYNLKRSALEILTLLSKSIPFVNLFPYKTQVVTDIIPCLDDKKRIVR REAQKCRNSWYILQK* >MMS19_pytUlt Pythium ultimum stramenopiles ADOS01001616 957 aa 4 ARM units 31% MFSLDAPLAPAIDAFVNPENDDNVHKTSLNTVVMQVHRKVSMEALIQALGLHLTSTDDKVRARAMQLLAEVLSRLPELPLTPNAVQLLVDFFADRLADYP SASACLQALLALESNHAKKIASPTVTIILIQKMVKVLHVPQLGQAMRKQCFELMQLALGQKVVVDVLVTAPESSSIDHGLLFAQAFLNAMEGEKDPRNLL LCLQIARELLAKLEVVFDRHDAVLQQYFDVVSCYFPITFTPPPNDPYGITSEELILSLRKAFAASDLLAKHVLPFLLEKLSSTVVEAKLDSLQTLVFCCE AYSINVALLHMLSIANALYHEVVKGEKKEVIEASLRAISRFSSVIGLAKTKAAGGAAYAWNKFVVELTTRAMSDLTGHATDSLVSVSAGQVLAALGKDSV LGFTHVLETSVPLLIQQFNESSTSTESKCEASLARLLLIVNTIDREVDQSASAQPMRPHALVLIDALVAFLSNNEALSTPTAKCSAIEALSHLVTYPPSP IVEIAQVKALVELFINFLLFDASPEVRRECLQSLRAISTIKQKATVKNYASLVMEIALTQLMDAVQLSAQNTKVAAVLASSGRDHPEFFNDVLDSITQLS QEASLFQATIVRLVDFCVVENQDSNKITFVANSSANGTQAHVDGILNAVAKIVELNADDKASMEFCVTSGGDNSIVFRLLKAVTTTAADAAAQNALLDDA KLASCARIFRTPMQNVSTETQQLLANAAISAFLTTQSTGASASHPAYLQLVPLFSAVINSANRNLNLPETSRVINTLLELAQSSTAVYHTTASTQQIEQI SSEAALSAAKSLASIVNKMSDGEEFDALIVLLLDQKLSQIIANEQKDVSVRVAALQIYVWIAKALVIRGHREHAPACLFFLCKFLTPETSDARSQIAMHV AKSFKLLVTEFPDVLNRKCGAFITVRQHKKKYAGILGNADLTFYFVCVVPVPSANV* >MMS19_sapPar Saprolegnia parasitica stramenopiles ADCG01000470 804 aa 1 large ARM repeat 31% MFSLDAPLQPAIDGFVDPENGEQQHTTHLNNVVMSVHRKTPIEQVIQGLGAHLTHVQDKRRARATLLLAEVLTRLPDLRLSSDTAHLLLTFFLERLKDGP SMAACLKALVALISLHAALLPANDAWTVCATCHAWCERAVVETLLNLPTPIASLSQSMRKQSFELLQLIVRRGALGDHEGRVLMDGFLRAMSGEKDPRNL LFCLRFAAELLTTYANVVDADVAKGFFDATSCYFPITFRPPPNDPYGITSEDLVLALRSVFVGHDSLAKHVLPMVLDKLSRTTVVEMTKDILETLAFCCA KYPLNRLLLHFTPVAAAVYHHVLHGDNTAVIAVAIDALKTITRAVSPPSKLPGMQALAWNKCIVYLVNQAVEDLAHQAPDSMVSTGAGHVLCAIASVGVA GFSHVLSSALALLLEQCAAQAGSPAEAATARLVQLLGCIDAEVDHSAPPLVPYVSAIQTTLVHGLETATSSRQQKLCLQGLRCLVLRPPSPLLDDASLEV LLQGWTSTVLSNPFPDVRDEATSTLQAIALKSPGLAQIVLTRCVPSFLQVLEQPAVLFFASWCGDMDDGLGQCSVWAVDRGHGARHPRGPHAALARPRHL SAPPAAVPDQLDAPVCDRDDGGRGRHCPRQQGLGRVHGLRHPRHSHFIVGALPPRRRARRARPDAVDRRPSDRAGDGQHERACHAVRVPPGANNAAHVDL AAGLDVGHFALAWPRPATATIERSTLLVRRGVDGARTRRAAAPLAAAVYARRAEQRRRREHSQGACGALQRPPRGHAAVRCVPKVDHVARCPPRRHGVAG GRHS* >MMS19_polPal Polysphondylium pallidum amoeba EFA86574 994 aa x HEAT 28% MSKANIDSYININNNDQTKQTSLNILLLEINANKLSIHQLVEYLGDYLQNTDSILRARGTLLLSEVLCRLPDLKLNEAQVEFLAAFYHDRLQDYACASEV VKGVYGLCVNHKVPYPHNQKMIRAIFQEVHPSTLVQTHRKMVLQLIEHLLEHNLTEIQELKGDFMAGFLQFIDGEKDPRNIIYTFRLIPRVILYIPEYKN FADSLFEILSCYFPISFNPKPGDPNSITKDDLVSSLLNCFGASTYFAEHCIPFLIDKICSNVVDTKIESLKTLLFCCSKYGPVALRPHLDDIWGTLRTQI LTQKSATVIDESKKTMFYLTRVLAADQETLQSFLSMVDKECLHHIKTSQDSKLAVSCASILFQTVSASVKSSRIVLSHILPTIIDFFKELSLHLSDDPIH KANEQLSIIGLFNDLLKANNISFQYNNENIDKEINPLEEYKDKLYDLFIGLLSNSSALVRTLAVDCLANLYVTRHIKTSVPITFVLDQEKRQSIIKDLGV WLLIQIFRNKSLEALMSITKLEQVEQMNLFAIPTLLQMINANQSKNVSESKHYLEAFSQLCTHQPLLQSVIPQIKTLLEHSIKKKYINNDEFENSLLVLQ SLENTFSNSIDEQTMTICYREILLPLVKELFEQVFSLDVNSQEQKDQVLGIMKPAISMIHSNNKKEAIELFINIYLNGDLSALQINKEFKPFSSDATEQA KLLIPIFTSVISQSKFELSTNKLLKEMLMSRALDSNVEESISNACAICYGSIINKQTDQTDLPLDHLEQLISSSSTNKTQALNLLIWIEKGLVTNGNPQS IKVGELLAQLITSENTEISQKAAKSFYILLSDHDTFDHKSGAIVKRQKNETVSSQFLVAITNLLRNVPKEVLLGELQEVLPIVLHSLHSNQRDLLNSSLQ TLMMLVDEASTSISSHLDSLIPTLIKISVNGESLTFRQSSLEILTRISRAIPYPKIYPFRNQVINGIVPALDDKKRLVRREATKCRNSWFILQ* >MMS19_entHis Entamoeba histolytica amoeba XP_651925 868 aa x HEAT 25% MSTPAQQLNEFIESPKVIKEGYEIIDQLMKNNYNVNSLVTDLGDTLPSEDERIRFRATSLLTYCLIKYPIKEESKDVFVDYLASRLVDAVCLEPILTALL QLVTKKPSDEIINEIAMAYSCMRTQLYTKEVRILVYQFYKVFINYYQATEVIDTCVQLIELERDPECLKEVFDLIKLVSQKNEIDADSAPLLFDCASAYF PILYPPKGDEALRIDLTNKILDAFVSAPIYAQFALPFLLDKLDADLSSIKLEALKAIYFCIQRFELKYVYAYFTHIWESIEQNISTVGVVEVNEFAFAIA SYFCSLDDFHSKNLMESIKMFCLRMMSETDEIIINFVNGLLEELTKKSEKFFKVFVPVFIQCFHDQLQDADDRPKEQFERELFIVRLIYQRIIEGMPLLD CVKGQVAWDLHRLATPLHPCFVSLLDIDVSLALLNLLGEQRMVPFQNAIELSENKCHDAIPILQRLYEKEEDVMISLLPANKIITNLELVSGIALHSPKL FEQLLKLIPTLQSNEYVPVFQSILSDALPFNCLDVYVNHCIPVFIVITNGVLSPLFNTLMNRLSRLHSILSSKKISELTEGVLSQLKEHSRLLLILPSLL QFYQPENLIVYLNEVQEVDKDTIAIYSLLISKLTNIIPHVLEQNKEYFNGYKTIQELDSHESNKQATPIFIEELCRMNNKAIECLKEMIVFDSINKKNEL HWKEELFNLVYERFIESHQVTTVEESHIMILLFSLLPTEKLLTYESTVLKIFNIICVPTSHLNEIDSVVVLLFNILPTVSQYPMSLIESELDSIITKLFN VLYINGTTIKYRCDIIDLLTRIRVVYGIDAIRPYQKNVIKKLLVPLDDNKRLVRRSAAICRNIWETTA* >MMS19_naeGru Naegleria gruberi early eukaryote: heterolobosea XP_002678884 1070 aa x HEAT 27% MQTSSNSNGEQELISLIDSLCNPTLPNTNKESLKSKLIEFVVNGTLTINEGIKLLGEYLNHATDDRIRGAAYAVLDLILENIPNNVGSETDETKQTTQLK LVASLLRFIGDRFYDFDCLATLLPCLFSLFKKWSSYISSEQAINVVLQFFENVNIQSIGSTSGVAHATKTRSLCFEWFSLLADRFPSIVRTIDFLNGFIQ SLEGERDPGNLLYCFNLIPKVIAIFDDSELSSKILSAVSDDLFDITSCYFPITYTPPANDTRGITREDLSRSLKLCFGCNKFFAPTLFPFLLEKLSSDLV DTKLETLDYLCYCIEKFGEVNSREYLTEIWSYIKAESVKTNSMDVMKKCYESITKIARIVIIPNDPSNKPFDIPNIEAILRTALLELKSKEPKFAAQYAR MIYACAVPTFEISMMVFNRVMPELVATLSESDTKDKLYGSLLMITQLLQAVAEQKGENQLPEVVFNLISQVQTVFLSIYEEEFSKNDKEMILVMVETISR IAIFRIPSTLLRDIYVSRILLKSYGEENKSFSLLHATSLEEYKERVIKDIAWIYKYAPDIVSEDILVPLFGALYGCENKSEHINRILSNISAVGKVCPSM TPSITHRLFERIESIPISESHYEHERVKVFETITSLDVSLIPAHDKVSYIQRIVKMSVTDSSSQMVDDSDTMDCSDSECAHVHHNQGNFSFLTLLLGRSL ENELQQVVLDSVLQYANSVPSTGLKNFISVLSAIVIACRPTVGMGNLITMTDSLLQMALKGEQPSQVTKCIAQLVGSVLNKLPLDSTEFQQLITICNATV FDAFSQMLTVYNGDSESAERYIEMVSWILKGLVMRGAYVPHADRYSSLLCGSLVFEYNSSKVNKKVAEGFLIAIGEDETSIHKENHAIIQVLYKQRFFAT NVRKLMDSINTVTQPHIIGSILLALSNLIHNVPTKVILSEVKNIFPIVLKFLEMRQILIEQDNNSEDLLYAAIKTTLTLLSDAKEEMSVHLSSIVPILLD TCKFKKSQAVRILSVEALLELTNGYKYYEIYPLKKDIIKGLEACLDDKKRKVRKAAVKCRNSYFVLSNNQ* >MMS19_phyInf Phytophthora infestans early eukaryote: stramenopiles 1114 aa x HEAT 33% MVSYEQLGSLPQKGSQNPVVNQKLEAIAMFSLDAPLAPAIDAFVDPENDDAAQKTGLNTVVMQVHRHVSMEALIQALGAYLTNGDDKVRSRATLLLAEVL TRLPELQLTPSAVQLLMTFFADRLADFPSASACLRALLALETHHAAQVQSPRTTVALIPKLGKTLHIPQLGQAMRKMCFDLMQLALMQSTVVELLLDSVP ASKDAQDASVDDAEQSEDLGRQLAQTFLSAMEGEKDPRNLLLCMQVARTLLSKLEPVFSRSDTLLQQYFDVVSCYFPIIFTPPPNDPYGITSEGLILSLR HAFAASDLLAPLVLPFLLKKLASTVVEAKLDAIQTLVFCGERYSVNALLLQMHAVATALYDEVLDGEKQEVIAEARQAISRFSGVVARAKAQDTPGAAYA WSKFVVDMTARAAGELRENAADSMVSVSAGQVLAALGRESSMSFAHVLKIAVPLLVEQLNNESSGSDSVPSKCEAALARILLLIDTIDREIDQSGQGQPM RPHAAALIDALVNFLSSDHDNQTKPGSSPTARCVAVEALCHLLTFPPSPIVAPAQVKALINLFTRMLLLDPVAEVRTACLQSLKEISTVSTASEGSTNSG EHPVTGGYAAFVVEISLARLMAAVSEGSDQEDDDDEEGTGVAAVLTASNRNFDSFFEEALLAITELCRESSIFQATIFLLIDLCVEKGDGKQSAIGFCEA EGDATRQRHVDCILDAVAKIVEINAGDRTSMEFCVKASSSASIIFRLLTAVETLAARATASSGYKSGLVDEVKLSACVRIFRAVMQNVSSATQQQLVDAV VPAFLRTNTSEPASLQFVPLFAAVINSAARDVALPDSSLVINRLLELAQSGATAVSESPPRQLQLVYTDAALSAAKSLASIVNKMSDGAEFDALIDLLLS RKLAVVISNSAESFTVRVAALQIYAWIAKALVIRGHKVHAPVCLRFLCSFLTPDGDVNMEQEGDDQHAAALRMEVAKTFKLLVSEYLDVLNRKCGAFITF LYRQRLFDLVFPVLLEYIRARIDEESSVAALVAFAQVIAHSPKAIYLPHLAQIFPLMVQALNTDDRELGSAAIQTFKPLLLESVESAKPFLKDVFPGLLK QAQFGYVVSCSDS* >MMS19_albLai Albugo laibachii early eukaryote: stramenopiles 1077 aa x HEAT 27% MFQLDAPLSPAIKKFIDSGASNDEETGQKTSLNAVVMHTHRIGSIETLIQELEPYLTDDCNDFARARATLLIAEVLTRLPDLPLSGNRIQVLNNFFCARL DDPPSIPASFQALLALQKHHSTEIPDSENMELVIRISDTLHVPQLNQPMRKRYYELVYLVIQQERMQKALSRSQQAQVFIRSFLNAMTGEKDPRNLKHCF QIAQTMMQKLEMVFQEAELSEQYFRVISCYFPITFTPPPNDPYGVTTEELIRSLRNVLTASDVLIHQMVPFLLEKLSGSMSEEAKVDALDTLGHCVETFS LKNLLLHIRSIGQVFYHEILNGERARVIETASNVLSRVSSVIGRAKVQGSSGSGFAWNAVVVTITNQAVEKLHENSVDSMSSASAGKVLASMSRESLVVS THVLNTSMPLLIEQVKHSFEASSSQCEAALDRLMLFVDTIDEEVEQISTIHPIHSHASPILEALVKFLEEDTPTSTPNAKRLSIRIISHLVIYPSTPVVR PSDVERIVRLFTRGFLSDASKHVRSEFLSSLKALSGAIKTPSTLQSVHCKREKTLQLYGTLLKEHCIAQLLALVQDGKSPEAETFQKSSCRTRKDFEQDT LAAITELSHDPVIFKEAVVHLLQSCFIDQDGLLIFRSFEVEHTLQFFQAVATIIELNASNASNMEFCASIDDQNGIAFKLLDAFVSMAMSNGQSKEQKFL PPNAIAFSTRILRTIMQNICFDTQQKLLDRAISRFHPILQTEESTPSQHLYQIVSAFSTVINSANRSLAFPKAYCVIDSLMAVSRSITTESHGYTNEIVL LISQSIGSILNKVRDKHFEAKVESLLTGLSQSIHNDQEQAQWHTSIEVYIWITKGLLLCGHPKYSSQSVAFLTQLLIHHSDKGVRGQVAEGVRVILTEFP NVLNRKCGASCNMLFRQRLFELVGPNLLAFISKHSEETTEALTGFCYIVAFSPKAAFISLISTIMPLVLRGLSSDHVELGAAAIKAYKIVSDTSIEHVKP FLKDVFHGLLQQAQHSANALDRKDALECIGMLTTLPYELIHSYKDRVLRQLLFCLDDRKRFVRYTAVRVRNKWSVL*
>CIAO1_homSap Homo sapiens O76071 length=339 3FM0_A PDB: 3FM0 MKDSLVLLGRVPAHPDSRCWFLAWNPAGTLLASCGGDRRIRIWGTEGDSWICKSVLSEGHQRTVRKVAWSPCGNYLASASFDATTCIWKKNQDDFECVTT LEGHENEVKSVAWAPSGNLLATCSRDKSVWVWEVDEEDEYECVSVLNSHTQDVKHVVWHPSQELLASASYDDTVKLYREEEDDWVCCATLEGHESTVWSL AFDPSGQRLASCSDDRTVRIWRQYLPGNEQGVACSGSDPSWKCICTLSGFHSRTIYDIAWCQLTGALATACGDDAIRVFQEDPNSDPQQPTFSLTAHLHQ AHSQDVNCVAWNPKEPGLLASCSDDGEVAFWKYQRPEGL* >CIAO1_droMel Drosophila melanogaster Q7K1Y4 MGRLILEHTLQGHKGRIWGVAWHPKGNVFASCGEDKAIRIWSLTGNTWSTKTILSDGHKRTIREIRWSPCGQYLASASFDATTAIWSKSSGEFECNATLE GHENEVKSVSWSRSGGLLATCSRDKSVWIWEVAGDDEFECAAVLNPHTQDVKRVVWHPTKDILASASYDNTIKMFAEEPIDNDWDCTATLTSHTSTVWGI DFDADGERLVSCSDDTTIKIWRAYHPGNTAGVATPDQQTVWKCVCTVSGQHSRAIYDVSWCKLTGLIATACGDDGIRIFKESSDSKPDEPTFEQITAEEG AHDQDVNSVQWNPVVAGQLISCSDDGTIKIWKVTE* >CIAO1_triAdh Trichoplax adhaerens B3RNR8 MTTVAFLPLSFPFLIFNFGQYIRIWAKNSDSDQWTCKSILTEGHTRTIRSVAWSPCGNYLASCSFDATICIWSKKDGDFECMATLEGHENEVKCVNWSSS GVYLASCSRDKSAWIWEFIEEDEEYECASVLTDHSQDVKHVVWSPKENALVSASYDNTIKIYKEVDDDWECSHTLIGHESTVWSLSFHSSGELFVSCGDD KVLKIWKCLKSGPSDVKWISICTIAGYHNRPIYDVDWSKLNNKIATACGDDAIRIFSIVRITISISNQLLFIAYYQAHNHDVNVVRWHPKVDNILASGSD DNCIKIWKVHSNN* >CIAO1_nemVec Nematostella vectensis A7RWD2 cnidarian MTGHEDRVWSVAWSPNGFVLASCGGDKTIRIWGKEGDKWICKTILEDGHQRTIRSLGWSPCGTFLASASFDATTCIWDQKSGEFECNATLEGHENEVKSV DWSVSGSLLATCGRDKSVWIWEVQEDDEYECASVIHSHTQDVKKVVWHPTKEILASCSYDDTIKLYKEDEDDWSCCDTLEGHESTVWSISFDGSGDRIVS CSDDKTVRIWKSYPPGNQEGVVVSGKHTKWKCVCVLSGYHDRTIYDVHWSKVSGLIATASGDDCIRIFKEDTNSDRNQPSFQLVATQRKAHSMDVNSICW HPKDENILATCSDDGTVKLWRFTPAEE* >CIAO1_sacCer Saccharomyces cerevisiae A6ZYM0 length=330 Cia1 YDR267C PDB: 2HES MASINLIKSLKLYKEKIWSFDFSQGILATGSTDRKIKLVSVKYDDFTLIDVLDETAHKKAIRSVAWRPHTSLLAAGSFDSTVSIWAKEESADRTFEMDLL AIIEGHENEVKGVAWSNDGYYLATCSRDKSVWIWETDESGEEYECISVLQEHSQDVKHVIWHPSEALLASSSYDDTVRIWKDYDDDWECVAVLNGHEGTV WSSDFDKTEGVFRLCSGSDDSTVRVWKYMGDDEDDQQEWVCEAILPDVHKRQVYNVAWGFNGLIASVGADGVLAVYEEVDGEWKVFAKRALCHGVYEINV VKWLELNGKTILATGGDDGIVNFWSLEKAA* >CIAO1_chlRei Chlamydomonas reinhardtii A8IZG4 MDPFTLEPIGALSGHDDRVWNVAWSPQGDMLASCSGDKTVRIWSRRQPRPSEQWYCSAILDQCHTRTIRSVAWSPTGRALATASFDATVAVWELSSGVWE QVAELEGHENEVKCVAWNPDGRLIATCGRDRSVWIWESMPGREFECVDVKQGHSQDVKAVTWHPSGELLVSAGYDDTIKLWTYDGDEWGCAQTLGGTGTG HESTVWDVCWDPVSRARLASCSDDLTLRLWESRAAPTSTPASAPAGAAAAGFVPSRPDLRCAVTLSGHHRRTVFSLDWAPTGLIATGDGDDSILAEEEAS GLLTQPGGQWGCWARVAKAHGADVNCVRWNPAEPRLLASCSDDGLIRLWWLR* >CIAO1_dicDis Dictyostelium discoideum XP_646229 amoebozoa MTDTTKNDKYNLKLIDSMQKEAPYDKVWNLAWHPNGEILATCANDKYIQIWSKDTNGKWGLVQSLEGHEKTVRRVAWSPCGRFLAGASFDASTSIWEKSK DELEFTHVSSLEGHTYEVKSVAWDSTGTLLATCSRDKSIWIWQMEDDNDFECLSINSGHGQDIKCVLWHPNEELLASSSYDDTIKFWKDIDGDWECINTL TGHESSIWDLAFNKDGDKLVSCGEDKLVLFWKFDKENEKWINIFKFKNENSRPIYSIDWSSLTNTIVTGSADDSIIFYEQESDDTPDKYKIILKKKNAHD SDVNCTKWNPKFKNILASCGDDGFIKIWELQDK* >CIAO1_polPal Polysphondylium pallidum EFA75350 amoebozoa MSLNEISVLSYDQPSKIWNIEWSPDGKLLASCGDDKTIHIWMEESENKWVVLQKLEAHEKTVRRIAWSPDGKYLAAASFDASTSIWEVNNGEFNHISTLE GHSFEVKSVAWDASGQLLATCSRDKSIWIWQMEDDQDFECISINNGHSQDVKCVRWHPSLEILASASYDDTIKMWQDTDGDWECIDTLSAHESTIWDIQF NASGNRLVSCSDDRSVCFWRLDSTTGRWKLLSRLESVHSRPIFSVDWSHNQELSPTEQLICTGGGDDSIIIYHQKQQQQQQQSDSSSSSSTTPNEIEQYE ILYKHEKAHKSDINSIRWNPKKPNILASSGDDSTIKIWSFVC* >CIAO1_tetThe Tetrahymena thermophila XP_001017221 alveolata MIEEKMEEQKEFVKCIGQLNGHTDKIWSVSWHPTLDIFATCSSDKTIKIWGLKENSENQYELKQTISDTHERTIRTLAFSPDGMMLACGSFDSTISIYAL NNGSFEFVSKLEGHEHEVKCVAWDSEGKFLASCSRDKTVWVWDYENGFDFSCYSVIDAHTQDVKHVKWIPGTNNLASTSFDDKLKLWEQEDDDWKCSATY SNHSATVWCVEFSKTGQYMASCGDDKQIKVYKKNENGAFSSPYIVETTIKNAHARTIYSLSFSEDATFLASVGADNTLNVYQKNMYVTTFEGQDNNLYEL LEKKVNCHFADINCVAFHPSKDILVTVSDDRQIKLWSVEINL* >CIAO1_phySoj Phytophthora sojae EGZ17716 stramenopiles fragment LGVRGHPRGRAGPHHPRLRYRARCRSPDGRYLASVSFDATTVIWEKQGSSYEVISSLEGHESEVKSVAWSPSGSYLATCSRDKSVWIWEADADTDFECIS VLHGHTQDVKFVAWHPTEDLLVSASYDDTVRIWAENDDDWYCKETLAGHTSTVWGVALNPQGTQMASVSDDTDVVVWQRDVNSKEVNEDGSPKEWKQAFT VSGCHERTIFSVDWSKHGDLLVTGAADNAIRVFQGQPTDSPSSFELAVQQKEAHASDINCVRWSPQLQEDGGKKALLLASASDDGLVRIWKLQLP* >CIAO1_thaPse Thalassiosira pseudonana XP_002294332 stramenopiles fragment EWKLIATIREGHSRTIRSVAFAPTSSTLGVPILASASFDGKVLIWEHFADEENHGTFEPIAQLEGHENEIKHLAWNQTGSLLASCGRDKTIWIWECFLPG TVGGSASGGGGDDEGEFECLAVLQGHEGDVKSIAFALSHGQWGEGDEILLSASYDNSIKVWAEESGDWYCAATLAVHTSTVWCLGINPGGVRFLSGSEDG SMAIWKMYTATERKRLFPREHAVSSTDGLWNSGHGRIASGGGDNCIQIYREETGGSGAGSSSDAPKFAIEAMAINAHDGDVNCVKWYPRDGTSLVSCGDD GAVRIWKYSQAG* >CIAO1_naeGru Naegleria gruberi XP_002680935 heterolobosea MTTNDDLAALTTQEIISAVEGNVSDHEESVWSIAWHPKYSNLLATCSSDKTVRLYYVRVLSPSGRLFAKCIDVLENQHNRTIRRVDWSLPSGNALACASF DGTSSIWILLQNKLQNHLQALEEESQNSKESSPTTSANLGLLKCVSTLEGHENEVKSVAWNYKSASLMDQSDDHDGEDGDCGLLATCGRDKTVWVWEAID KVGFSDFDCNSVCSGHTQDVKFVAWHPLTRSNMPSLLYSASYDNTIRIWKEGGFEEDHDRQSDEWKCVGILRGHTSTVWGLAFEPQLSSDDPEYPQYMVS VGDDKSLILWREDVVGNYIDMNVTQVQTISDVHTRTIYYVDWCVYKHPSTGQSISLVATAGGDNTIAIYQFDTTTRQLKLLTKIANAHDSDINCCIWNKN EFGLLSSCSDDGAVKFWKLKM*