Selenoprotein evolution: introduction: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| (60 intermediate revisions by the same user not shown) | |||
| Line 62: | Line 62: | ||
.... <span style="color: #FF0000;">CRLC UFCK Monosiga brevicollis (choanoflagellate)</span> | .... <span style="color: #FF0000;">CRLC UFCK Monosiga brevicollis (choanoflagellate)</span> | ||
=== | === SEPW: odd paralog SEPV gained in placentals === | ||
Selenoprotein | Selenoprotein SEPW is one of the shortest known mammalian proteins at 87 aa. With its CxxU motif, it is likely limited to simple redox reactions. Curiously, despite its small size, the protein still has 5 coding exons. One of these is of relatively recent origin because chondrichtyes and telost fish have the second and third exons fused (which, given the phylogenetic tree and rarity of intron gain/loss and assuming parsimony, must be the ancestral condition. | ||
Like many genes on the densely packed human chromosome 19, | Like many genes on the densely packed human chromosome 19, SEPW seems to have given rise to a segmental duplication, here during the placental stem (ie, is absent marsupial and earlier diverging vertebrates). The second copy, called SELV, retains the same intron placements and phases but with greatly expanded and compositionally anomalous exon 1 which is quite prone to large indels (from replication slippage). | ||
This long exon retains some conservation at its distal end to the last 5 residues of | This long exon retains some conservation at its distal end to the last 5 residues of SEPW (unsurprising because the cysteine of the CxxU motif is at the end). There is observable conservation also at the amino terminus but otherwise the exon is evolving unconstrained. Possibly the initial methionine of SEPW was lost at the time of duplication but the terminal residues and splice donor were retained. Transcription far upstream lead to a remote methionine serving as replacement. | ||
This is a very odd gene but transcripts in a half dozen species and moderate conservation (75% within placental mammals) in exons 2-5 over hundreds of millions of years witin placentals proves that it is evolving under selective constraint and so is functional. Distal exons however retain only 50% conservation to those of | This is a very odd gene but transcripts in a half dozen species and moderate conservation (75% within placental mammals) in exons 2-5 over hundreds of millions of years of branch length witin placentals proves that it is evolving under selective constraint and so is functional. Distal exons however retain only 50% conservation to those of SEPW, far too little to support ongoing gene conversion. | ||
=== RDX12: selenocysteine from echinoderm to teleost fish === | |||
This gene has also been denoted C17orf37 (human) and SEPW2A (zebrafish). It is a [http://www.ncbi.nlm.nih.gov/pubmed/17503775 weak paralog] of SEPW and SELV with a related [http://www.jbc.org/cgi/content/full/282/51/37036 thioredoxin-like fold]. | |||
It shares but a single intron with SEPW1, the phase 21 intron that splits the cysteine codon in the CxxC or CxxU, suggesting gene duplication took place in early eukaryotes. As in so many selenoproteins, an ancestral selenocysteine codon that persists to living fish, sharks, lamprey, cephalochordates, urochordates, and echinoderms is lost to cysteine in tetrapods, with the timing still unknown in the understudied lobefinned fish. | |||
The length of the first exon is highly variable, causing considerable difficulty in gene recovery. This variability seems to result from an wandering start codon rather than indels or splice donor read-through. In mammals however, both length and sequence are well conserved. Smaller than the smallest known enzyme, RDX12 probably functions as an auxillary redox protein, though where exactly in metabolism is not known. There are no known disease alleles. | |||
>RDX12_homSap C17orf37 NM_032339 >SEPW1_homSap | |||
0 MSGEPGQTSVAPPPEEVEPGSGVRIVVEY<font color="#FF0000">C</font> 2 0 MALAVRVVY<font color="#FF0000">C 2</font> | |||
1 EP<font color="#FF0000">C</font>GFEATYLELASAVKEQYPGIEIESRLGGT 1 1 GA<font color="#FF0000">u</font>GYKSK 0 | |||
2 GAFEIEINGQLVFSKLENGGFPYEKD 0 0 YLQLKKKLEDEFPGRLDI 0 | |||
0 LIEAIRRASNGETLEKITNSRPPCVIL* 0 0 CGEGTPQATGFFEVMVAGKLIHSKK | |||
0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
Alignment of ~34% identity: | |||
RDX12 VRIVVEYCEPCGFEATYLELASAVKEQYPG-IEI--ESRLGGTGAFEIEINGQLVFSKLENGGF 83 | |||
||+| || |+++ ||+| +++++|| ++| | || ||+ + |+|+ || + |+ | |||
SEPW1 VRVV--YCGAuGYKSKYLQLKKKLEDEFPGRLDICGEGTPQATGFFEVMVAGKLIHSKKKGDGY 66 | |||
SELV VLIRVTYCGLuSYSLRYILLKKSLEQQFPNHLLFEEDRAAQATGEFEVFVNGRLVHSKKRGDGF | |||
* * | |||
<font color="#0066CC" >>RDX12_homSap MSGEPGQTSVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGETLEKITNSRPPCVIL* | |||
>RDX12_panTro MSGEPGQTSVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_gorGor MSGEPGQTSVAPPPEEVEPGSGVRMVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_ponAbe MSGESGQTFVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_rheMac MSGEPGQTSVAPPPEEVEPGSGVRIMVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASKGEPLEKITNSRPPCVIL* | |||
>RDX12_calJac MSGEPGQTSVAPLPGEVEPGSGVHIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASTGEPLEKITNSRPPCIIL* | |||
>RDX12_otoGar MSGEPGHSSVAPPPGEVEPGSGVRIMVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCIIL* | |||
>RDX12_tupBel MSGESGETSVAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGT-------------------------------------------TNSRPPCVIL* | |||
>RDX12_musMus MSGEPAPVSVVPPPGEVEAGSGVHIVVEYCKPCGFEATYLELASAVKEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASNGEPVEKITNSRPPCVIL* | |||
>RDX12_ratNor MSGEPGQVSVVPPPGEVEAGSGVHIVVEYCKPCGFEATYLELASSLEEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_dipOrd MSGEPGEVSLAPPPGEAEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_cavPor MSGEPGELSVARPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIQIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_oryCun MSGEPGPTSAAPSPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCTIL* | |||
>RDX12_ochPri MSGEPGPTSAAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELANAVKEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_vicPac MSGETGPTSVAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_turTru MSGETGPASSAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_bosTau MSGDTGTTSVAPPPGETEPGHGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_equCab MSGEPGLTSVAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_canFam MSGEPGPTSEAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_myoLuc MSGDAGPVSAAPHPGELEPGSGVRIVVEYCEPCGFEATYLELASAVKEQFPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_pteVam MSGDLGPTSAAPHPREIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCIIL* | |||
>RDX12_eriEur MSGEPGPTAVAPPPGEVEPGSGVRIVVEYCEPCGFEATYQELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_sorAra MSG----LAVAPPPGEGEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_loxAfr MNGEPGPISLAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_proCap MNGEPGPVSLAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_echTel MSGEPGPVSLAPPPGEAEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITSSRPPCAIL* | |||
>RDX12_monDom MSGESGAELVAPLPGEALPGSGVRIVVEYCEPCGFESTYLELASAVKEEYPGIKIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASSGEPLEKITNSCPPCVIL* | |||
>RDX12_macEug MSGEPGADLEEPLP.....GSGVRIVVEYCEPCGFESTYLELASAVKEEYPGIQIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASSGEPLQKITNSRPPCVIL* | |||
>RDX12_ornAna ---------------------------------------------------------------AFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASLGEPLEKITNSRPPCSIL* | |||
>RDX12_galGal AVGTESEAGDGDGFGSDSGSERVHIMVEYCEPCGFGATYEELASAVREEYPDIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCTIL* | |||
>RDX12_melGal AAAGTETEAGDGFVSGSGSERRVHIMVEYCEPCGFGATYEELASAVREEYPDIEIESRLGGTGAFEIEINGQLIFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCVIL* | |||
>RDX12_taeGut MSGGTGDGTGDGNGAERRVRIVVEYCEPCGFEATYQELASAVRDEYPDIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCVIL* | |||
>RDX12_anoCar MSDGSGEPAAEAPPATEGGVRIVVEYCKPCGFESAYLELANAVKEEYPDVEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRKAINGEPLEKITKSRPPCVIL* | |||
>RDX12_xenTro MSVSIVVEYCEPCGFKSHYEELASAVLEEFPDVTIDSRPGGTGAFEIEINGQLVFSKLELGGFPYAKDLIEAIRKASNGEPVEKITNSQAPCVIL* | |||
>RDX12_ambMex MAAVTIVVEYCKHCGFESHYLELESAVKEEFPDVVIESRCGETGTFEIKINGQIVFSKLELGGFPYEKDLMEAIKRASNGEPVERITNSRAPCVIL*</font > | |||
<font color="#990099" >>RDX12_tetNig MAVKIIVEFCGGuGYGPRYEELARVVKAEFLDADVSGFVGRLGSFEIVINEQLVFSKLETGGFPYEDDVLQVIQCAYDGKPVEKLTKSRPPCVIM* | |||
>RDX12_fugRub MGVTIRVEYCGGuGYGPRYEELARVVRAEFPDADVSGFVGRMGSFEIQINEQLIFSKLETGGFPYEDDVMHAIQCVSDGKPVEKITKSRPPCVIM* | |||
>RDX12_gasAcu MGVTMRVEYCGNuGYEPRYQELRTAVKQDFPDADVTGFVGRRGSFEIVLNGQLIFSRLESWGFPHVEDVLDAVKKAADGKPVDKITISRAPCVIM* | |||
>RDX12_oryLat MGVKIDVEYCGRuGYEPRYQDLASTVKDEFPEAEVSGFVGRSGSFEIQINGQLVFSKLELGGFPYEDDVLNAVQNAHDGKPVQKITKSRAPCVIM* | |||
>RDX12_danRer MGVQIKVEYCGGuGYEPRYQELKRVVTAEFTDADVSGFVGRQGSFEIEINGQLIFSKLETSGFPYEDDIMGVIQRAYDGQPVEKITKSQPPCVIL* | |||
>RDX12_oreMas MGVKVRVEYCGGuGYEPRYRELARVVKGEFSDADVTGVVGRTGSFEIEINGQLVFSKLETGGFPYEDDVMDAIHNAYDGKPLQKITKSRAPCVIM* | |||
>RDX12_calMil ..........GAuGYEPRYQKLAIVIKDEFPDADVSGKVGRTGSFEIEINGQLIFSKLETGGFPYENDISEAVQKANNGEELQKIENSRPPCVIL* | |||
>RDX12_squAca MGVKIHFEYCGAuGYKPRYQELANTIMGTFPDAAISGDVGRTGSFEIEINGQLVFSKLETSGFPYEDDIMDAVQKASAGDDVQKIVKSRPPCVIL* | |||
>RDX12_torCal MSVKVHVEYCGAuGYGNRYQDLANNILKTFPDADISGDVGRKGSFEVEINGQLVFSKLETKGFPFENDIITAVQNASNGVEMQQITNSRASCVIL* | |||
>RDX12_petMar MAVNVNVEYCGGuGYEPRYQELAANILKQAPGVEVIGQVGRSGSFEVTINGELIFSKLECGGFPFAEDIIAEVKKVQGGEKVGKVTKSQAPCVIL* | |||
>RDX12_cioInt MADSNKVKVEIEYCGSuGYYGRFMDLKNDLESGCPNALVSGFVGREGSFEVSINGKQIFSKLETFGFPYPNDLIEAMKTAQNGEEVAAINNSQSPCTIL* | |||
>RDX12_braFlo MNYSRSKMAPKVEVEYCGGuGYAPRYWELANQIKTAVPDAEVTGVVGRSSSFEIIVDGQLLFSKLESGGFPQEQEILEALSNYKEGEKVEQVTNIQFPWCTIL* | |||
>RDX12_strPur MAQKKVTIEYCGSuGYYPRYRELHEMIESAIKGVDVSGKRGRPSSFEVKLNGQVLYSKLKNGGFPDLDAIVEAIEDYKGEGTVTPVEKKAASCIIL* | |||
>RDX12_astPec MATRKVEIEYCGGuGFYPRFMDLHNDIKSGVPGVAVDGRVGRSTSFEVTLNGKLLFSKLKTGGFPINKEIVTAIKEYKGEGDVTPVTNTASNCVLL*</font > | |||
---- | |||
=== DIO1, DIO2, and DIO3: a curious history === | === DIO1, DIO2, and DIO3: a curious history === | ||
The iodothyronine deiodinases of selenoproteins have been thoroughly studied because of their role in thyroid hormone metabolism. The three paralogs seen in mammals diverged long ago -- the percent identity today is about 45% and exon breaks and phases are completely unrelated. One scenario is that DIO2 and DIO3 in mammals arose as an initial retroprocessed gene that then duplicated again, with DIO2 subsequently acquiring an intron and DIO3 remaining a single coding exon. The critical era for exon acquisition and loss is poorly represented in | The iodothyronine deiodinases of selenoproteins have been thoroughly studied because of their role in thyroid hormone metabolism. The three paralogs seen in mammals diverged long ago -- the percent identity today is about 45% and exon breaks and phases are completely unrelated. One scenario is that DIO2 and DIO3 in mammals arose as an initial retroprocessed gene that then duplicated again, with DIO2 subsequently acquiring an intron and DIO3 remaining a single coding exon. The critical era for exon acquisition and loss is poorly represented in database species. | ||
DIO2 and DIO3 are found on the same chromosome arm of chr 14 in human but separated by 22 Mbp, rather far for a tandom duplication. This structure is conserved in the chicken genome with only 8.7 Mbp intervening. Again, it is hard to sort out coincidence from correlation. Note the SECIS element might well accompany both retroprocessing and tandem duplications. | DIO2 and DIO3 are found on the same chromosome arm of chr 14 in human but separated by 22 Mbp, rather far for a tandom duplication. This structure is conserved in the chicken genome with only 8.7 Mbp intervening. Again, it is hard to sort out coincidence from correlation. Note the SECIS element might well accompany both retroprocessing and tandem duplications. | ||
| Line 171: | Line 242: | ||
=== SELH: rapid evolution === | === SELH: rapid evolution === | ||
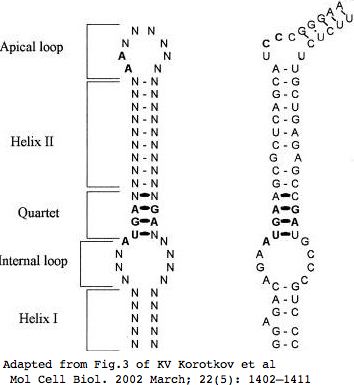
SELH is another small selenoprotein with a conserved CxxU redox motif split by a phase 21 intron. The introns in mammalian SELH are exceedingly short (eg, 93 and 162 bp in human with gene coding span very short at 619 bp) but the level of transcription is not remarkable so provides no explanation for the lack of retroposons. Some species like orangutan have processed pseudogenes, implying transcription in germ-line tissues. Zebrafish has a diverged duplicate gene with tryptophan at the seleocysteine site in addition to an intronless transcribed gene with TGA selenocysteine -- it appears that this has displaced the normal three-intron gene (seen in other fish). That can work in selenoproteins since retropositioning begins at the 3' end of transcripts, meaning the SECIS likely accompanies the coding region. | SELH has been given the highly unsatisfactory official name [http://www.genenames.org/data/hgnc_data.php?hgnc_id=18251 C11orf31] by HGNC. It is another small selenoprotein with a conserved CxxU redox motif split by a phase 21 intron. The introns in mammalian SELH are exceedingly short (eg, 93 and 162 bp in human with gene coding span very short at 619 bp) but the level of transcription is not remarkable so provides no explanation for the lack of retroposons. Some species like orangutan have processed pseudogenes, implying transcription in germ-line tissues. Zebrafish has a diverged duplicate gene with tryptophan at the seleocysteine site in addition to an intronless transcribed gene with TGA selenocysteine -- it appears that this has displaced the normal three-intron gene (seen in other fish). That can work in selenoproteins since retropositioning begins at the 3' end of transcripts, meaning the SECIS likely accompanies the coding region. | ||
[[Image:Selh.png|left|]] | [[Image:Selh.png|left|]] | ||
<br clear="all"> | |||
Protein conservation is below average -- SELH percent identities (to human) drop to the low 80's within Laurasiatheres, to 72% with marsupial, and 57% with chicken. Further, rodents exhibit significant residue loss upstream of the CxxU motif, very unusual in such a short protein but indicative of an inessential structural region. Indeed observed conservation is primarily centered in the middle of the protein. No phylogenetic conversion of selenocysteine to cysteine is observed within vertbrates | Protein conservation is below average -- SELH percent identities (to human) drop to the low 80's within Laurasiatheres, to 72% with marsupial, and 57% with chicken. Further, rodents exhibit significant residue loss upstream of the CxxU motif, very unusual in such a short protein but indicative of an inessential structural region. Indeed observed conservation is primarily centered in the middle of the protein. No phylogenetic conversion of selenocysteine to cysteine is observed within vertbrates | ||
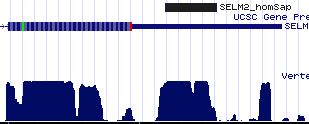
=== | === SELM2: perinuclear but ER retention signal === | ||
Another selenoprotein of unknown function, SELM2 with CxxU motif, surfaces during Blastp searches using selenoprotein SELM2 (aka SEP15 which oddly has a CxU motif) as query. A 3' UTR SECIS element cannot be located with [http://genome.unl.edu/SECISearch.html SECISearch 2.19] yet it seemingly must be located in the comparative genomic peaks of conservation lying between SELM and converently transcribed nearby neighboring gene SMTN. Indeed, that has been validated by [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=134693 direct experiment.] | |||
[[Image:SELM2_SECIS.jpg|left|]] | |||
<br clear="all"/> | |||
[[Image: | The SECIS element has CC in place of supposedly critical apical loop AA. The evolutionary origin of this divergence has not been dated and would be difficult to do. This SECIS element might be recognized by KIAA0256 rather than SECISBP2. | ||
[[Image:SELM2_anomSECIS.jpg|left|]] | |||
Since the protein is quite short with 5 introns, complete SELM sequences are best recovered from cDNAs and genomic alignments in the UCSC 28way. This is an ancient protein recoverable from vertebrates, amphioxus, sea urchin, shrimp, mite, moths, and hydra, and plants. Moths (silkworm and hawkmoth) -- but not their outgroups -- encode cysteine in a CxxC motif in place of CxxU. No loss of selenocysteine is seen in 22 species of phylogenetically dispersed vertebrates. | Since the protein is quite short with 5 introns, complete SELM sequences are best recovered from cDNAs and genomic alignments in the UCSC 28way. This is an ancient protein recoverable from vertebrates, amphioxus, sea urchin, shrimp, mite, moths, and hydra, and plants. Moths (silkworm and hawkmoth) -- but not their outgroups -- encode cysteine in a CxxC motif in place of CxxU. No loss of selenocysteine is seen in 22 species of phylogenetically dispersed vertebrates. | ||
SELM | SELM begins with an experimentally established signal peptide. There are no glycosylation sites nor additional conserved cysteines beyond the CxxU motif. SuperFamily finds no similarity to proteins of known 3D structure. The terminal residues appear to be a phylogenetically conserved KDAL-class endoplasmic retention signal though it is reported perinuclear. | ||
In whole-mount zebrafish embryos, SELM is expressed within the notochord and anterior somites, axial fin fold, dorsal spinal chord neurons, then in lateral line neuromasts. SelM is located in the ER/Golgi as is its distant homolog Sep15 (found associated with UDP-glucose glycoprotein glucosyltransferase, an ER-resident protein involved in quality control of protein folding). | In whole-mount zebrafish embryos, SELM is expressed within the notochord and anterior somites, axial fin fold, dorsal spinal chord neurons, then in lateral line neuromasts. SelM is located in the ER/Golgi as is its distant homolog Sep15 (found associated with UDP-glucose glycoprotein glucosyltransferase, an ER-resident protein involved in quality control of protein folding). | ||
<br clear="all"/> | |||
=== MSRB123: methionine sulfoxidases === | === MSRB123: methionine sulfoxidases === | ||
MSRB1 is a short odd protein, rich in cysteines, with two CxxC motifs, a near-amino terminal cysteine and a more distal selenocysteine in a motif with serine. | MSRB1 is a short odd protein with HGNC gene name [http://www.genenames.org/data/hgnc_data.php?hgnc_id=14133 SEPX1], rich in cysteines, with two CxxC motifs, a near-amino terminal cysteine and a more distal selenocysteine in a motif with serine. MSRB1 is now known to be a stereospecific methionine-R-sulfoxide reductase repairing oxidative damage to methionine in native proteins. The two pairs of cysteines bind a zinc atom. The all-beta structure has been determined in a bacterial homolog with a internal structural duplication so weak it needs an xray determination to be revealed. Humans have 9 such domains in 9 proteins according to SuperFamily. These could potentially encode other selenoproteins at least in some species. | ||
The fold exists as a small family of paralogs, three in mammals, with specialization to cell compartment -- cytosol, mitochondria and endoplasmic reticulum (via KAEL* signal), respectively.). All contain catalytic zinc. This multiplicity of MSRB contrasts with a single non-homologous methionine-S-sulfoxide reductase (not a selenoprotein in any species). | The fold exists as a small family of paralogs, three in mammals, with specialization to cell compartment -- cytosol, mitochondria and endoplasmic reticulum (via KAEL* signal), respectively.). All contain catalytic zinc. This multiplicity of MSRB contrasts with a single non-homologous methionine-S-sulfoxide reductase (not a selenoprotein in any species). | ||
| Line 247: | Line 326: | ||
This gene family has a curious evolutionary history: every additional level of phylogenetic resolution adds another twist -- even 29 genomes from cnidarian to placental do not provide adequate genomic saturation. The original single-copy gene (itself an ancient internal tandem duplicatation fold with active site on dimeric cleft) procedes uneventfully from ur-bacteria to hagfish/lamprey divergence. | This gene family has a curious evolutionary history: every additional level of phylogenetic resolution adds another twist -- even 29 genomes from cnidarian to placental do not provide adequate genomic saturation. The original single-copy gene (itself an ancient internal tandem duplicatation fold with active site on dimeric cleft) procedes uneventfully from ur-bacteria to hagfish/lamprey divergence. | ||
However, prior to teleost fish divergence, block duplication of the 8 exon gene took place. No flanking synteny persists in extant species. The copy that would remain closest to the ancestral gene then lost its selenocysteine to nucleophile | However, prior to teleost fish divergence, block duplication of the 8-exon gene took place. No flanking synteny persists in extant species. The copy that would remain closest to the ancestral gene then lost its selenocysteine to nucleophile threonine, its 3' UTR SECIS insertion element decayed, and the old function strayed to salvaging selenide from free selenocysteine (arising from selenoprotein catabolism or diet). The copy we call SEPHS2 diverged rapidly ( rather unconstrained amino terminus in both length and sequence) but with conservation about the selenocysteine active site motif and beyond. | ||
Between monothere and marsupial divergence, a fully processed retrogene (no introns) carrying the 3' UTR SECIS insertion element became functionally established, remarkably displacing the previous 8-exon gene (now an undetectable decayed pseudogene or deletion), to the extent that every species of placental mammal retains only the processed retrogene and threonine copies. Opossum, but not platypus nor elephant or tenrec, retains all 3 gene copies. | Between monothere and marsupial divergence, a fully processed retrogene (no coding region introns) carrying the 3' UTR SECIS insertion element became functionally established, remarkably displacing the previous 8-exon gene (now an undetectable decayed pseudogene or deletion), to the extent that every species of placental mammal retains only the processed retrogene and threonine copies. Opossum, but not platypus nor elephant or tenrec, retains all 3 gene copies. | ||
Here the processed retrogene mechanism (begin at 3' end of UTR but not extend necesarily to initial methionine or 5' regulatory regions) brings along the SECIS selenocysteine insertion loop making a new selenoprotein feasible and while explaining non-homologous start peptides. Humans have an assortment of fragmentary and processed pseudogenes for which SEPHS1 is usually the parent, most notably a mis-spliced processed | Here the processed retrogene mechanism (begin at 3' end of UTR but not extend necesarily to initial methionine or 5' regulatory regions) brings along the SECIS selenocysteine insertion loop making a new selenoprotein feasible and while explaining non-homologous start peptides. Humans have an assortment of fragmentary and processed pseudogenes for which SEPHS1 is usually the parent, most notably a mis-spliced processed pseudogene omitting exon 7 (build 35, chr7:63756925-63757879). | ||
=== SELO: rapid change in last exon === | === SELO: rapid change in last exon === | ||
| Line 279: | Line 358: | ||
VHLLQKTLRHPFHKQREAEEA GYSSRPPLWARELRVSCSS* SELO_calMil Callorhinchus milii (elephantfish) | VHLLQKTLRHPFHKQREAEEA GYSSRPPLWARELRVSCSS* SELO_calMil Callorhinchus milii (elephantfish) | ||
== Selenoprotein | == Selenoprotein pseudogenes: SELW == | ||
Processed pseudogenes of selenoproteins are of special interest. First, they represent fossil transcripts within germline tissue (somatic cell insertions would not be heritable) offering clues to ancestral functionality, continuity of germline expression and potentially validation of contemporary alternative splicing. Second, retrogenes occasionally retain expression and functionality in descendent clades, thus extending the selenoproteome repertoire. Finding a complete set of selenoproteins in each species has long been a critical research goal. | |||
Since the mechanism of processed retrogene insertion begins with the 3' end, the critical SECIS element and polyadenylation signal would be brought along. Although the process does not always extend to full 5' length, many selenoproteins are quite short, so the prospects of retropositioning a complete intronless copy with a suppressible TGA codon are rather favorable. However regions necessary for initiation of transcription are left behind, making expression and its control subject to properties of pre-existing sequence upstream of the insertion site. | |||
The status of a given retroprocessed locus is not always clear, especially for recent insertions for which insufficient time has elapsed for accrual of definitive pseudogene signatures (such as in-frame non-TGA stop codons, pronounced Kd/Ks ratio, or structurally implausible substitutions at conserved residues). Conventional gene duplicates commonly diverge rapidly away from the parent gene as they neo- or sub-functionalize. | |||
Because few vertebrate genome projects are accompanied by an extensive transcript program (eg mouse, human), absence of supporting expression is not a reliable guide to pseudogene character. Even in human, some well-established coding genes still lack GenBank transcripts as of December 2008, presumably because these are rare or restricted to cell types not represented in studied libraries. Conversely, untranslated transcripts covering non-genes can be found, indeed most of the genome is transcribed at some level despite only 1-2% coding for protein. | |||
Two processes further complicate the characterization of pseudogenes: they become obliterated past the point of reliable recognizability by point mutations and small indels over a few tens of millons of years and they are subject to repeat element insertion (eg Alu elements) that may follow initial rounds of speciation, complicating the establishment of orthologous position especially in intergenic regions not providing stable flanking markers. | |||
These Alu insertions result in pseudo exons which in general lack GT-AG splice sites, conserved reading frame, or parental gene position but nonetheless cause the pseudogene to appear segmentally duplicated or incompletely retro-processed. These factors limit ability to date an initial event and track it forward (reliably interpreted) in all descendent clades. | |||
Each of the 25 human selenoproteins can be used as initial Blat query. Typically the secondary matches (if any) are retrogenes, though a recent pseudogene may score higher because introns of authentic selenogenes interrupt alignment and so lower score. This process tallies the number of pseudogenes back to the phylogenetic depth commensurate with Blat sensitivity -- use of Blast or profile querying may detect still older debris. | |||
Blat suffices here because the primary purpose is finding functional retrogenes paralogous to well-characterized genes. The target genome can be varied to place the event between consecutive phylogenetic tree divergences. Both query protein and target genome can be changed to search for retrogenes in other clades (eg artiodactyl or marsupial). | |||
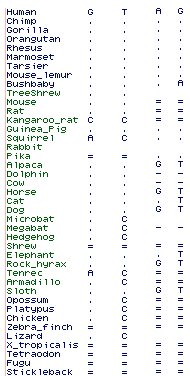
By this method, some 81 retroprocessed loci can be found for 11 human selenoproteins, suggesting the other 14 selenoproteins are not as frequently expressed in germline tissue. Most of these loci have so many deleterious accrued changes that they can be immediately classified as pseudogenes. Others are not so clear and require detailed case-by-case curation: | |||
SELW gives a single secondary match on a different chromosome from the parental gene, at coordinates chr1:31,339,873-31,341,525 of NCBI assembly 36. By examining syntenic regions from other species established by whole genomic multiz alignment, the event can be dated to post-marmoset, pre-macaque. The retrogene in all old world apes has two apparent exons whereas the parent gene has five. | |||
The RepeatMasker track shows an AluSg retroposon interrupting the reading frame, suggesting (invoking parsimony) a single insertion event prior to macaque divergence. Close examination of the putative exons shows they wholly lack potential splice donor and acceptors (GT-AG or less common alternatives). Consequently the AluSg cannot be spliced out. However if it were included in a transcript, that would introduce non-TGA stop codons as well as grossly disrupt protein structure with extraneous residues (as Alus derive from 7S RNA). | |||
Consequently this feature is a pseudogene in all apes, despite constituting nearly a full length copy of SELW with just 7 conservative substitutions, only frame-preserving indels, Ka/Ks ratios mildly indicative of positive selection, and a SECIS element capable of a stem-loop (though missing two key bases GA at position 78-79). | |||
In support of this, none of numerous ape transcripts align to this feature. However GenBank does contain an unusual macaque cDNA from placenta, CN804216, with two internal stop codons and only 67% identity to SELW from that species. While that might reflect a previously functional selenoretrogene with continuing relic transcription, it fails to match any of the 31 million genomic traces from macaque. Further, the private contractor providing the 2004 sequence reports only 97 Q20 bases. Consequently the sequence is an artefact, though an instructive one. | |||
tBlastn alignment of SELW of M. mulatta with cDNA CN804216 | |||
SELW MALAVRVVYCGACGYKSKYLQLKKKLEDEFPGRLDICGEGTPQATGFFEVMVAGKLIHSKKKGDGYVDTESKFLKLVAAIKAALAQG* | |||
MAL V VYCGA Y SKYLQL KKL++EFPGRLDIC E T A+GF E++ G LI S KKGDGYV T S FLKLVA IKAAL G | |||
CN804216 MALVVSCVYCGA*CYYSKYLQLPKKLKNEFPGRLDICCESTRWASGFVEILGTGTLIRSTKKGDGYVHT*STFLKLVADIKAALG*G* | |||
Macaque contains a second contiguous retroprocessed SELW locus at chr11:99257376-99257637 that can be detected by Blast but not Blat. As expected, it is considerably more decayed (3 stop codons, a frameshift, numerous substitutions) than the retroprocessed feature discussed above. This feature is found from marmoset to human but is missing in tarsier, lemurs, and earlier diverging mammals Using the UCSC 44-species genomic alignment). Consequently it is an older feature predating new world monkey divergence. | |||
SELW MALAVRVVYCGACGYKSKYLQLKKKLEDEFPGRLDICGEGTPQATGFFE----VMVAGKLIHSKKKGDGYVDTESKFLKLVAAIKAALAQG | |||
M L +++YCGA G KSKY QLKK LED+ P LDIC TP TGFFE MVAG L KK DGY++ ESKFLKLVA IKA LAQG | |||
Retro2 MPLMFQIIYCGA*G*KSKYFQLKK-LEDKSPRYLDICSNRTP*VTGFFEGMVVFMVAGSLQEDKKISDGYMNGESKFLKLVATIKAGLAQG | |||
Clade-specific SELW pseudogenes are most conveniently survey by tBlastn of NCBI database wgs, which contrary to its name contains assembled contigs from all genome projects, including short contigs omited from final genome assemblies. Restricting the query to mammals, many species exhibit retroprocessed products of SELW including Dasypus (4), Choloepus (2), Loxodonta, Echinops, Tupaia, Bos (2), Felis (2), Myotis, Rattus, Microcebus (2), and Tarsius in addition to the ape retroprocessed loci discussed above. While not all of these situations can be definitively declared pseudogenes, it can be concluded that SELW is transcribed in germline tissue in a wide variety of extant placentals and thus in their last common ancestor. | |||
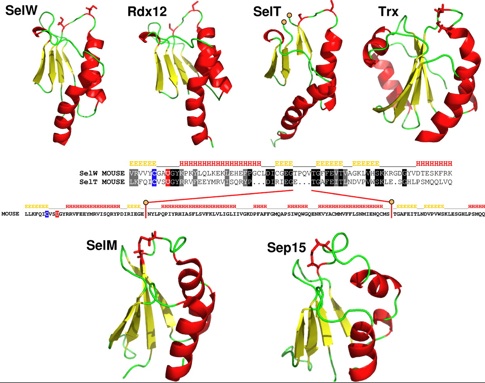
The locus in armadillo contig AAGV020179194 is noteworthy. It is either a very recent pseudogene or functional (being 85% identity to sloth SELW). A retrogene under selection should have a subsitution site mismatch with conserved residue profile because the latter sites would still mostly be important despite putative re-functionalization. The structure of mouse SELW has recently been [http://www.jbc.org/cgi/content/full/282/51/37036 determined] to be a thioredoxin-like fold (PDB: 2NPB), similar to SelV, SelT, SelH and Cys-containing Rdx12 (aka C17orf37) as shown in Fig.4 of that paper (below). | |||
SELW MALAVRVVYCGAUGYKSKYLQLKKKLEDEFPGRLDICGEGTPQATGFFEVMVAGKLIHSKKKGDGYVDTESKFLKLVAAIKAALAQG 87 | |||
MALAVR+VYCGA GYK KY+QLKK+LEDEFPG LDI GEGTPQ TGFFEVMVAGKL+HSKK+GDG+VD ESKFL+LV AIKA LA G | |||
dasNov MALAVRMVYCGA*GYKPKYIQLKKRLEDEFPGCLDISGEGTPQTTGFFEVMVAGKLVHSKKRGDGFVDMESKFLRLVPAIKATLAHG 619 | |||
[[Image:SELW pdb.jpg]] | |||
Microarrays for human SELW show it predominantly expressed in heart. Available macaque transcripts (tBlastn of est_others restricted to Macaca) have been recovered from lens, thymus, kidney, liver, brain, ovary, lymphocytes, ileum, jejunum and testis (DR771274). | |||
= | <br clear = "all"> | ||
[[Image:SELW expr.jpg]] | |||
== Alternative splices of selenoproteins: introduction == | |||
In the search for significant alternate splice forms of selenoproteins, it is very important not to take transcript data at face value. Even if multiple transcripts support a given alternative splice, that does not imply that such transcripts exist in vivo in healthy cells (millions of GenBank entries come from cancer cell lines), nor that any such protein is ever made (nonsense-mediated decay), nor if translated that the protein is stably folded (rather than immediately degraded at the protosome), nor that stable protein fragments (such as selenoproteins missing their redox site) are functional. Due to post-translational trimming of amorphous termini, many alternative splices in distal regions would be irrelevent to mature protein. | |||
1 | On the experimental quality side, human and mouse transcript entries at GenBank contain millions upon millions of defective entries, notably from the early years of EST factories. Transcripts can be riddled with direct sequencing error and not align satisfactorily to any assembled genome. Others are accurate but represent cross-species laboratory contamination. A vast number do not correspond to plausible coding genes, remain unvalidated by later work, and seem merely to have been primed off naturally occuring non-genic genomic polyA. It appears that many partially processed transcripts (5' caps not required, intron-retaining) entered the sequencing queue. Consequently, each alternative splice transcript must be individually evaluated for various parameters affecting its quality. | ||
Despite vast expenditures on proteomics mass spectroscopy, not 1 in 10,000 proposed alternatively spliced products has been validated to date in vertebrates. Indeed for fully half of all human proteins, no product -- much less an alternative splice form -- has ever been reported (according to SwissProt, November 2008). Some protein structure scientists question whether the vast majority alternative splice forms can stably fold as it makes no sense from a folding energetics perspective to omit a beta strand from a sheet or insert a helix into an existing sandwich. | |||
> | In this view, <font color="#990099">vertebrate transcription is inherently an error-prone and wasteful process with imperfect splicing accuracy mitigated downstream</font> by a series of quality controls. This is perhaps inevitable give intrinsically weak splice signals (the core GT and AG occur by random every 16 bp) dwarfed by the vast expansion of many internal introns resulting from a retroposon explosion that masks splice donors and acceptors with the effect of intron retention and exon skipping, respectively. For example the 79 coding exons of dystrophin (the largest human gene) occupies 11 kbp of a 2,400,000 bp transcript meaning introns average some 30,000 bp. | ||
2 | |||
Alternate splicing is often invoked to explain why humans are "special" yet this process is vastly more extensive in fruit flies and early-diverging life forms. Human proteins are among the slowest evolving and least innovative of any mammalian species, evolving at a third the rate of mouse. Take any cartilaginous fish protein, tblastn against all vertebrate genomes (thus avoiding annotation effort bias), and find human is very often the top hit. Consequently anthropomorphic justification of alternate splicing is wholly unwarranted. | |||
Thus screening the nominal selenotranscriptome for functionally significant alternative splices requires multiple types of strong supporting data to overcome the null hypothesis (artefact). It is extremely time-consuming to pursue candidates further experimentally, so prioritization by quality is imperative. | |||
Because documented human innovation past chimpanzee divergence is so minimal, the vast majority of valid alternate splices should exhibit strong comparative genomics support at significant phylogenetic depth. Unlike transcript support, whose availability and quality differ radically by species, comparative genomics support is uniformly available in a large set of phylogenetically spaced genomes and readily intepretable. | |||
Alternative splicing in 5', coding and 3' regions are separate topics. Of these, coding has the best prospects for bioinformatic validation especially when a determined 3D structure permits modelling of the alternatively spliced product and can supplement phylogenetic aligment. However comparative genomics is still applicable in some other cases at least to the extent of dating the phylogenetic origin of and persistence of the splice donor and acceptor. | |||
=== Evaluating an alternative 3' splice of SELS === | |||
> | SELS has a well-known 3' non-coding alternative splice (RefSeq NM_203472) which skips the SECIS element to terminate downstream in a spearate non-coding exon. Since the selenocysteine is near-terminal position, a nearly full-length non-functional protein could be produced, yet the transcript might be subject to nonsense-mediated decay, seemingly a very wasteful and atypical way (compared to regulation at the upstream promoter) of down-regulating functional SELS production. Another option is that SELS forms an essential homodimer -- having one monomer <font color="#0066CC">sparing a need for selenium</font> could provide a selective advantage (even as a quarter of the dimers would be duds). | ||
[[Image:SELS GTAG.jpg|left]] | |||
It might be inferred from extensive and available microarray data how SELS is expressed, yet that data is irrelevent if NMD is operative (making transcript level an invalid proxy for protein level). Since the function of SELS is not known, experimental conditions in which a mouse model might down-regulate SELS are unclear; the alternate splice donor could be knocked out in mouse yet no phenotypic effect might be observable. | |||
Conservation depth of the splice donor and acceptor are readily established at the UCSC 44-species whole genome alignment. Here we find that the GT donor in homologous position applies to most euarchontoglires and laurasiatheres (5 exceptions) but earlier diverging species lacks it. The extent of 3' sequence homology (chimp, macaque, bar, mouse, cow) extends to the region down to the SECIS but not strikingly to the alternate acceptor region. The AG acceptor site is similarly only feasible in euarchontoglires (at precise homological position). | |||
Consequently, comparative genomic support is fairly shallow, not greatly beyond random conservation expectations. There is more to splice junctions than just GT-AG and other species need further testing with predictive tools (eg GeneScan) for junctional validity. It is worth noting that a Papio anubis EST GE878368 dated 2008-10-19 supports this splice (as do numerous human transcripts): | |||
> | 3' UTR donor and acceptor splice match of baboon transcript GE878368 to human SELS chr15:99630478-99629374 | ||
<font color="#0066CC">GGCGGATGAgGCTAAgAATCTT</font><font color="#FF0000">gt</font>tagtg...atctcctgc<font color="#FF0000">ag</font><font color="#0066CC">GTAGAATATTCC</font> Papio | |||
<font color="#0066CC">GGCGGATGAGGCTAAGAATCTT</font><font color="#FF0000">gt</font>tagtg...atctcctgc<font color="#FF0000">ag</font><font color="#0066CC">GTAGAATATTCC</font> Homo | |||
Overall, this raises more questions than answers. The alternate splice cannot be dismissed as an artefact yet cross-species support for it is underwhelming. Supposing the selected function is to down-regulate this gene product, would not other mammals have a similar need? If they utilized another mechanism present for many million years of last common ancestor, euarchontoglires then lost that well-established mechanism and gained a novel one over a fairly brief evolutionary span. | |||
> | <br clear="all"> | ||
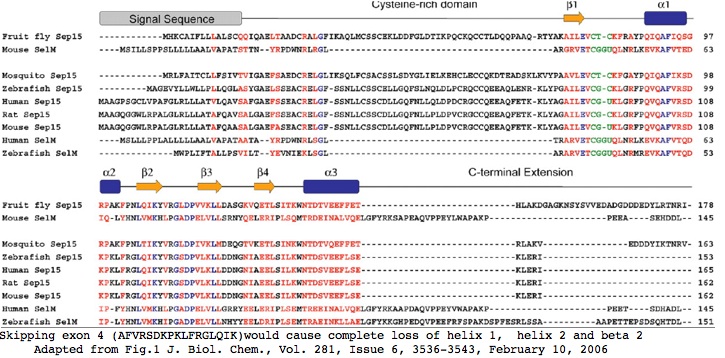
=== Evaluating an exon-skipping, frame-shifting alternative splice of SEP15 === | |||
> | The human SEP15 locus has two recognized splice forms at RefSeq resulting in proteins that differ markedly in distal sequence. The main splice form yields five exons. These exons, the seleoncysteine position, and the conventional stop codon are extraordinarily conserved back to lamprey, indeed throughout bilatera. This is clearly the ancestral form of SEP15. | ||
0 | |||
1 | <font color="#0066CC">>SEP15A_homSap length=165 NM_004261 | ||
2 | 0 MAAGPSGCLVPAFGLRLLLATVLQA 0 | ||
0 VSAFGAEFSSEACRELGFSSNLLCSSCDLLGQFNLLQLDPDCRGCCQEEAQFETKK 0 | |||
0 LYAGAILEVCGuKLGRFPQVQ 1</font> | |||
<font color="#FF0000">2 AFVRSDKPKLFRGLQIK 0</font> | |||
0 YVRGSDPVLKLLDDNGNIAEELSILKWNTDSVEEFLSEKLERI* 0 | |||
<font color="#0066CC">>SEP15B_homSap length=124 NM_203341 | |||
0 MVAMAAGPSGCLVPAFGLRLLLATVLQA 0 | |||
0 VSAFGAEFSSEACRELGFSSNLLCSSCDLLGQFNLLQLDPDCRGCCQEEAQFETKK 0 | |||
0 LYAGAILEVCGuKLGRFPQVQ 1</font> | |||
<font color="#FF0000">2 VCPWFRPCIKAFGRQWEHC* 0</font> | |||
The second splice form skips the fourth exon, jumping from the splice donor of the third exon to the splice acceptor of the fifth exon. The donor phases are such that the reading frame is shifted, causing the final exon to have altogether different amino acid sequence from the main splice form. The new reading frame reaches a stop codon after 20 amino acids but much earlier in 6 other species of mammals including old world monkeys and lemurs: | |||
Premature stop codon in macaque final exon of exon-skipping splice form | |||
Homo VCPWFRPCIKAFGRQWEHC* | |||
VCPWFRP IKAFG QWEHC | |||
Macaca VCPWFRPYIKAFG*QWEHC | |||
A goodly proportion of human ESTs skip exon 4 and a 2002 Riken mouse transcript BY235176 provides some comparative genomics support. However exon-skipping is not observed in a very large number of other mammalian transcripts: | |||
391 of 392 mammalian transcripts in 9 species do not skip exon 4 | |||
Mus musculus (mouse) 198 (only BY235176 skips) | |||
Papio anubis (Doguera baboon) 85 | |||
Macaca mulatta (rhesus macaque) 30 | |||
Macaca fascicularis (cynomolgus monkey) 22 | |||
Cavia porcellus (guinea pig) 2 | |||
Spermophilus tridecemlineatus (squirrel) 2 | |||
Bos taurus (cow) 11 | |||
Ovis aries (domestic sheep) 34 | |||
Equus caballus (equine) 8 | |||
The structure of the non-skipping form of SEP15 was recently [http://www.jbc.org/cgi/content/full/281/6/3536 determined]. Omission of exon 4 would obliterate almost all secondary structure, causing complete loss of helix 1, helix 2 and beta strand 2; the reading frameshift would take out almost all the rest (though the structure if any of the short form in this region is not known). The short form would be left only with its signal peptide, a single (ie dysfunctional) beta strand, and the cysteine-rich region. | |||
[[Image:SEP15 secStr.jpg]] | |||
Clearly the long form of SEP15 is the only relevent protein species at this locus. It is not feasible to have selection acting simultaneously on different translated reading frames; it is implausible that the short form could fold stably but wholly differently given a very brief evolutionary history. Conceivably the cysteine-rich region could still function though the protein would likely be subject to very rapid decay. The role of the selenocysteine without its supporting tertiary structure would be unclear. | |||
Thus the alternative splice forms here, though too abundant to be experimental artifact, are misleading. They do not in all likelihood encode an alternative functioning protein. Instead, they may merely reflect a human polymorphism in the intron preceding exon 4 that has the effect of weakening it as a splice acceptor (alternatively a polymorphism enhancing the splice acceptor preceding exon 5). This amounts to the final splice acceptor out-competing the fourth exon acceptor in a substantial fraction of transcripts. | |||
The net effect of is a certain inefficiency but nothing particularly detrimental to the cell: functional SEP15 selenoprotein is still produced and the fragmentary mis-spliced polypeptide is rapidly recycled. While this down-regulates SEP15 production, as an evolved adaptive mechanism, it lacks any depth of comparative genomics support. | |||
=== An AluJb-translating splice form of primate SEPN1 === | |||
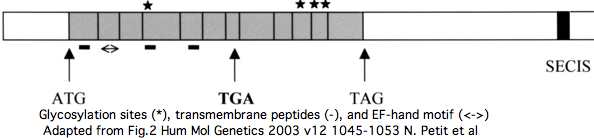
Two splice forms of SEPN1 have gained prominence, the 12 exon NM_206926 and the 13-exon NM_020451. Both have GenBank status of "Reviewed RefSeq: This record has been curated by NCBI staff" but that process has never been described and is never accompanied by explanatory comments. The difference between the two RefSeqs is an alternately included exon 3 which supposedly contains an additional TGA selenocysteine codon. | |||
However, far more likely, this is gross annotation error. As noted but not genetically analyzed in a [http://hmg.oxfordjournals.org/cgi/content/full/12/9/10452003 publication], the extra exon is nothing but an AluJb retroposon that inserted in minus strand orientation, with the polyA stretch serving as the pyrimidine stretch (polyT) augmenting a random GT to create a splice acceptor. This introduced a downstream inframe stop codon that happened to be TGA and so was "written off" as a disabling mutation because the later valid selenocysteine codon would be accompanied by a SECIS insertion element. As a further coincidence, a GT donor past the stop codon (but still well within the 296 bp AluJb) restored reading phase. | |||
The insertion event is readily dated using the 44-species genomic alignment at UCSC to the post-lemur, pre new world monkey stem. This implies that homologous amino acids will not exist in the SEPN1 genes of any other primate (or vertebrate). That is readily validated by blast searches of all genomes and transcripts at GenBank. | |||
[[Image:SEPN1 AluJb.jpg]] | |||
Although this new exon could represent a dramatic primate innovation, other lines of reasoning strongly suggest this extra exon is not functional. Retroposons are indeed occasionally co-opted for functional purposes; however 99.999% have no remotely plausible gene role and simply represent junk dna (as proven by various engineered megabase deletions in mice and massively reduced but still satisfactorily advanced fugu genome). | |||
Alu-inactivating mutations are far more commonly a source of genetic disease; however this particular AluJb does not inactivate the gene but simply results in a minor level of transcriptional noise from the occasional mis-splice. It cannot be argued that natural selection would have rapidly removed it from the genome in view of the other 1.1 million SINE elements in the human genome -- and these continue to accrue faster than they are being deleted. Statistical arguments concerning Ks/Ka cannot be applied to a mere 96 bp because nothing is known about evolutionary processes or rates in this very localized region of chromosome 1. | |||
AluJb SINE elements arise from 7S RNA, not proteins as in LINE elements. This distinction is exceedingly important. It means the extra exon was never vetted by selection for its compositional or folding properties. Indeed the reading frame here arose randomly from the three available. Random peptides of 32 amino acids have near-zero propensity for useful structural properties. | |||
Normally transcripts containing an inframe stop codon resulting from occasionally spicing in an Alu would be dismissed as junk rapidly degrading via nonsense-mediated decay. However an inframe TGA stop codon here must be given special consideration because SEPN1 already has a long-established selenocysteine site in exon 9 accompanied by a SECIS element in 3' UTR. There is precedent for a single SECIS element to substitute selenocystein at multiple TGA sites, notably the selenocysteine storage protein. | |||
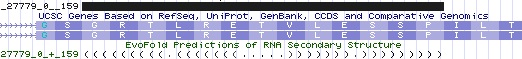
However there is much more to a selenocysteine-substitutable site than just TGA. It appears that a cis-acting [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=17456565 selenocysteine redefinition element (SRE)] is also required for any efficiency. This small stem-loop immediately downstream has been [http://www.ncbi.nlm.nih.gov/pubmed/15791204 specifically studied in SEPN1] exon 9 where it was determined to be a phylogenetically conserved stem–loop structure that starts 6 bp downstream from the UGA codon of exon 9 of SEPN1. It provides measurable but low levels of read-through via both selenocysteine and cysteine tRNAs. No such SRE can be detected in the AluJB-derived exon region. | |||
Although SRE elements are [http://www.ncbi.nlm.nih.gov/pubmed/16628248 successfully predicted] by genomewide bioinformatic EvoFold searches for conserved RNA folds, no such element is anticipated at the putative novel AluJb site. Selenocysteine insertion is inefficient to begin with. A second site lacking an SRE would result in multiplicative inefficiency, resulting in hardly any production of full length protein even if transcripts escaped the nonsense-mediated decay machinery. Since only a fraction of the transcripts encode the AluJb exons, it is implausible that significant in vivo levels of alternatively translated protein would occur. | |||
offset 0 1 2 3 | |||
hg18.chr1 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC | |||
monDom4 GGGCGGACTCTCCGGGAAACAGTCCTGGAAAGTTCGCCC | |||
echTel1 -GGCGGACTCTCCGGGAGACAGTCCTGGAGAGCCCG-CC | |||
loxAfr1 GGGCGGACTCTCCGGGAGACTGTCCTGGAGAGTTCGCCC | |||
dasNov1 GGGCGGACTCTCCGGGAGACCGTCCTGGAAAGTTCGCCC | |||
canFam2 GGGCGGACTCTCCGGGAGACCGTCCTGGAAAGTTCGCCC | |||
bosTau2 GGGCGGACTCTCCGGGAGACCGTCCTGGAAAGTTCGCCC | |||
oryCun1 GGGCGAACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC | |||
mm8 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGCCCGCCC | |||
rn4 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGCCCGCCC | |||
rheMac2 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC | |||
panTro1 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC | |||
galGal2 GGGCGAACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC | |||
xenTro1 GGGCGAACTCTCCGGGAGACCGTCCTAGAGAGTTTGCCC | |||
danRer3 GGGCGGACTCTTCGGGAGACAGTTCTCGAGAGTTCGCCC | |||
tetNig1 GGGCGGACTCTCCGGGAGACGGTGCGGGAGAGTTCGCCC | |||
fr1 GGGCGAACTCTCCGGGAGACAGTCCTGGAAAGTTCGCCC | |||
SS anno (((((((((.(((((((.....))))))).))))))))) | |||
pair symbol abcdefghi jklmnop ponmlkj ihgfedcba | |||
score 999999999799999989999989999997999999999 | |||
The UGA codon of exon 9 of SEPN1 is remarkable for occuring at the very end of the exon (human ...DDQSCu 12 GSGRTLRETVLESSP... denotes a complete TGA codon followed by 1 bp of the next codon which is completed to glycine by the first 2 bp of the next exon). The SRE, here a 39 bp stemloop occurs just after the splice acceptor and initial 2 residues of the next exon. The stemloop nucleotides thus selectively compete with whatever role the encoded 13 amino acids have, though there is opportunity to evolve independently at 11 third codon positions. | |||
2 | |||
2 | |||
[[Image:SEPN1 evoFold.jpg]] | |||
The fold of SEPN1 is unclear. It has no known Pfam domain other than an [http://www.expasy.org/uniprot/SELN_HUMAN EF hand at position 67-102], no paralogs within vertebrate genomes and no resemblance to any previously determined 3D structure by Blastp against PDB. A thioredin domain cannot be detected in the expected SCUG region by SuperFamily, SMART, or Pfam search. | |||
The primary sequence is moderately well conserved and full-length orthologs (all selenocysteine encoding in exon 9) are readily curated back to lamprey. The intron pattern is completely invariant and never contains supplemental exons in any lineage. The selenocysteine is evidently critical because no species has substituted cysteine, though homologs outside of deuterostomes are quite difficult to locate and do not encompass fungi or bacteria. The earliest diverging species, sponge, does not have muscle cells. None of the species are good model organisms; the gene appears to have been lost in drosophila and nematode. | |||
Nematostella vectensis (cnidaria) TGA-confirmed selenocysteine and phase 1 of exon 9 | |||
homSap VSYLPFTEAFDRAKAENKLVHSILLWGALDDQSCCGSGSGRTLRETVLESSPILTLLNESFISTWSLVKELE | |||
V YLPF EAF+RAKAE KLVH I+LWGALDDQSC G+GSGRTLRE LES P+L LL +++++ WSLV+EL+ | |||
nemVec VDYLPFKEAFNRAKAEKKLVHHIVLWGALDDQSC*GNGSGRTLREGPLESRPVLQLLQDNYVNCWSLVEELK | |||
Aplysia californica (mollusc) TGA-confirmed selenocysteine | |||
homSap QFVFEEIKWQQELSWEEAARRLEVAMYPFKKVSYLPFTEAFDRAKAENKLVHSILLWGALDDQSCCGSGRTLRETVLESSPILTLLNESFISTWSLVKELE | |||
+F +++I W+QE+S +EA LE+ ++PFK+V Y +EAF A+AE+KLVH I+LWGALDDQSC GS RTLRE+ L+ ++ LL++ F+S+W+L+ +L+ | |||
aplCal EFSWDDIAWEQEISEQEARDALELKLFPFKQVNYHNLSEAFTVAQAESKLVHCIVLWGALDDQSC*GSARTLRESALQGPKVMALLHQRFVSSWTLLVDLK | |||
Ixodes scapularis (tick) TGA-confirmed selenocysteine | |||
aplCal QVNYHNLSEAFTVAQAESKLVHCIVLWGALDDQSCXGSARTLRESALQGPKVMALLHQRFVSSWTLLVDLK | |||
QV YHNL+ A +A+ E K+VH +VLWG LDDQSC GS RTLR L PKV LL + F+S W++ LK | |||
ixoSca QVPYHNLTTAAALARQERKMVHSVVLWGNLDDQSC*GSGRTLRRGPLASPKVQELLLKHFISMWSITAQLK | |||
Oopsacas minuta (sponge) TGA-confirmed selenocysteine | |||
homSap FEEIKWQQELSWEEAARRLEVAMYPFKKVSYLPFTEAFDRAKAENKLVHSILLWGALDDQSCCGSGRTLRETVLESSPILTLLNESFISTWSLVKELEELQNN | |||
FE W E + EEA L+ +YPFK++ Y+ +A D A+ ENKLVHSI++WG+LDDQSC GSGRTLR+ L S + L E+FIS+WSL+ LE+L N | |||
oopMin FEIPAWDHEKTKEEATIELDRVLYPFKRIPYIDIRDAVDTARVENKLVHSIIMWGSLDDQSC*GSGRTLRDGPLASDVVQLTLKENFISSWSLIVHLEDLMAN | |||
[[Image:SEPN1 features.jpg]] | |||
Because the region containing the alternatively spliced exon is strongly conserved over many billions of years of vertebrate branch length, it is exceedingly likely to have a precisely defined stable functional fold. Because intron position almost never corresponds to natural domain boundaries (even in proteins with multiply interated domains such as actinin), introducing 32 gratuitous residues via a new exon is very likely to be structurally disruptive. Here the final two residues of the calcium responsive EF hand are disrupted. | |||
High conservation of exons 2-3 flanking the putative new exon in 22 species of vertebrate: | |||
SEPN1_homSap ELALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT GSTPAASCEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG | |||
SEPN1_panTro ELALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT GSTPVASYEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG | |||
SEPN1_ponAbe ELALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT GSTPAASYEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG | |||
SEPN1_calJac ESALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT GSTPEASYEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG | |||
SEPN1_micMur ESALKALGTEGLFLFSSLDTDQDMYISPEEFKPIAEKLT GSTPAASYEDEGLPADPSEETLTIEARFQPLLLETMTKSKDGFLG | |||
SEPN1_tupBel EAARKVLGTDGLFLFSSLDTDRDMYISPEEFRPIAEKLT GSTPAASYEE--PAPDPSEETLTIEARFQPLLPETMTKSKDGFLG | |||
SEPN1_musMus ESALKVLGTDGLFLFSSLDTDQDMYISPEEFKPIAEKLT GSVPVANYEEEELPHDPSEETLTIEARFQPLLMETMTKSKDGFLG | |||
SEPN1_ratNor ESALKVLGTDGLFLFSSLDTDQDSYISPEEFKPIAEKLT GSVPVASYEEEELPHDPSEETLTIEARFQPLLTETMTKSKDGFLG | |||
SEPN1_cavPor ESALKALGTEGLFLFSSLDTDQDMRLSPEEFKPIAEKLT GSTLTASYEEEELPLNPSEETLTIEARFQPLLPETMTKSKDGFLG | |||
SEPN1_ochPri ETVRKTLGTKSFFLFSSLDTDQDLYISPEEFKPIAEKLT GSAAAPSYEEEELTADPSEETLTIEARFQPLRPETMTKSKDGFLG | |||
SEPN1_turTru DSALKALGTEGLFLFSSLDTDGDMYLSPEEFKPIAEKLT GSTPSASYEEEDLPPDPSEETLTIEARFQPLLPESMTKSKDGFLG | |||
SEPN1_bosTau DSALKTLGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT GSTPTANYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG | |||
SEPN1_equCab ESALKALGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT GSTPAASYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG | |||
SEPN1_canFam ESALKALGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT GSTPAASYEEEELPLDPSEETLTIEARFQPLIPESMTKSKDGFLG | |||
SEPN1_myoLuc ESAMKALGTEGVFLFSSLDTDRDLYLSPEEFGPLAEKLT GSTPTTSYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG | |||
SEPN1_eriEur KSARKVLGTEGLFLFSSLDTDQDMSINPEEFRPIVEKLI GSTPKPSYEEEELSSDPREEKLTIETRFQPLLLNSMTKSNSRFPG | |||
SEPN1_loxAfr --ALKALGTEGLFLFSSLDTDQDMYISPEEFKPIAEKLT GSTPATSYEDEDLPPDPSEETLTIEARFQPLLPETMTKSKDGFLG | |||
SEPN1_proCap ESALKALGTEGLFLFSSLDTDQDMYISPEEFKPIAAKLT GSTPATSYEDEELPPDPSEETLTIEARFQPLIPETMTKSKDGFLG | |||
SEPN1_galGal ELALKSLGSEGLFLFSSLDTNNDLYLSPEEFKPIAEKLT GVTPVSDFEEDA--PDPNGETLSIVAKFQPLVMETMTKSKDGFLG | |||
SEPN1_xenTro EAALRTLGAEGLFLFSSLDTDNDMHISPEEFKPISEKLT GISTTSDYEEEE-LLDPNGETLSVASRFQPLLMETMTKSKDGFLG | |||
SEPN1_danRer EAGLKALGADGLFFFSSLDTDHDLYLSPEEFKPIAEKLT GVAPPPEYEEEI-PHDPNGETLTLHAKMQPLLLESMTKSKDGFLG | |||
SEPN1_petMar EMALRTLGNDGLFLFTSLDTNMDMQISPEEFRPIVDKII GPPPSEYE--GTQEADPQGEGLTMLARFEPLLMETMSKSRDGFLG | |||
While the function of SEPN1 (an endoplasmic reticulum glycoprotein with early developmental expression) is still obscure, it evidently involves a redox role for the selenocysteine (which occurs [http://www.pnas.org/content/97/11/5854.full thioredoxin-like] immediately adjacent to an invariant cysteine as SCUG rather than as UxC or UxxC). Since the redox function is already met, what purpose is served by introducing a gratuitous additional selenocysteine at a site far removed from the active site or evolved interaction sites with other enzymes? Although the new site is of the UxxC form common in other functional selenoproteins, this is likely an accidental outcome of ancestral AluJb translation (rather than subsequently evolved) because it is present in all descendent clades. | |||
Finally it's worth noting that SEPN1 is a much-studied disease gene,[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=606210 rigid spine with muscular dystrophy type 1]. None of the known disease alleles (notably M1V, G273E, G315S, R466Q, H293R, N340I, U462G, U462x, W453S, R466Q) map to the region of the putative new exon. While two separate disease alleles modify the selenocysteine codon in exon 9 none effect the putative new selenocysteine in exon 3. | |||
No physical evidence has ever been put forward for the existence of the extra exon in stable protein -- it is all based on genomic inference and transcripts. There is however [http://hmg.oxfordjournals.org/cgi/content/full/12/9/1045 solid experimental evidence] for the short form of the protein. The long form cannot even be detected under transfection conditions in which it should be greatly over-expressed. | |||
In summary, the evidence clearly refutes folklore of AluJb co-optively recruited as a coding exon of an alternatively spliced protein. Instead of representing a primate innovation, the AluJb instead creates a subclinical disease state, partially knocking out an essential gene by causing a fraction of its expression to be mis-spliced to a deviant untranslatable transcript. The GenBank entry NM_020451 -- which cites but fails to read the key 2003 paper -- is incompetently annotated: no 590 residue protein with two selenocysteines exists. This error has propagated irrevocably into hundreds of bioinformatic databases as mis-numbering of disease alleles and related problems. | |||
== Selenoprotein sequences available from genome project species == | |||
...... DIO1 DIO2 DIO3 GPX1 GPX2 GPX3 GPX4 GPX5 GPX6 GPX7 MSRB1 MSRB2 MSRB3 SELH SELI SELK SELM1 SELM2 SELO SELS SELT SELU1 SELU2 SELU3 SELV SEPHS1 SEPHS2 SEPN SEPP1 SEPP2 SEPW TXNRD1 TXNRD2 TXNRD3 | |||
homSap DIO1 DIO2 DIO3 GPX1 GPX2 GPX3 GPX4 GPX5 GPX6 GPX7 MSRB1 MSRB2 MSRB3 SELH SELI SELK SELM1 SELM2 SELO SELS SELT SELU1 SELU2 SELU3 SELV SEPHS1 SEPHS2 .... SEPP1 ..... SEPW TXNRD1 TXNRD2 TXNRD3 | |||
panTro DIO1 .... .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... SELS .... ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... | |||
ponPyg DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
macMul DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... SELK ..... ..... .... SELS SELT ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... | |||
calJac DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
tarSyr .... .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
otoGar DIO1 DIO2 .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... SELS SELT ..... ..... ..... SELV ...... ...... .... ..... ..... .... ...... ...... ...... | |||
micMur DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
tupBel DIO1 DIO2 .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
musMus DIO1 .... DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 MSRB2 MSRB3 SELH .... SELK ..... SELM2 SELO SELS SELT ..... ..... ..... SELV SEPHS1 SEPHS2 .... SEPP1 ..... SEPW ...... ...... ...... | |||
ratNor DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... SELK ..... SELM2 SELO SELS SELT ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... | |||
cavPor DIO1 .... DIO3 .... GPX2 .... .... .... .... .... MSRB1 ..... ..... .... .... SELK ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
speTri DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
dipOrd DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
oryCun DIO1 .... .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
ochPri DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
canFam DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 MSRB2 MSRB3 SELH .... SELK ..... SELM2 .... SELS SELT ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... | |||
felCat DIO1 .... DIO3 GPX1 GPX2 .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
equCab DIO1 DIO2 .... GPX1 GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... SEPP1 ..... .... ...... ...... ...... | |||
myoLuc DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... SELS .... ..... ..... ..... SELV ...... ...... .... ..... ..... .... ...... ...... ...... | |||
pteVam DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... .... ...... ...... ...... | |||
bosTau DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 MSRB2 ..... SELH .... SELK ..... SELM2 .... SELS SELT ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... | |||
oviAri .... .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
susScr DIO1 DIO2 DIO3 .... .... .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... SELM2 .... SELS .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
turTru DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
vicVic DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
eriEur DIO1 .... .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... SELS .... ..... ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
sorAra .... DIO2 .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... | |||
dasNov .... .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
choHof DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... SELH .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... | |||
loxAfr DIO1 .... .... .... GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... SELV SEPHS1 SEPHS2 .... ..... ..... .... ...... ...... ...... | |||
proCap DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... .... ...... ...... ...... | |||
echTel DIO1 DIO2 DIO3 .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
monDom DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... .... ..... ..... .... SELS SELT SELU1 ..... ..... .... SEPHS1 SEPHS2 .... ..... SEPP2 SEPW ...... ...... ...... | |||
macEug .... .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... SELU1 ..... ..... .... ...... SEPHS2 .... ..... SEPP2 .... ...... ...... ...... | |||
ornAna DIO1 DIO2 DIO3 GPX1 GPX2 .... GPX4 .... .... GPX7 MSRB1 MSRB2 MSRB3 SELH SELI SELK SELM1 SELM2 SELO SELS SELT SELU1 ..... ..... .... SEPHS1 SEPHS2 .... SEPP1 SEPP2 SEPW ...... ...... ...... | |||
tacAcu .... .... .... GPX1 .... .... GPX4 GPX5 .... GPX7 ..... ..... ..... SELH .... SELK SELM1 ..... SELO SELS .... SELU1 ..... ..... .... SEPHS1 SEPHS2 .... SEPP1 ..... .... ...... ...... ...... | |||
galGal .... DIO2 DIO3 GPX1 GPX2 GPX3 .... .... .... .... MSRB1 ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... | |||
taeGut .... .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... .... .... SELU1 ..... ..... .... ...... ...... .... ..... SEPP2 .... ...... ...... ...... | |||
anoCar DIO1 DIO2 .... GPX1 .... .... .... .... .... .... MSRB1 ..... ..... .... .... SELK ..... ..... .... .... .... SELU1 ..... ..... .... ...... ...... .... ..... SEPP2 SEPW ...... ...... ...... | |||
xenTro DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 MSRB2 ..... .... .... SELK ..... SELM2 .... SELS .... ..... ..... ..... .... SEPHS1 SEPHS2 .... SEPP1 SEPP2 SEPW ...... ...... ...... | |||
danRer DIO1 DIO2 DIO3 GPX1 .... .... .... .... .... .... MSRB1 ..... ..... SELH .... .... ..... SELM2 .... SELS .... ..... ..... ..... .... SEPHS1 ...... .... SEPP1 SEPP2 SEPW ...... ...... ...... | |||
tetNig .... DIO2 DIO3 GPX1 .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... SEPP2 .... ...... ...... ...... | |||
takRub DIO1 DIO2 DIO3 GPX1 .... .... .... .... .... .... ..... ..... ..... SELH .... .... ..... ..... .... .... .... SELU1 ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... | |||
gasAcu .... DIO2 DIO3 GPX1 .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... SEPP2 .... ...... ...... ...... | |||
oryLap .... DIO2 DIO3 GPX1 .... .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... SEPP2 .... ...... ...... ...... | |||
calMil DIO1 DIO2 DIO3 .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... SELO SELS .... SELU1 ..... ..... .... SEPHS1 ...... .... ..... SEPP2 SEPW ...... ...... ...... | |||
petMar .... DIO2 .... .... GPX2 .... .... .... .... .... ..... ..... MSRB3 .... .... .... SELM1 ..... SELO SELS SELT ..... ..... SELU3 .... SEPHS1 ...... SEPN ..... ..... SEPW ...... ...... ...... | |||
braFlo .... .... DIO3 .... .... .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... .... .... SELU1 ..... ..... .... SEPHS1 ...... .... ..... ..... .... ...... ...... ...... | |||
== Reference set of 656 vertebrate selenoproteins == | |||
The sequences below reflect the full range of deuterostome species for which reliable full-length orthologs could be collected. The protein sequences display position and phase of each exon. The selenocysteine is displayed as lower case u for visibility. That residue is often displaced in tetrapods on with cysteine, typically as phyloSNP though sometimes with erratic persistence within mammals. | |||
> | === SELH: 25 vertebrate sequences === | ||
0 | |||
<pre> | |||
1 | >SELH_homSap Homo sapiens (human) NP_734467 Selenoprotein H exons chr11 80% identity musMus | ||
0 MAPRGRKRKAEAAVVAVAEKREKLANGGEGMEEATVVIEHC 2 | |||
1 TSuRVYGRNAAALSQALRLEAPELPVKVNPTKPRRGSFEVTLLRPDGS 1 | |||
2 SAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 | |||
> | >SELH_ponPyg Pongo pygmaeus (orang_sumatran) | ||
0 | 0 MAPRGRKCKAEATVVAVAEKrEKLTNGGEGMEEATIVIEHC 2 | ||
1 TSuRVYGRNAAALSQVLCLEAPELPVKVNPTKPRRGSFEVTLLRPDGS 1 | |||
1 | 2 SVELWTGIKKGPPCKLKFPEPQEVVEKLKKYLS* 0 | ||
> | >SELH_macMul Macaca mulatta (rhesus) | ||
0 | 0 MAPRGRKRKAEAAMVAAAEKQEKLANSGEGMEETTVVIEHC 2 | ||
1 TSuRVYGRNAAALSQALRLEAPELPVKVNPSKPRRGSFEVTLLRPDGS 1 | |||
1 | 2 SAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 | ||
> | >SELH_micMur Microcebus murinus (mouse_lemur) | ||
0 | 0 MAPRGRKRKAEASVVATAEKREKLENGGEAVEEATVVIEHC 2 | ||
1 TSuRVYGRNAAALSQALRLEAPELPVKVNPAKPRRGSFEVTLQRPDGS 1 | |||
1 | 2 SAELWTGIKKGPPRKLKFPEPQVVVKELKKYL.* 0 | ||
> | >SELH_tupBel Tupaia belangeri (tree_shrew) | ||
0 | 0 MAPRGRKRKAEAAVVATAEKQEKLQNGGEGVKEASIVIEHC 2 | ||
1 TSuRVYGRNAAALSQALRLEAPELPVKVNSAKPRRGSFEVTLLRPDGS 1 | |||
1 | 2 SVELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 | ||
> | >SELH_musMus Mus musculus (mouse) | ||
0 | 0 MAPHGRKRKAGAAPMETVDKREKLAEGATVVIEHC 2 | ||
1 TSuRVYGRHAAALSQALQLEAPELPVQVNPSKPRRGSFEVTLLRSDNS 1 | |||
1 | 2 RVELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 | ||
> | >SELH_ratNor Rattus norvegicus (rat) TGA verified 77% | ||
0 | 0 MAPLGRKRKAGAAPIESADKREKLAEGAAVVIEHC 2 | ||
1 TSuRVYRRHAAALSQALQLEAPEISVQVNRSKPRRGSFEVTLLRPDNS 1 | |||
1 | 2 RVELWTGIKKGPPRKLKFSEPQEMVEELKKYLS* 0 | ||
> | >SELH_speTri Spermophilus tridecemlineatus (squirrel) | ||
0 | 0 MAPRVRKRKAEAAAVSTSEKREKLENGKEQVEEAVVIEHc 2 | ||
1 TSuRVYGRNAAALSQALRLEAPELPVKVNPSKPRRGSFEVTLLRRDGTS 1 | |||
1 | |||
1 | |||
> | >SELH_oryCun Oryctolagus cuniculus (rabbit) | ||
0 | 0 MAPGKRKRKAEAAPVASAEKREKLANGGQGVEEIVIEHc 2 | ||
1 tSuRVYGRNAAALSQALRLQAPELPVTVNPSKPRRGSFEVTLLRPDGS 1 | |||
1 | 2 gAELWTGIKKGPPRKLKFPEPQQVVEELKKYLS* 0 | ||
> | >SELH_ochPri Ochotona princeps (pika) | ||
0 | 0 MAPNRRKRKAEAVADAAAEKREKQAKQANGVGGGEEIVIEHc 2 | ||
1 TSuRVYGRNAAALSQALRLEAPELPVKVNPAKPRRGSFEVTLQRPDGS 1 | |||
1 | 2 SAELWTGIKKGPPRKLKFPEPQQVVEELKKYLS* 0 | ||
2 | |||
> | >SELH_canFam Canis familiaris (dog) | ||
0 | 0 MASRGRKRKAEAAGVAAAEKRDKPASGRKAVEEATVVIEHC 2 | ||
1 TSuRVYGRNAAALSQALRLETPELPVEVNPAKPRRGSFEVTLLRPDGS 1 | |||
1 | 2 SVELWTGIKKGPPRKLKFPEPQEVVKALKQHLS* 0 | ||
> | >SELH_bosTau Bos taurus (cow) TGA verified | ||
0 | 0 MASRGRKRKAEAALAAAAEKREKPAGGQEGGVEGPSVVIEHC 2 | ||
1 TSuRVYGRNAAALSQALRLQAPELTVKVNPARPRRGSFEVTLLRADGS 1 | |||
1 | 2 SAELWTGLKKGPPRKLKFPEPHVVLEELKKYLS* 0 | ||
> | >SELH_oviAri Ovis aries (sheep) | ||
0 | 0 MASRGRKRKAEAALAAAAEKREKPAGSREGEVAGPSVVIEHC 2 | ||
1 TSuRVYGRNAAALSQALRLQAPELAVKVNPSRPRRGSFEVTLLRADGS 1 | |||
1 | 2 AELWTGLKKGPPRKLKFPEPHVVLEELKKYLS* 0 | ||
> | >SELH_susScr Sus scrofa (pig) | ||
0 | 0 MASRGRKRKAETALGAAAEKQETPASGRKGMEEPSVVIEHC 2 | ||
2 | 1 TSuRVYGRNAAALSQALRVEAPELPVRVNPTKPRRGSFEVTLMRPDGS 1 | ||
1 | 2 SAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 | ||
>SELH_equCab Equus caballus (horse) | |||
0 MASRGRKRKAEAVVAVAEKREKLTSGGKGVEEVTVVIEHc 2 | |||
1 TSuRVYGRNAAALSQALRLEAPELPVKVNPAKPRRGSFEVTLLRPDGS 1 | |||
2 sAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 | |||
> | >SELH_choHof Choloepus hoffmanni (sloth) | ||
0 | 0 MASRGRKRKAEAVVLAAAEKQEKVASGKEGEKEAVVVIEHC 2 | ||
1 TSuRVYGRNAAALSQALRLEAPEIPVKVNFAKPRRGSFEVTLLRPDGS 1 | |||
1 | 2 TELWTGIKKGPPRKLKFLQPLKVVEELKKYLS* 0 | ||
> | >SELH_loxAfr Loxodonta africana (elephant) | ||
0 | 0 MASRGRKRKAEAAVAAAAAEKREKPVGGQAAAEEVVVIIEHC 2 | ||
1 TSuRVYGRNAAALSQALRLEAPELSVKVNPSKPRRGSFEVTLQRPDGS 1 | |||
1 | 2 GAELWTGIKKGPPRKLKFPEPQEVVEELKKYL* 0 | ||
> | >SELH_monDom Monodelphis domestica (opossum) | ||
0 | 0 MAPRGRKRKADVAAAALTEKPEKLAQGGEEGAGEARVVIEHC 2 | ||
1 QSuRVYARHAEAVGQALRLARPGLPVLLNPAKPRRSSFEVTLLRPDGS 1 | |||
1 | 2 RVELWSGIKKGPPRKLKFPEPAQVVEELKARLV* 0 | ||
> | >SELH_ornAna Ornithorhynchus anatinus (platypus) E5DI3CH10F50JR | ||
0 | 0 DAGGAEVGEGLHVVIEHC 2 | ||
2 | 1 RSuGVYGRRAEALSRALSLAAPDLPVLLNPTKPRRNSFEVTLLRPDGT 1 | ||
1 | 2 RTELWSGIKKGRP | ||
>SELH_galGal Gallus gallus (chicken) tga confirmed | |||
0 MAPRGRKRAARRPAEPEARADPPEKRPRDEAEGSPGDAGGPRVVIEHC 2 | |||
1 RSuRVYGRNAAALSEALRGAVASLAVEINPRQPRRNSFEVSLVKEDGS 1 | |||
2 TVQLWSGIGKGPPRKLKFPEPAAVVEALRSSLA* 0 | |||
>SELH_danRer Danio rerio (zebrafish) AI877878 BM037625 CKSU tga verified exons fused | |||
0 MATRGKSARKRKADSDEKEKLDDAKKEKLEDKDEETGLRVVIEHC 2 | |||
> | 1 KSuRVYGRNADVVREALADSHPELKVMINPHKPRRNSFEITLMDGERADVLWSGIKKGPPRKLKFPEPAEVVTALKQALEKE* 0 | ||
0 | |||
1 | |||
> | >SELH_takRub Takifugu rubripes (fugu) tga confirmed, no Ests 3 exons | ||
0 | 0 MTPRVLMTGRRGTKRKAEEDEKPKEEKKEKQREDDQGGPRVAIEHC 2 | ||
1 KSuRVYGRNAEAVKSALLAAHPGLTVVLNPEKPRRNSFEITLLDEGK 1 | |||
2 ETSLWTGIKKGPPRKMKFPQPDVVVTALQEALKTE* 0 | |||
1 | |||
2 | |||
> | >SELH_ictPun Ictalurus punctatus (catfish) CB940790 tga verified | ||
0 | 0 MATRAKAGRGAKRKADVIAAAEPVAKQDKGNKGEREDDEGQRVIIEHC 2 | ||
1 KSuRVYGRNADAVREALLSAHPELHVVLNPEKPRRNSFEVTLIEGK 1 | |||
1 | 2 KELVLWTGLKKGPPRKLKFPEPAEVVTALEEALKSK* 0 | ||
0 | >SELH_salSal Salmo salar (salmon) CA043802 tga verified | ||
1 | 0 MASRNKAGRVLKRKASVKEESVEEKRGKGEDDQPEIVTEGRRVVIEHC 2 | ||
2 | 1 KSuRVYGRNAEGVRVALLAACPDLTVVLNPQKPRSKSFEVILVEGE 1 | ||
2 KEVCLWSGIKKGPPRKLKFPEPEVVVSALEKALKTE* 0 | |||
> | >SELH_oryLat Oryzias latipes (medaka) BJ026077 tga verified | ||
0 2 | 0 MASKAGRRGTKRKVEAKKEEDKTSTEEKKARGENAHEEAGLKVLIEHC 2 | ||
1 | 1 KSuRVYGRNAEEVKSALLAARPELTVVCNPEKPRRNSFEITLLDGAK 1 | ||
2 ETSLWTGIKKGPPRKLKFPQPDDVVAAFKDALKTE* 0 | |||
> | >SELH_oncMyk Oncorhynchus mykiss (trout) BX312781 | ||
0 | 0 MASLTKAGRVLKRKVETEESSVEGKRGKGEDDHPEIVTEGQRVVIEHC 2 | ||
1 | 1 KSuRVYGRNAEGVRVALLAACPDLTVVLNPQKPRSKSFEVILFEGE 1 | ||
2 KEVCLWSGIKKGPPRKLKFPEPEVVVSALEKALKTE* 0 | |||
</pre> | |||
> | === SELL: 10 pre-tetrapod sequences === | ||
0 | <pre> | ||
>SELL_danRer Danio rerio (zebrafish) 9 exons note illegal gene name, serious genome misassembly | |||
1 | 0 MAEVDESTLVGALKELTIFGRTSLEYVEKDTAN 1 | ||
1 | 2 GCTLQEVNGKVLPLLGLMSAGAKFCNS 2 | ||
0 | 1 LGVKNQKEAESLWERFFQ 2 | ||
1 | 1 NTEVRESVEGLLQLE 0 | ||
0 VEWDRFLKRLDVQMQMSDTVLSQHPAAQTLSSDMRFLHVQTKE 2 | |||
1 NVSLGQYLGKGENLLLVLLRHFGuLPuRDHLTELKNSK 0 | |||
0 MLLEAQSLRVLVVSYGSLEGATFWLEQTGFEFDMLLDTERT 0 | |||
0 | 0 VYKMFGLGSSMSKVMKFKLMFHYSEIMAMNRTLPEMPPQFIEDLFQ 0 | ||
0 MGGDFVLEQDGKVIFSYRCKSPVDRPSATQILATVSAHYAH* 0 | |||
NVSLGQYLGKGENLLLVLLRHFGuLPuRDHLTELKNSK Danio rerio | |||
NVSLGQYLGKGENLLLVLLRHFGcLPcRDHLTELKNSK Tetraodon nigroviridis | |||
SVTLGEFLGQNQKLLLVLIRHFGuLPuRDHVAELEASE Oryzias latipes | |||
SVTLGRYLGQGQKPLLVLIRHFGuLPuRDHVAELEANQ Gasterosteus aculeatus | |||
SVSLGRYFGKGENLLLVLIRHLGuLPuRDHLAQMETNQ Osmerus mordax | |||
NVTLGLYFGKGENLLLVLIRHFGuLPuREHVAELATQQ Salmo salar | |||
NVTLGMYFGKGENLLLVLIRHFGuLPuREHVAELATQQ Oncorhynchus kisutch | |||
EVLLEEFLQRQEKLLLVLLRHFAuLPuRDHLAQLQKNQ Squalus acanthias | |||
EIQLEEFLHRREKLLLVLLRHFAuLPuRDHLVQLQAQQ Leucoraja erinacea | |||
EVLLNNYLGKEYLVLLVMLRHLAuVPuREHVAQLQAHQ Eptatretus burgeri | |||
FDGNLLFSSNHRNCLVILLRHFAuLPuRKHVTEIQEKQ Ciona intestinalis | |||
AVEL-RSLWREHACVVAGLRRFGCVVCRWIAQDLSSLA Homo sapiens C1orf93 | |||
RVPF-GALFRERRAVVVFVRHFLCYICKEYVEDLAKIP Homo sapiens C9orf21 | |||
TFKA-KELWEKNGAVIMAVRRPGCFLCREEAADLSSLK Homo sapiens C10orf5 | |||
</pre> | </pre> | ||
=== SELI: 10 deuterostome sequences === | |||
=== | |||
<pre> | <pre> | ||
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_homSap Homo sapiens (human) | |||
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_ponPyg Pongo pygmaeus (orang_sumatran) | |||
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_macMul Macaca mulatta (rhesus) | |||
VKQLSSHFQIYPFSLKKPNSDuLGMEEKNIGL* SELI_otoGar Otolemur garnettii (bushbaby) | |||
VKQLSSHFQIYPFSLRKPNSDuLGVEEKSIGL* SELI_tupBel Tupaia belangeri (tree_shrew) | |||
VKQLSRHFQIYPFSLRKPNSDuLGMEEQNIGL* SELI_musMus Mus musculus (mouse) | |||
VKQLSRHFQIYPFSLRKPNSDuLGLEEKSIGL* SELI_ratNor Rattus norvegicus (rat) | |||
VKQLSSHFQIYPFSLRKPNSDuLGTEEKNIGL* SELI_cavPor Cavia porcellus (guinea_pig) | |||
VKQLSSHFQIYPFSLKKPNSDuLGMEEKNIGL* SELI_speTri Spermophilus tridecemlineatus (squirrel) | |||
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIAL* SELI_oryCun Oryctolagus cuniculus (rabbit) | |||
VKQLSSHFQIYPFSLRKPNSDuLGMEAKNIVV* SELI_ochPri Ochotona princeps (pika) | |||
VKQLSNHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_canFam Canis familiaris (dog) | |||
VKQLSNHFQIYPFSLRKPNSDuLGTEEKSIGL* SELI_felCat Felis catus (cat) | |||
VKQLSNHFHIYPFSLRKPNSDuLGVEEKNIGL* SELI_bosTau Bos taurus (cow) | |||
VKQLSSHFQIYPFSLRKPNSDuLGM.......* SELI_susScr Sus scrofa (pig) | |||
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_equCab Equus caballus (horse) | |||
VKQLSSHFHIYPFSLRKPNSDuLGAEE-ITGL* SELI_eriEur Erinaceus europaeus (hedgehog) | |||
VNQLSNHFQIYPFSLKKPNSDuLEMEEKNIGL* SELI_sorAra Sorex araneus (shrew) | |||
VKQLSSHFQIYPFSLKKPNSDuLGVEEKKIGL* SELI_dasNov Dasypus novemcinctus (armadillo) | |||
VKQLSSHFQIYPFSLRKPNSDuLGMEVYKIRQ* SELI_loxAfr Loxodonta africana (elephant) | |||
VYQLSNHFKILPFSLKKPSSDuLEVEEEKIGLQSTEVL* SELI_monDom Monodelphis domestica (opossum) | |||
VSQLSRHFNIRPFSLKKPTPDuLGMEEEKISLRSAEVL* SELI_galGal Gallus gallus (chicken) | |||
VSQLSQHFHIRTFSLRKPSSDuLGVEEAMAAGAEEKVALRSAEEL* SELI_anoCar Anolis carolinensis (lizard) | |||
VNQLSKHFNILPFSLRKPSTDuL--EEEKIGLQSAQVL* SELI_xenTro Xenopus laevis (frog) | |||
VKQLSDHFNIFAFSLKKPNSDuQ--DEEKIGLKAAEV* SELI_danRer Danio rerio (zebrafish) | |||
VQQLSDHFNIKAFSLKKPSADuQ--EEERIGLTEAEV* SELI_tetNig Tetraodon nigroviridis (pufferfish) | |||
VQQLSNHFSILAFSLKKPNSDuQ--EEDQIGLKAAEV* SELI_gasAcu Gasterosteus aculeatus (stickleback) | |||
VQQLSKHFNIFAFSLKKPSSDuQ--EEERIGLTGAEV* SELI_oryLap Oryzias latipes (medaka) | |||
VNQLSKHFNICAFSLKKPSTEuLEEEQKICLQASEVL* SELI_leuEri Leucoraja erinacea (skate) | |||
VIQMADHFNINIFTLKKQ--DuPKVQTTKYLLYVCGVMSWQC... SELI_braFlo Branchiostoma floridae (amphioxus) | |||
VNKLAAHFGIKVFSIKENS-DuHAAPSVAANAAVHAGV* SELI_cioInt Ciona intestinalis (tunicate) | |||
> | >SELI_homSap Homo sapiens (human) chr2 | ||
0 | 0 MAGYEYVSPEQLAGFDKYK 0 | ||
2 | 0 YSAVDTNPLSLYVMHPFWNTIVK 0 | ||
2 | 0 VFPTWLAPNLITFSGFLLVVFNFLLMAYFDPDFYAS 1 | ||
0 | 2 APGHKHVPDWVWIVVGILNFVAYTL 1 | ||
0 | 2 DGVDGKQARRTNSSTPLGELFDHGLDSWSCVYFVVTVYSIFGRGSTGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0 | ||
0 TISFVYIVTAVVGVEAWYEPFLFNFLYRDLFTAMII 1 | |||
2 GCALCVTLPMSLLNFFR 2 | |||
1 SYKNNTLKLNSVYEAMVPLFSPCLLFILSTAWILWSPSDILELHPRVFYFMVGTAFANST 0 | |||
0 CQLIVCQMSSTRCPTLNWLLVPLFLVVLVVNLGVASYVESILLYTLTTAFTLAHIHYGVRV 0 | |||
0 VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* 0 | |||
> | >SELI_macMul Macaca mulatta (rhesus) | ||
0 | 0 MAGYEYVSPEQLAGFDKYK 0 | ||
2 | 0 YSAVDTNPLSLYVMHPFWNTIVK 0 | ||
2 | 0 VFPTWLAPNLITFSGFLLVVFNFLLMAYFDPDFYAS 1 | ||
0 | 2 APGHKHVPDWVWIVVGILNFVAYTL 1 | ||
0 | 2 DGVDGKQARRTNSSTPLGELFDHGLDSWSCVYFVVTVYSIFGRGSTGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0 | ||
0 TISFVYIVTAVVGVEAWYEPFLFNFLYRDLFTSMII 1 | |||
2 GCALCVTLPMSLLNFFR 2 | |||
1 SYKNNTLKHNSVYEAVVPLFSPCLLFILSTAWILWSPSDILELHPRVFYFMVGTAFANST 0 | |||
0 CQLIVCQMSSTRCPTLNWLLVPLFLVVLVVNLGVASYVESILLYTLTTAFTLAHIHYGVRV 0 | |||
0 VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* 0 | |||
> | >SELI_bosTau Bos taurus (cow) NM_001075257 | ||
0 | 0 MASYEYVSPEQLAGFDKYK 0 | ||
2 | 0 YSAVDTNPLSLYVMHPFWNTVVK 0 | ||
2 | 0 VFPTWLAPNLITFSGFLLVVFNFLLLAYFDPDFYAS 1 | ||
0 | 2 APGHKHVPDWVWIVVGILNFMAYTL 1 | ||
0 | 2 DGVDGKQARRTNSSTPLGELFDHGLDSWSCVYFVVTVYSIFGRGSSGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0 | ||
0 TISFVYIVTAVVGVEAWYEPFLFNFLYRDLFTAMIF 1 | |||
2 GCALCVTLPMSLLNFFR 2 | |||
1 SYKNNTLKHNSVYEAVVPFFSPCLLFILSTAWILLSPSDILELHPRVFYLMVGTAFANST 0 | |||
0 CQLIVCQMSSTRCPTLNWLLLPLFVIVMVVNVGVTPYVESILLFTLTAAFTLAHIHYGVRV 0 | |||
0 VKQLSNHFHIYPFSLRKPNSDuLGVEEKNIGL* 0 | |||
> | >SELI_monDom Monodelphis domestica (opossum) | ||
0 | 0 MAGAIENRSMGIRDDL 0 | ||
2 | 0 YRAVDTNPLSLYIMHPFWNAIVK 0 | ||
2 | 0 IFPTWLAPNLITLSGFLLLIFNFLLMAYFDPDFFAS 1 | ||
0 | 2 APGGKHVPDWVWILVGILNFVAYTL 1 | ||
0 | 2 DGVDGKQARRTNSSTPLGELFDHGLDSWSCVYFVVTVYSIFGRGSTGVSVVVLYILLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0 | ||
0 TISLVYIVTAIVGVEAWYEPFLFNFLYRDLFTAMII 1 | |||
2 GCASCVTLPMSLYNFFK 2 | |||
1 AYKNNSLKHSSVYETMLPFISPCLLFILSTTWVLWSPSDILETHPRLFYYMVGTAFANIT 0 | |||
0 CRLIVCQMSNTRCQTLNLLLIPLALVVLLVRLELPLHVEPILLYLLSIIITLAHIHYGIQV 0 | |||
0 VYQLSNHFKILPFSLKKPSSDuLEVEEEKIGLQSTEVL* 0 | |||
> | >SELI_ornAna Ornithorhynchus anatinus (platypus) 53% LOC100086265 | ||
0 | 0 MAGYEYVSAEQLAGFDKYK 0 | ||
2 | 0 YSALDTNPLSLYVMHPFWNTIVKV 0 | ||
2 | 0 IFPTWLAPNLITFSGFLLLVFNFLLMAYFDPDFYAS 1 | ||
0 | 2 APGQKHVPDWVWIVVGILNFTAYTL 1 | ||
0 | 2 DGVDGKQARRTNSSTPLGELFDHGLDSWACVYFVVTVYSIFGRGRTGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDMSQV 0 | ||
0 TISIVYIVTAVVGVEAWYEPFLFNFLYRDLFTTMII 1 | |||
2 GCAVTVTLPMSLYNFYR 2 | |||
1 NKTLKYSSVYETMLPFVSPCLLFTLSTTWIFLSPSNILETHPRLFYFMVGTLFANIT 0 | |||
0 CQLIVCQMSNTRCQPLNWLLMPLALVILVVHSGLAPHSETFLLYSLTALVTVAHIHYGIRV 0 | |||
0 VNQLSKHFKILPFSLRKPSSDuLGLEEEKIGL* 0 | |||
> | >SELI_galGal Gallus gallus (chicken) | ||
0 | 0 MEYVTAEQLAGFSKYK 0 | ||
2 | 0 YSAVDSNPLSLYVMHPFWNTIVK 0 | ||
2 | 0 IFPTWLAPNLITFSGFLLLVFNFFLMAYFDPDFYAS 1 | ||
0 | 2 APDHQHVPNGVWVVVGLLNFIAYTL 1 | ||
0 | 2 DGVDGKQARRTNSSTPLGELFDHGLDSWACVYFVVTVYSTFGRGSTGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0 | ||
0 TISIVYIVTAIVGVEAWYAPFLFNFLYRDLFTTMII 1 | |||
2 ACALTVTLPMSLYNFYK 2 | |||
1 AYKNNTLKHHSVYEIMLPLVSPVLLFALCTTWIFVSPMDILEVHPRLFYFMVGTAFANIS 0 | |||
0 CQLIVCQMSSTRCQPLNWMLLPIALVLFMVMSGFAPSSETLLLYLLTAFLTLAHIHYGVVV 0 | |||
0 VSQLSRHFNIRPFSLKKPTPDuLGMEEEKISLRSAEVL* 0 | |||
> | >SELI_xenTro Xenopus tropicalis (frog) | ||
0 | 0 MAWEYVSPEQLAGFDKYK 0 | ||
2 | 0 YSAVDTNPLSLYVMHPFWNSIVR 0 | ||
2 | 0 FFPTWLAPNLITFSGFLLLVFTFLLMAFFDPDFYAS 1 | ||
0 | 2 APGQEHVPNWVWIVAGLLNFTAYTL 1 | ||
0 | 2 DGVDGKQARRTNSSTPLGELFDHGLDSWACIFFVVTVYSIFGRGETGVSAVVLYFLLWVVLFSFILSHWEKYNTGILFLPWGYDLSQV 0 | ||
0 TISCVYLVTAVVGAETWYQPVVFNILYRDLFTTMIV 1 | |||
2 GCAVCVTLPMSLYNVFK 2 | |||
1 GYRNNTLKHTSLYEAMLPIISPVLLFILSTLWILASPSKILEHHPRLFYFMVGTTFANIT 0 | |||
0 CQLIVCQMSNTRCRPLSWLLIPLTLAVVVSLSTVTPRDETLALVLLTVLVTLAHIHYGVNV 0 | |||
0 VNQLSKHFNILPFSLRKPSTDuLEEEKIGLQSAQVL* 0 | |||
> | >SELI_danRer Danio rerio (zebrafish) | ||
0 | 0 MALYHYVTQEQLSGFDKYK 0 | ||
2 | 0 YSAVDSNPLSIYVMHPFWNSVVK 0 | ||
2 | 0 VLPTWLAPNLITFTGFMFLVLTFTMLSFFDFDFYAS 1 | ||
0 | 2 GEGHTHVPSWVWIAAGLFNFLAYTL 1 | ||
0 | 2 DGVDGKQARRTNSSTPLGELFDHGLDSWACVFFVATVYSVFGRGETGVSVVTLYYLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0 | ||
0 TISIVYIITAVVGVETWYKPVFLNIHYRDLFTVMII 1 | |||
2 GCLFVVTLPMSLYNVFK 2 | |||
1 AYRNNTLKHSSLYEALLPFFSPVLLFVLSTLWISFSPTNIVQKQPRIFYLMVGTAFSNVT 0 | |||
0 CKLIVCQMSNTRCKPLSLLLLPMSIVVLLVISGTVQGGESLVLFIWTAVVLLTHIHYGVSV 0 | |||
0 VKQLSDHFNIFAFSLKKPNSDuQDEEKIGLKAAEV* 0 | |||
> | >SELI_petMar Petromyzon marinus (lamprey) seleno exon not available | ||
0 | 0 MAAEHAYITAEQLAGFDKYK 0 | ||
2 | 0 YSAVDTNPLSVYILHPFWNFVVK 0 | ||
2 | 0 LFPKWVAPNLLTFSGFLLLVFNFFILAYYDWDFYAS 1 | ||
0 | 2 APGQVHVPGWVWILAGILSFLAYTL 1 | ||
0 | 2 GIDGKQARRTGSSTPLGELFDHGLDSWSCLFFGSTVYSIFGRGQTGVSLATMHMLLWVVLFSFVLSHWEKYNTGVLFLPWGYDISQV 0 | ||
0 TISIVYIVTAVVGVEAWHQDLPFGLLYRDLFLFMLL 1 | |||
2 GGAFLVTLPQSLFNLHK 2 | |||
1 AHKAGTLKKDSLWEAMLPMVSPTLLFVLTSAWVWLSPERVVDSQPRVFYWLVGTLFSNIT 0 | |||
0 CKLIVCQMSSTRCQPLNWLLAPLACAVAFVATFQPKVEVETAVLFGMTALVTAAHIHYGVCVV 0 | |||
0 * 0 | |||
> | >SELI_braFlo Branchiostoma floridae (amphioxus) frag | ||
0 | 0 RLPRWLAPNLMTFVGFLLLIFNFALLTWYDPQFRAS 12 SSQHPDDPLVPNWVWLVCGFAHFLSHTL 1 | ||
2 | 2 DGVDGKQARRTNSSTPLGELFDHGLDSWATMFFPTAIYSMFGRGTEYGITVTQMYMVVWVVMIGFIMSHWEKYNTGTLFLPWGYDMSQV 0 | ||
2 | 0 CMLVMYIAAFFCGQDLWRTHLPMGLRFHYLFLFSFY 1 | ||
0 | 2 GGFLFISLPHTIGNLYR 2 | ||
0 | 1 STSEGTGRNLSLYEGMLPLLSPVLLFLLCVAWLKMSPTDILEQQPRLFYFMSGTVFANIA 0 | ||
0 CRLIIAQMSNTRCEGVNALLWPLMLIVAGVCAVPLGQWELVLLIGYTVMATLLHIHFGVCV 0 | |||
0 VIQMADHFNINIFTLKKQDuPKVQTTKYLLYVCGVMSWQC | |||
> | >SELI_cioInt Ciona intestinalis (tunicate) 1 exon fusion, 1 split | ||
0 | 0 MYITEQELSGFDTYK 0 | ||
2 | 0 YSCKDTSPLSNYVMHPFWNEAVK 0 | ||
2 | 0 LFPRWLAPNVMTFGGFVLLILQYILLWYFDPTYHAS 12 TTDIAYPSIPTWVWWFSLFAQFFSHTL 1 | ||
0 | 2 DGCDGKQARRTGTSSPLGELFDHGIDSWCVSLFTLNVLSVFGRDLAP 1 | ||
0 | 2 VSLMYSVQCMTLMAFLLSHWEKYNTGILFLPWAYDLSQV 0 | ||
0 LMAIVYLITAVSGVGVWRGTLLGYKVTTWFKGVLYV 1 | |||
2 ATFGFAVPMCLYNIYI 2 | |||
1 ANKNGTGKNLTFYEGSLPLLSPLLSFTILTTWIYISPSNVLSTNPRIFMLIISVLFSNIL 0 | |||
0 CRLIVSQMSNTRCQVFNKFVFFIGAGFACTLYFNNPVIENYILLALAIGLTVAHIHYGITV 0 | |||
0 VNKLAAHFGIKVFSIKQDSDuHAAPSVAANDAVHAGV* 0 | |||
</pre> | |||
> | === SELK: 32 vertebrate sequences === | ||
0 | <pre> | ||
2 | >SELK_homSap Homo sapiens (human) chr3 last coding bp and stop codon split off into 5th exon | ||
0 MVYISN 1 | |||
2 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 | ||
1 FKTLLQQDVKKRRSYGNSSDSRYDDGRG 2 | |||
2 | 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 | ||
1 * 0 | |||
2 | |||
> | >SELK_panTro Pan troglodytes (chimp) | ||
0 1 | 0 MVYISN 1 | ||
2 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 | ||
2 | 1 FKTLLQQDVKKRRSYGNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 | |||
1 * 0 | |||
> | >SELK_ponPyg Pongo pygmaeus (orang_sumatran) | ||
0 | 0 MVYISN 1 | ||
2 | 2 AQVLDSWSQSPWRLSLITDFFWGIAEFVVLF 2 | ||
2 | 1 FKTLLQQDLKKRRSYGNSSDFRYDDDGRG 2 | ||
1 PPGNPPGRMGPINHLGGPSPPPMAGGuGR 2 | |||
1* 0 | |||
> | >SELK_macMul Macaca mulatta (rhesus) | ||
0 | 0 MVYISN 1 | ||
2 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 | ||
2 | 1 FKTLLQQDVKKRRSYGNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 | |||
1 * 0 | |||
> | >SELK_otoGar Otolemur garnettii (bushbaby) | ||
0 | 0 MVYISN 1 | ||
2 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 | ||
2 | 1 FKTLLQQDVKKRRGYGNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRINHLQGPSPPPMAGGuGR 2 | |||
1* 0 | |||
> | >SELK_micMur Microcebus murinus (mouse_lemur) | ||
0 1 | 0 MSYISN 1 | ||
2 1 | 2 GQLLDSRSQSPWRLSLITDFFWRKPEFVVLF 2 | ||
2 | 1 FKTLLQQDVKKRRGCGNSSDSRYDDGRG 2 | ||
1 PPGNPFQRMGQINHLHGPSPPMAGGuGR 2 | |||
1* 0 | |||
> | >SELK_tupBel Tupaia belangeri (tree_shrew) | ||
0 | 0 MVYISN 1 | ||
2 | 2 GRVSDSWSQSPWRLSLITDFFWGMAKFVVLS 2 | ||
2 | 1 FKTLLQQDVRKRKGYGNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRISHLRGPSPPPVAGGuGR 2 | |||
1* 0 | |||
>SELK_musMus Mus musculus (mouse) | |||
0 MVYISN 1 | |||
2 GQVLDSRNQSPWRVSFLTDFFWGIAEFVVFF 2 | |||
1 FKTLLQQDVKKRRGYGSSSDSRYDDGRG 2 | |||
1 PPGNPPRRMGRISHLRGPSPPPMAGGuGR 2 | |||
1 * 0 | |||
>SELK_ratNor Rattus norvegicus (rat) | |||
0 MVYISN 1 | |||
> | 2 GQVLDSRNQSPWRLSFITDFFWGIAEFVVFF 2 | ||
0 | 1 FKTLLQQDVKKRRGYGGSSDSRYDDGRG 2 | ||
1 | 1 PPGNPPRRMGRISHLRGPSPPPMAGGuGR 2 | ||
1 | 1 * 0 | ||
> | >SELK_speTri Spermophilus tridecemlineatus (squirrel) | ||
0 | 0 MVYISK 1 | ||
1 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVILF 2 | ||
1 | 1 FKTLLQQDVKKRRGYGNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGGINHLQGPSPPPMGGGuGR 2 | |||
1* 0 | |||
> | >SELK_cavPor Cavia porcellus (guinea_pig) | ||
0 | 0 MVYISN 1 | ||
1 | 2 GQVLDNRSQSPWRLSLITDFFWGIAEFVVLF 2 | ||
1 | 1 FKTLLQQDVKKGRGYRNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRINHLQGPSPPPMAGGuGR 2 | |||
1 * 0 | |||
> | >SELK_oryCun Oryctolagus cuniculus (rabbit) | ||
0 | 0 MVYISN 1 | ||
1 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLL 2 | ||
1 | 1 FKTLLQQDVKRGRGYRNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 | |||
1 * 0 | |||
> | >SELK_ochPri Ochotona princeps (pika) | ||
0 | 0 MVYISN 1 | ||
1 | 2 GQVLDSRSQSPERLSLITDFFWGIAEFVVLF 2 | ||
1 | 1 FKTLLQQDVKRRRDYRSSSDSRYDDGRG 2 | ||
1 LPGNPPRRMGRIYHLRCPSPPPPPGGGuGR 2 | |||
1* 0 | |||
> | >SELK_canFam Canis familiaris (dog) | ||
0 | 0 MVYISN 1 | ||
1 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 | ||
1 | 1 FKTLLQQDVKKRRGYGNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 | |||
1 * 0 | |||
> | >SELK_neoVis Neovison vison (mink) | ||
0 | 0 MVYISN 1 | ||
1 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 | ||
1 | 1 FKTLLQQDVKKRRGYGSSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 | |||
1* 0 | |||
> | >SELK_felCat Felis catus (cat) | ||
0 | 0 MVYISN 1 | ||
1 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLL 2 | ||
1 | 1 FQTLLQQDVKKRRGYGNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 | |||
1 * 0 | |||
> | >SELK_equCab Equus caballus (horse) | ||
0 | 0 MVYISN 1 | ||
1 | 2 GQVLDSRSQSPWRLSFITDFFWGIAEFVVLF 2 | ||
1 | 1 FKTLLQQDVKKRRGYGSSSDSRYDDGRG 2 | ||
1 PPGNPRRRMGRINHLHGPSPPPMAGGuGR 2 | |||
1 * 0 | |||
> | >SELK_myoLuc Myotis lucifugus (microbat) | ||
0 | 0 MVYIEN 1 | ||
1 | 2 RQVLDSRSQSPwRLSLITDFFWGIAEFVVLF 2 | ||
1 | 1 FKTLLQKDMKKGRGYGNSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRIDHLRGPSPPPMAGGuGR 2 | |||
1* 0 | |||
> | >SELK_bosTau Bos taurus (cow) | ||
0 | 0 MVYISN 1 | ||
1 | 2 GQVLDSRSQSPWRLSFITDFFWGIAEFVVLF 2 | ||
1 | 1 FRTLLQQDVKKRRGYGSSSDSRYDDGRG 2 | ||
1 PPGNPPRRMGRINHLQGPNPPPMAGGuGR 2 | |||
1 * 0 | |||
>SELK_susScr Sus scrofa (pig) | |||
0 MVYISN 1 | |||
> | 2 GQVLDSRSQSPWRLSFITDFFWGIAEFVVLF 2 | ||
0 | 1 FRTLLQQDVKKRRGYGGSSDSRYDDGRG 2 | ||
2 | 1 PPGNPPRRMGRINHLRGPNPPPMAGGuGR 2 | ||
1 * 0 | |||
1 | |||
> | >SELK_dasNov Dasypus novemcinctus (armadillo) | ||
0 | 0 MVYISN 1 | ||
2 | 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVILL 2 | ||
1 FRTLLQQDVKKKRGYGSSSDSRYDDGRG 2 | |||
1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 | |||
1 | 1 * 0 | ||
> | >SELK_loxAfr Loxodonta africana (elephant) | ||
0 | 0 MVYISN 1 | ||
2 | 2 GQVLDSRSQSPWRLSFITDFFWGIAEFVVLF 2 | ||
1 FKTLLQQDVKKRGGYGSSSDFRYDDGRG 2 | |||
1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 | |||
1 | 1 * 0 | ||
2 | |||
> | >SELK_ornAna Ornithorhynchus anatinus (platypus) EUEMSW407EBWK3 454 | ||
0 | 0 MVYISN 1 | ||
2 | 2 GHVLNGQNRSLWSLSFIKDFFWGILDFIIMF 2 | ||
1 FKSMIHPNVKRGCRNSSSDSKYDDGRG 2 | |||
1 PPGYPRRGMGRINHSNGPSPPPMAGGGuGR 2 | |||
1 | 1 * 0 | ||
> | >SELK_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01DCGU7 454 | ||
0 | 0 MVYISN 1 | ||
2 | 2 GHVLNGQNRSPWSLSYIKDFFWGILDFIIMF 2 | ||
1 FKSMIHPNVKRGCRNSSSDSKYDDGRG 2 | |||
2 PPGYPRRGMGRINHSNGPNPPPMAGGuGR 2 | |||
1 | 1 * 0 | ||
> | >SELK_anoCar Anolis carolinensis (lizard) | ||
0 | 0 MVYISN 1 | ||
1 | 2 GQVLDNQSRAPWSFSSITDFFWGIADFVVMF 2 | ||
2 | 1 FQSIIHPDLTRGSTSSSSSRYDDGRG 2 | ||
1 PPGLPRRRMGRISHFGGPSPPPMAGGuGR 2 | |||
1 * 0 | |||
> | >SELK_galGal Gallus gallus (chicken) | ||
0 | 0 MVYISN 1 | ||
2 | 2 GQVLDNRSRAPWSLSAITDFFWSIADFVVMF 2 | ||
1 FQSIIQPDLRRRGYTSSSYLGQSDGRG 2 | |||
1 PPGNPRRRMGRINHWGGGPSPPPMAGGGuGR 2 | |||
1 | 1 * 0 | ||
> | >SELK_taeGut Taeniopygia guttata (finch) | ||
0 | 0 MVYISN 1 | ||
2 | 2 GQVLDSQSRAPWRLSSITDFFWSIADFVVLF 2 | ||
1 FQSIIQPDLRRRGYTSSSYSGYHDRRG 2 | |||
1 PPANPRRRMGRINHWGGGPSPPPMAGGGuGR 2 | |||
1 | 1* 0 | ||
> | >SELK_xenTro Xenopus tropicalis (frog) | ||
0 | 0 MVYISN 1 | ||
2 | 2 GQVLDGQSRSPWRLSFLTDMFWGITDFVVMF 2 | ||
1 FQSIIHPNVTRRGCQNSSSSTRYDDGRG 2 | |||
1 PPGHPRRMGRINHGSGPSAPPMAGGGGuGR 2 | |||
1 | 1 * 0 | ||
2 | |||
> | >SELK_oncMyk Oncorhynchus mykiss (trout) | ||
0 | 0 MVYVAN 1 | ||
2 | 2 GQVLDNRSQSPWRMSFLTDLFWGAVEFIGLF 2 | ||
1 FQSLVQPDLTKRGNSGSSSTGYSDGRG 2 | |||
1 PPGPPGGRRRMGRINHGGGPSAPPMGGGGuGR 2 | |||
1 | 1 * 0 | ||
> | >SELK_salSal Salmo salar (salmon) | ||
0 | 0 MVYVAN 1 | ||
2 | 2 GQVLDNRSQSPWRMSFLTDLFWGAVEFIGLF 2 | ||
2 | 1 FQSLVQPDLTKRGNSGSSSAGYSDGRG 2 | ||
1 PPGPPGGRRRMGRINHGGGPSAPPMGGGGuGR 2 | |||
1 | 1 * 0 | ||
1 | |||
> | >SELK_oryLap Oryzias latipes (medaka) BJ003636 tag confirmed | ||
0 | 0 MVYVSN 1 | ||
2 | 2 GQVLDSRAQSPWRLSLLVDLFWDALEFFRLF 2 | ||
1 FKTMFHPDLTKDGNSASSRFSDGRG 2 | |||
1 PPGPPGGRRRIGRINHGAGPNAPPMGGGGuGR 2 | |||
1 | 1 * 0 | ||
> | >SELK_braFlo Branchiostoma floridae (amphioxus) tga confirmed | ||
0 | 0 MVYVTG 1 | ||
2 | 2 GQMLENRSPWRLSIIPEIFWGFINFIVLF 2 | ||
1 FQTMYNPSLSKHGSDTPSSYSRGDGRG 2 | |||
1 PPPPPPRRRFGGVRRGGAPGPPPMAGGGuGR 2 | |||
1 | 1 * 0 | ||
</pre> | |||
> | === SELO: 6 vertebrate sequences === | ||
0 | <pre> | ||
2 | >SELO_homSap Homo sapiens (human) tga-sel taa-stop | ||
0 MAVYRAALGASLAAARLLPLGRCSPSPAPRSTLSGAAMEPAPRWLAGLRFDNRALRALPVEAPPPGPEGAPSAPRPVPGACFTRVQPTPLR | |||
0 | QPRLVALSEPALALLGLGAPPAREAEAEAALFFSGNALLPGAEPAAHCYCGHQFGQFAGQLGDGAAMYLGEVCTATGERWELQLKGAGPTPFSR 2 | ||
1 | 1 QADGRKVLRSSIREFLCSEAMFHLGVPTTRAGACVTSESTVVRDVFYDGNPKYEQCTVVLRVASTFIR 2 | ||
2 | 1 FGSFEIFKSADEHTGRAGPSVGRNDIRVQLLDYVISSFYPEIQAAHASDSVQRNAAFFRE 0 | ||
0 VTRRTARMVAEWQCVGFCHGVLNTDNMSILGLTIDYGPFGFLDR 2 | |||
1 YDPDHVCNASDNTGRYAYSKQPEVCRWNLRKLAEALQPELPLELGEAILAEEFDAEFQRHYLQKMRRKLGLVQVELEEDGALVSKLLETMHLT 1 | |||
2 GADFTNTFYLLSSFPVELESPGLAEFLARLMEQCASLEELRLAFRPQMDPR 2 | |||
1 QLSMMLMLAQSNPQLFALMGTRAGIARELERVEQQSRLEQLSAAELQSRNQGHWADWLQAYR 2 | |||
1 ARLDKDLEGAGDAAAWQAEHVRVMHANNPKYVLRNYIAQNAIEAAERGDFSE 0 | |||
0 VRRVLKLLETPYHCEAGAATDAEATEADGADGRQRSYSSKPPLWAAELCVTuSS* 0 | |||
>SELO_musMus Mus musculus (mouse) | |||
0 MAVYRAALGASLAAARLLPLGRCSPSPAPRSTLSGAAMEPAPRWLAGLRFDNRALRALPVEAPPPGPEGAPSAPRPVPGACFTRVQPTPLR | |||
> | QPRLVALSEPALALLGLGAPPAREAEAEAALFFSGNALLPGAEPAAHCYCGHQFGQFAGQLGDGAAMYLGEVCTATGERWELQLKGAGPTPFSR 2 | ||
1 QADGRKVLRSSIREFLCSEAMFHLGVPTTRAGACVTSESTVVRDVFYDGNPKYEQCTVVLRVASTFIR 2 | |||
1 FGSFEIFKSADEHTGRAGPSVGRNDIRVQLLDYVISSFYPEIQAAHASDSVQRNAAFFRE 0 | |||
0 VTRRTARMVAEWQCVGFCHGVLNTDNMSILGLTIDYGPFGFLDR 2 | |||
1 YDPDHVCNASDNTGRYAYSKQPEVCRWNLRKLAEALQPELPLELGEAILAEEFDAEFQRHYLQKMRRKLGLVQVELEEDGALVSKLLETMHLT 1 | |||
2 GADFTNTFYLLSSFPVELESPGLAEFLARLMEQCASLEELRLAFRPQMDPR 2 | |||
1 QLSMMLMLAQSNPQLFALMGTRAGIARELERVEQQSRLEQLSAAELQSRNQGHWADWLQAYR 2 | |||
1 ARLDKDLEGAGDAAAWQAEHVRVMHANNPKYVLRNYIAQNAIEAAERGDFSE 0 | |||
0 VRRVLKLLETPYHCEAGAATDAEATEADGADGRQRSYSSKPPLWAAELCVTuSS* 0 | |||
> | >SELO_ratNor Rattus norvegicus (rat) | ||
0 MASFRAAFGASLAVARTRPQCVGLELQSSAPWSAWAAAMEPTPRWLARLRFDNRALRALPVETPPPGPEDSLSTPRPVPGACFSRARPAPLR | |||
QPRLVALSEPALALLGLEVSEEAEVEAEAALFFSGNALLPGTEPAAHCYCGHQFGQFAGQLGDGAAMYLGEVCTAAGERWELQLKGAGPTAFSR 2 | |||
1 QADGRKVLRSSIREFLCSEAMFHLGIPTTRAGACVTSESTVMRDVFYDGNPKYEKCTVVLRIAPTFIR 2 | |||
1 FGSFEIFKPPDELTGRAGPSVGRNDIRVQMLDYVISSFYPEIQAAHTCDTDNIQRNAAFFRE 0 | |||
0 VTRRTARMVAEWQCVGFCHGVLNTDNMSIVGLTIDYGPFGFLDR 2 | |||
1 YDPDHVCNASDNAGRYTYSKQPQVCRWNLQKLAEALEPELPLVLAEAILKEEFDTEFQRHYLQKMRKKLGLVRVEKEDETLVAKLLETMHQT 1 | |||
2 GADFTNTFCVLSSFPAEPSDTAEFLTQLTSQCASLEELKLAFRPQMDPR 2 | |||
1 QLSMMLMLAQSNPQLFALIGTQANVTKELERVEHQSRLEQLSPSELQSKNRDHWETWLQEYR 2 | |||
1 ERLDKEKEGVGDIAAWQAERVRIMHANNPKYVLRNYIAQKAIEAAENGDFSE 0 | |||
0 VRRVLKLLESPYHSEEEATGPEAVARTTDEQSSYSSRPPLWAAELCVTuSS* 0 | |||
> | >SELO_ornAna Ornithorhynchus anatinus (platypus) uc003bjx.1 | ||
0 2 | |||
1 HADGRKVLRSSIREFLCSEAMFHLGIPTTRAGTCVTSESAVIRDVHYDGNPKYEKCAVVLRIASTFLR 2 | |||
1 FGSFEIFKPRDEHTGRQGPSVGRNDIRIRMLDYVIGTFYPEIEEANADDTVRRNAAFFRE 0 | |||
> | >SELO_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01DJRM3 454 | ||
0 YGGHQFGTWAGQLGDGRAHLIGIYTNRHGEQWELQLKGSGRTPYSR 2 | |||
1 NGDGRAVFRSSVREFLGSEAMHYLRIPTSR | |||
> | >SELO_galGal Gallus gallus (chicken) | ||
0 MQRSGGVLRRGRADTERGETGGGWLSALRFDNLAMRSLPVDPFEDCAPRAVPGACFARVRPTPLRNPRLVAMSAPALALLGLEAGGPEAERE | |||
AEAALYFSGNRLLPGSEPAAHCYCGHQFGSFAGQLGDGAAIYLGEVRGPRGARWELQLKGAGITPFSR 2 | |||
1 QADGRKVLRSSIREFLCSEAMFHLGIPTTRAGTCVTSDSEVVRDIFYDGNPKKERCTVVLRIASTFIR 2 | |||
1 FGSFEIFKPPDEYTGRKGPSVNRNDIRIQMLDYVIGTFYPEIQEAHADNSIQRNAAFFKE 0 | |||
0 ITKRTARLVAEWQCVGFCHGVLNTDNMSIVGLTIDYGPFGFMDR 2 | |||
1 YDPEHICNGSDNTGRYAYNRQPEICKWNLGKLAEALVPELPLEISELILEEEYDAEFEKHYLQKMRKKLGLIQLELEEDSKLVSELLETMHLT 1 | |||
2 GGDFTNIFYLLSSFSVDTDPSRLEDFLEKLISQCASVEELRVAFKPQMDPR 2 | |||
1 QLSMMLMLAQSNPQLFALIGTKANINKELERIEQFSKLQQLTAADLLSRNKRHWTEWLEKYR 2 | |||
1 VRLHKEVESISDVDAWNTERVKVMNSNNPRYILRNYIAQNAIEAAENGDFSE 0 | |||
0 VRNVLKLLENPFQETEDSTEMETKEEEATATAAACAQATRSRLSYCSKPPLWASELCVTuSS* 0 | |||
> | >SELO_calMil Callorhinchus milii (elephantfish) frag tgt-cys tga-sel tga-stop? P taa-stop | ||
0 | 0 LNFDNLALRSLPVDSSGERSCRRVPGACFSLAGATPVDNPRLVASSR 0 | ||
2 | 0 QSDGRKVLRSSIREFLCSEAMFHLGIPTTRAGTCVTSDTKVLRDVFYDGNSKHENCTIILRIAPTFLRF 2 | ||
1 FGSFEILKPEDELTGRQGPSSNRNDIRIQMLDYVIGTFYPEAQQAHPENQVQRNAAFFRE 0 | |||
0 | 0 VTQRTARLVAEWQCVGFCHGVLNTDNMSIMGLTIDYGPFGFMDR 2 | ||
1 | 1 FDPSYICNASDNRGRYAYNQQPEICKWNLGKLAEVLVPELPLKDSQSIIDEEYDTEFQRHYLQKMRKKLGLLQCEQEDDDKLVSELLDIMYRT 1 | ||
2 GADFTNTFYLLSSFPVELESPGLAEFLARLMEQCASLEELRLAFRPQMDPR 2 | |||
1 QLSILLMLSQSNPQLFEVIGSKEGIAKELDLIERSSKLQQATAEDIHSNNAKVWTEWLQKYR 2 | |||
1 SRLATEAEGVDDVDEQNAERVKVMNLNNPKFILRNYIAQNAIEAAEKGDFSE 0 | |||
0 VHLLQKTLRHPFHKQREAEEAGYSSRPPLWARELRVSCSSuR* 0 | |||
> | >SELO_leuEri Leucoraja erinacea (skate) DT725503 frag sel-tga taa-stop | ||
1 NPQLFELIGDKRGVAKEMDRIERFSELQQMTTEELLSKLKNAWTDWLQKYR 2 | |||
1 SRLENEAEGVDDVAELKAERVKVMNANNPKFILRNYIAQNAISAAEDGDFSE 0 | |||
0 VRRVLKLLEHPYSEDACVEEELMQEATVEAPASCEPLSNKRIPYHSKPPDWSGKLLVTuSS* 0 | |||
1 | |||
0 | |||
> | >SELO_petMar Petromyzon marinus (lamprey) frag | ||
0 GASMCSLYVLPLGRN 1 | |||
2 APRQVPGAVFSRVRPSPVERPRVVAISVPALRLLGLRDPEAEAARPEAAEFLSGNRVPPGAQPAAHCYCGHQFGSFAGQLGDGAAMYLGEVQPGPGQRWEVQLKGAGPTPYS 2 | |||
1 HSDGRKVLRSSLREFLCSEAMHHLGVPTTRAGSCVTSHSTVLRDVHYDGNARPEQCSVVLRIAPSFLR 0 | |||
1 FGSFEIFKSTDKDTGRTGPSAGREDIKVTMLDYVIDTFYPELLEGHGDGASHKYTAFFRE 0 | |||
0 VVRRTAHLVAEWQCVGFCHGVLNTDNMSILGLTIDYGPFGFMDR 2 | |||
1 GADFTNSFRLLSRLTLEPGSVEELATLLCQQCATVEEMKRAYKPRIDPR 2 | |||
0 | |||
2 | |||
1 | |||
0 | |||
0 | |||
1 | |||
2 | |||
</pre> | </pre> | ||
=== | === SEPN1: 14 metazoan sequences === | ||
<pre> | <pre> | ||
> | >SEPN1_homSap Homo sapiens (human) | ||
0 | 0 MGRARPGQRGPPSPGPAAQPPAPPRRRARSLALLGALLAAAAAAAVRVCARHAEAQAAARQ 0 | ||
0 | 0 ELALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT 1 | ||
2 | 2 GSTPAASCEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG 0 | ||
2 | 0 VSRLALSGLRNWTAAASPSAVFATRHFQPFLPPPGQELGEPWWIIPSELSMFTGYLSNNRFYPPPPKGKE 0 | ||
0 | 0 VIIHRLLSMFHPRPFVKTRFAPQGAVACLTAISDFYYTVMFR 2 | ||
0 | 1 IHAEFQLSEPPDFPFWFSPAQFTGHIILSKDATHVRDFRLFVPNHR 2 | ||
0 | 1 SLNVDMEWLYGASESSNMEVDIGYIPQ 0 | ||
0 | 0 MELEATGPSVPSVILDEDGSMIDSHLPSGEPLQFVFEEIKWQQELSWEEAARRLEVAMYPFKK 0 | ||
1 | 0 VSYLPFTEAFDRAKAENKLVHSILLWGALDDQSCu 1 | ||
0 | 2 GSGRTLRETVLESSPILTLLNESFISTWSLVKELEELQ 0 | ||
0 | 0 NNQENSSHQKLAGLHLEKYSFPVEMMICLPNGTV 0 | ||
0 | 0 VHHINANYFLDITSVKPEEIESNLFSFSSTFEDPSTATYMQFLKEGLRRGLPLLQP* 0 | ||
0 | |||
>SEPN1_musMus Mus musculus NM_029100 | |||
0 MGQARPAARRPHSPDPGAQPAPPRRRARALALLGALLAAAAAVAAARACALLADAQAAARQ 0 | |||
0 ESALKVLGTDGLFLFSSLDTDQDMYISPEEFKPIAEKLT 1 | |||
2 GSVPVANYEEEELPHDPSEETLTIEARFQPLLMETMTKSKDGFLG 0 | |||
0 VSRLALSGLRNWTTAASPSAAFAARHFRPFLPPPGQELGQPWWIIPGELSVFTGYLSNNRFYPPPPKGKE 0 | |||
0 VIIHRLLSMFHPRPFVKTRFAPQGTVACLTAISDSYYTVMFR 2 | |||
1 IHAEFQLSEPPDFPFWFSPGQFTGHIILSKDATHIRDFRLFVPNHR 2 | |||
1 SLNVDMEWLYGASETSNMEVDIGYVPQ 0 | |||
0 MELEAVGPSVPSVILDEDGNMIDSRLPSGEPLQFVFEEIKWHQELSWEEAARRLEVAMYPFKK 0 | |||
0 VNYLPFTEAFDRARAEKKLVHSILLWGALDDQSCu 1 | |||
2 GSGRTLRETVLESPPILTLLNESFISTWSLVKELEDLQ 0 | |||
0 TQQENPLHRQLAGLHLEKYSFPVEMMICLPNGTV 0 | |||
0 VHHINANYFLDITSMKPEDMENNNVFSFSSSFEDPSTATYMQFLREGLRRGLPLLQP* 0 | |||
>SEPN1_bosTau Bos taurus NM_001114976 | |||
0 MGRTRPGERGPPGSGPAAPPAPPPRRARALALLGALLAAAAAAAAARAFARYSEAQAAARQ 0 | |||
0 DSALKTLGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT 1 | |||
2 GSTPTANYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG 0 | |||
0 VSRLALSGLRNWTTAASPSAVFAARHFRPFLPPQGHVELGEPWWIIPSELSVFTGYLSNNRFYPPPPKGKE 0 | |||
0 VIIHRLLSMFHPRPFVKTRFAPQGAVACLTAISDFYYTVMFR 2 | |||
1 IHAEFQLSEPPDFPFWFSPGQFTGHVVLSKDATHVRDFRLFVPNHR 2 | |||
1 SLNVDMEWLYGASESSNMEVDIGYIPQ 0 | |||
0 MELEALGPSVPSVILDEDGNLIDSRLPSGEPLQFVFEEIQWQQELSWEEAARRLEVAMYPFKK 0 | |||
0 VTYLPFTEAFDQAKAKNKLVHSILLWGALDDQSCu1 | |||
2 GSGRTLRETVLESSPILALLNESFISTWSLVKELEDLQ 0 | |||
0 NNQENPSHQKLAGLHLEKYSFPVEMMICLPNGTV 0 | |||
0 VHHINANYFLDITSMKPEDIENHVFSFSSTFEDPSTATYMQFLKEGLRRGLPLLQP* 0 | |||
>SEPN1_equCab Equus caballus NM_001917067 | |||
0 MEKAGVCCAQ 0 | |||
0 ESALKALGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT 1 | |||
2 GSTPAASYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG 0 | |||
0 VSRLALSGLRNWTTAASPSAVFAARHFQPFLPPRGHVELGEPWWIIPSELSVFTGYLSNNRFYPPPPKGKE 0 | |||
0 VIIHRLLSMFHPRPFVKTRFAPQGAVACLTAISDFYYTVMFR 2 | |||
1 IHAEFQLSEPPDFPFWFSPGQFTGHIILSKDATHVRDFRLFVPNHR 2 | |||
1 VSDVDMEWLYGASESSNMEVDIGYIPQ 0 | |||
0 MELEALGPSVPSVILDEDGNLIDSRLPSGEPLQFVFEEIKWQQELSWEEAARRLEVAMYPFKK 0 | |||
0 VTYLPFTEAFDQAKAKNKLVHSILLWGALDDQSCu 1 | |||
2 GSGRTLRETVLESSPILALLNESFISTWSLVKELEDLQ 0 | |||
0 NNQDNPSHKKLADLHLEKYSFPVEMMICLPNGTV 0 | |||
0 VHHINANYFLDITSMKPEDIESNVFSFSSTFEDPSTATYMQFLKEGLRRGLPLLQP* 0 | |||
>SEPN1_galGal Gallus gallus NM_001114972 | |||
0 MAVPGAAPSRLALALAALAALAAVKYYRDAEAARQQ 0 | |||
0 ELALKSLGSEGLFLFSSLDTNNDLYLSPEEFKPIAEKLT 1 | |||
2 GVTPVSDFEEDAPDPNGETLSIVAKFQPLVMETMTKSKDGFLG 0 | |||
0 ISHVALSGLRNWTAPVSPKSVMLARQFKAFLPPKNKLDLGDPWWIIPSELNIFTGYLSNNRFYPPPPKGKE 0 | |||
0 IIIHRLLSMFHPRPFVKTRFAPQGSVACIQAISTYYYTIAFR 2 | |||
1 IHAEFQLNEPPDFPFWFSPGQFTGYIVLSKDSSHVREFKLFVPNKR 2 | |||
1 SLNVDMEWLYGASEGSNMEVDIGYLPQ 0 | |||
0 MELESTGPSVPSVIYDENGNVIDSRDPSGEPIQFVFEEITWQQEIPWEEAAQKLEVAMYPFKK 0 | |||
0 VSYLPFTEAFERAKAEKKLVHSILLWGALDDQSCu 1 | |||
2 GSGRTLRETVLESSPILALLNESFISSWSLVKELEELQ 0 | |||
0 TNRENEFYSKLADLHLEKYNFPVEMIICLPNGTV 0 | |||
0 IHHINANYFLDITSMKPEDVESSIFSFSANFDDPSTATYLQFLKEGLQRAKAYLQN* 0 | |||
>SEPN1_xenTro Xenopus tropicalis NM_001077433 | |||
0 MSADLRKRKKDTTADNDAPQEAQAEEENKKEKPSSCRYRFLWKLLLGLLLIALLFGIKIHRDNELVRQQ 0 | |||
0 EAALRTLGAEGLFLFSSLDTDNDMHISPEEFKPISEKLT 1 | |||
2 GISTTSDYEEEELLDPNGETLSVASRFQPLLMETMTKSKDGFLG 0 | |||
0 ITHSALSGLRNWTTPVVPNNVFYAGQFKAFLPPKNKLEVGNPWWIIPSELSIFTGYLPNNRIYPPPPKGKE 0 | |||
0 VIIHKLLSMFHPRPFIKTRFAPQGSVACIRAISDFYYDIVFR 2 | |||
1 IHAEFQLNEPPNFPFWFSPGQFTGNIVISKDAAHVRHFKLFVPNNR 2 | |||
1 TLNVDMEWLYGASESSNMEVDIGYLPQ 0 | |||
0 MEIESLGPSIPSTIYDENGNIMESRNAEGEAIEFVFEDINWQSEMTFEEAARKLEVTMYPFKR 0 | |||
0 VSYYPFPEAFDRALAEKKLVHSVLLWGALDDQSCu 1 | |||
2 GSGRTLRETVLESLPVLALLNESFISTWSLVKELEELQ 0 | |||
0 SNKDSYATFASLHLEKYNFPVEMMICLPNGTV 0 | |||
0 VHHINANYFLDITSLKPDEVETNLFSFSDGVEDLSTVTYIKFLKEGLEKAKPFLHS* 0 | |||
>SEPN1_danRer Danio rerio NM_001004294 | |||
0 MAADVDKTPAGEQKDDHEDRGTPSSRRARSRFTHISSLFIIAAIPVIGFCIKYCLDIQFVKRH 0 | |||
0 EAGLKALGADGLFFFSSLDTDHDLYLSPEEFKPIAEKLT 1 | |||
2 GVAPPPEYEEEIPHDPNGETLTLHAKMQPLLLESMTKSKDGFLG 0 | |||
0 VSHSSLSGLRSWKRPAISSSTFYASQFKVFLPPSGKSAVGDTWWIIPSELNIFTGYLPNNRFHPPTPRGKE 0 | |||
0 VLIHSLLSMFHPRPFVKSRFAPQGAVACIRATSDFYYDIVFR 2 | |||
1 IHAEFQLNDVPDFPFWFTPGQFAGHIILSKDASHVRDFHIYVPNDK 2 | |||
1 TLNVDMEWLYGASETSNMEVDIGYLPQ 0 | |||
0 MELSAEGPSTPSVIYDEQGNMIDSRGEGGEPIQFVFEEIVWSEELKREEASRRLEVTMYPFKK 0 | |||
0 VPYLPFSEAFSRASAEKKLVHSILLWGALDDQSCu 1 | |||
2 GSGRTLRETVLESSPVLALLNQSFISSWSLVKELEDLQ 0 | |||
0 GDVKNVELSEKARLHLEKYTFPVQMMVVLPNGTV 0 | |||
0 VHHINANNFLDQTSMKPEDEGPGLSFSAGFEDPSTSTYIRFLQEGLEKAKPYLES* 0 | |||
>SEPN1_petMar Petromyzon marinus (lamprey) tga-sel taa-stop frag | |||
0 0 | |||
0 EMALRTLGNDGLFLFTSLDTNMDMQISPEEFRPIVDKII 1 | |||
2 GPPPSEYEGTQEADPQGEGLTMLARFEPLLMETMSKSRDGFLG 0 | |||
0 VGQSCLAGLRGWKKAEAPSQHFGANQFKVFLPPKSDLELGEAWWLVPNDLNLFTGYLPNSRYYPPPPVAKE 0 | |||
0 IIIFKLLSMFHPRPFVKSRFAPQGSVACIRAQSDMYYDIVFR 2 | |||
1 VHAEFQLNEPPAFPFWFTPAQFTGHVTIARDSSHVRAFHMFVPNNR 2 | |||
1 SLNVDMEWLFGSMDQGNMEVDIGFMPK 0 | |||
0 MELVAEGPSVPALIYDENGNAINTSDPDVEPIQFVFENIEWRSEISFQEAYRQMEVAMYPFKK 0 | |||
0 IQYHPFTEAFEKAKAEDKLVHSILLWGALDDQSCu 1 | |||
2 KVLTQSLKFDFNITSPVLALLSENFISSWSLVKDLEDL 0 | |||
0 KQEEQAEHAKWATLHLAKYTFPVEMMIALPNGTV 0 | |||
0 VHCINANDFLDATAVKAEDLTPDLPAEFLDPTSTTYLKFLKEGLQKAQTYLQA* 0 | |||
>SEPN1_braFlo Branchiostoma floridae genomic fragment ABEP01021665 TGA verified | |||
0 MASEGKRDPPDKRGGGGDAAGTDHRPSVTTKRSTLRSCCFWVCAPAAVLLAALGVGMYLDYRRWVRK 0 | |||
0 EVGMMTLGPDATSLFLAYDTNEDGYLSMEEFKPLLDRLQ 1 | |||
2 GNETSYPVDEEIDQAGEIIVLKTHFEPFVMQSMSKNKNPFGFLG 0 | |||
0 NSNLQGLREWREPVRELASFGANHFAPFLPPPAAYSVLGAAYEIIPSTLTPFSGTLSSNRYFPPKVSNKE 0 | |||
0 VILHRLLGLFHPRPFVHMRFAPQGAVATVRAENERWVDVVFR 2 | |||
1 IHAEFQLNEPPLYPFWFTPAQFTGNIVLSRDASRVRHFHLYVPTNRSLNV 2 | |||
1 DMEWLHGPGHENMEVDIGYIPR 0 | |||
0 MELTSVAPSTPVLIYQEDGSYIDQREEELSEVCSVYEPTIFLEEDSGDVRWNSEISMEEAMKSLEVKMYPFKT 0 | |||
0 VPYYNFTEAFSRAEAENKLVHSILLWGALDDQSCu 1 | |||
2 GSGRTLREGPLESSPIVKMLMDHFISTWVLVAELEGIK 0 | |||
0 FPVEMMVSLPNGTVVHHLNANDLLDMSSGESFDVASNM 0 | |||
0 * 0 | |||
>SEPN1_cioInt Ciona intestinalis (tunicate) XR_053040 TGA confirmed for selenocysteine | |||
MLRNRKSRIKENVKREEKVTANDEKQQADKPTEVPHSISENKKKEP | |||
NDKCLTFVVVFAAAVIAIALGLQVYVYSIIQENAKNAAVRQLGQHGLKLFKEYDINKDMQ | |||
IDPKEFRQLYDFILSEDILKASHEVEEEITKEIFDPSTDEIVQLKADMIPLVLSSMSQTN | |||
KNPAFGTPLFHYSKDFSGLVAWKSVKTEQKDLYAKEFKAFLPNNSSQLVGEPYWLIQRPN | |||
NPEELTSNRYRYPIPKTKIEKLLHNLLVMFHPRPFVTMRFSPQGAAAVIRAQNQVYLEIV | |||
FRFHSEFQLNEPPYLPYWFTPAQFTGRLIIARDGTHVRGFQLYVPNNKKLNVDMEWETND | |||
PNAPENNMEVDIGFLPLMTINQTQPSSPMILNNDDGTSIDRRELIKQGKDIVVEFDEIQW | |||
KDDAITTQYAHGQLEKLFFPFKKVDYLPFNEAFKRASVENKLIHQVVLWGALDDQSC-GS | |||
GRTLRETALESSPVIQLLNQSFISTWSLLKDLEVISNDKQSPLSNVANLSIEAYKFPVQM | |||
MVILPNGTVISSLNANDWLAQANGEVPVDEKWLSLGSKDYNTFLSNALEASKNL* | |||
>SEPN1_strPur Strongylocentrotus purpuratus (urchin) TGA confirmed for selenocysteine | |||
MMPSKDRKKPPDRGEGRSAASAAGANCVATATQTTEHYPPNFGRSNRGRCGWKSLTFLSLFVAVVAVMAGNLA | |||
NRIMMETFVMEEMKLTIGEEGVSIFNAYDRDGDGHLNVIEFEPLLERLNIEDTAERGDFT | |||
YGDEDLDPAEEVLTIEANFIPLQQDTMSKAKDQSFLPLLTHAEFQLNEPPSHPFWFTPAQ | |||
FIGHLIIRKDASHVKYFNMHVPSNRSLNVDMEWMNGPNEVENMEVDIGFMPKMELNAPNP | |||
SSQVTIYSESGEVLYEPSNQRETSSGIIHWDEEISMEEALVLLEKEMYPFKKIEYMPLNA | |||
AFKRAHAEKKLVHHVLLWGALDDQSCuGSGRTLRETSLESPPVLQLLGSHYVSSWSLVAE | |||
LEVIKQNKSDPEYAALASMSLEKYNFPVEMLVSLPNGTVIHHINANDLLDASNTENTSIL | |||
DTFTDPLVTNYAKFLKEGISKAEAVGF* | |||
>SEPN1_nemVec Nematostella vectensis (cnidaria) some exon differences TGA confirmed and phase 1 | |||
0 MTESDEDDGDTATSSKEKVNGSPFEEESVLSSRTKVLTVSLLVALLSISCYVFFIEHPILEGTGDKIEEEIMGLFVSYDTNADGRLDPYEFVPVAHRILGQK 0 | |||
0 ESRSEKQAVVHQNWDDSGEHLTMKALFKPLELSTMTKTSDSYF 0 | |||
0 LSDSALTGLKSWKVVSIPSMTLPVNAFAAFLPPSHASTGMLGTPWFIVEPQIAKF 1 | |||
2 SPQLSSNRYNPPDVSGNNAVIHQLLAMFHPRPFIFTRFGPQGTAACIEAQNNEYLAIQFR 2 | |||
1 IHAEFQLNEPPLHPFWFSPAQFAGRLVISRDGKHIASFHMGVPTNKSLNV 1 | |||
2 DMEWLLGKPQVTTKQ 1 | |||
2 ENDASETTSSNMGVDI 1 | |||
2 GFLPQMEIVDQGESCSVTAGGCDSGTSGKHSDNIEWNDQIPNWAAEKKLEQHFYPFKK 0 | |||
0 VDYLPFKEAFNRAKAEKKLVHHIVLWGALDDQSCu 1 | |||
2 GSGRTLREGPLESRPVLQLLQDNYVNCWSLVEELK 0 | |||
0 KISKDGSDQEMANIAQISLDNYRFPVESMVMHANGSL 0 | |||
0 VARMNANEMMEVPEHDSLPKKIVETTFHQGFEDDISYTYFNFLKNSLRTE* 0 | |||
>SEPN1_oopMin Oopsacas minuta (sponge) AM761280 transcript TGA confirmed | |||
ARGEIYNSNRYNAPQPSGREIVFFKLLQQFHHNVFLQNRFGPRGGVAMIRAKAGPIIEVV | |||
FRIHAEYQLNEHPLHPFWFSPAHFTGNLVFNTDTDEVLQFELYLPSIRKLNVDMEWITGS | |||
DELEGEKMEVDIGYTSAMKLSTNQTFEIPAWDHEKTKEEATIELDRVLYPFKRIPYIDIR | |||
DAVDTARVENKLVHSIIMWGSLDDQSCuGSGRTLRDGPLASDVVQLTLKENFISSWSLIV | |||
HLEDLMANSSVKQEVKDL | |||
>SEPN1_oscCar Oscarella carmela (sponge) EC367053 not quite to TGA | |||
ENSNLEGLRTWKKPHKRFNTYNAPNFQSFLPYRGASLGEVYVIVAKPIQFGNQINSVKYF | |||
PPYPEGREVLMHMLLNQFHRRPFVRMRFEPQGTIGLIRAENEEYVEIMFRCHAEFQLNEP | |||
PMFPFWFTPGQFTGHVIMKKNGSHVRHFKMFVPTNKSLNVDMEWMTGPATSLDPNDPDGS | |||
FQGNNQVDIGYMAEMRLESTRSSYFPMLPDENEEDYFKRASKEDEEIKWNKEISRQEALE | |||
KLERYFFPFKKSHTCLSKSVSWLS | |||
</pre> | </pre> | ||
=== | === SELS: 18 vertebrate sequences === | ||
< | <pre> | ||
> | >SELS_homSap Homo sapiens (human) | ||
0 | 0 MERQEESLSARPALETEGLRFLHTT 1 | ||
2 | 2 VGSLLATYGWYIVFSCILLYVVFQKLSARLRALRQRQLDRAAAAV 1 | ||
2 | 2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLKQ 0 | ||
0 | 0 LEEEKRRQKIEMWDSMQEGKSYKGNAKKPQ 0 | ||
1 | 0 EEDSPGPSTSSVLKRKSDRKPLRGG 1 | ||
0 | 2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 | ||
0 | |||
>SELS_panTro Pan troglodytes (chimp) | |||
0 | 0 MERQEESLSARPALETEGLRFLHTT 1 | ||
0 | 2 VGSLLATYGWYVVFSCILLYVVFQKLSARLRALRQRQLDRAAAAV 1 | ||
0 | 2 EPDVVVKRQEAVAAARLKMQEELNAQVEKHKEKLKQ 1 | ||
0 | 0 LEEEKRRQKIEMWDSMQEGKSYKGNAKKPQ 0 | ||
0 EEDSPGPSTSSVLKRKSDRKPLRGG 1 | |||
2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 | |||
>SELS_macMul Macaca mulatta (rhesus) | |||
0 MEREEDSLSARPALETEGLRFLHVT 1 | |||
2 VGSLLASYGWYIVFSCILLYVVFQKLSARLRALRQRQLERAAASV 1 | |||
2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLKQ 0 | |||
0 LEEEKRRQKIEMWDSMQEGKSYKGNAKKPQ 0 | |||
0 EEDSPGPSTSSVVKRKSDRKPLRGG 1 | |||
2 GYNPLSGEGGGSCSWRPGRRGPSSGGuS* 0 | |||
>SELS_otoGar Otolemur garnettii (bushbaby) | |||
0 MEEEAEPLPARPALEAEGLRFLHVT 1 | |||
2 VGSLLAAYGWYIVFSCVLLYVVFQKFSTRLRALRQRQLDRAAAAV 1 | |||
2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLKQ 0 | |||
0 LEEEKRRQKIEMWDSMQEGKSYKGNTKKPQ 0 | |||
0 EEDNSGPSVIPKRKSDRKPLRGG 1 | |||
2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 | |||
>SELS_musMus Mus musculus (mouse) | |||
0 MDRDEEPLSARPALETESLRFLHVT 1 | |||
2 VGSLLASYGWYILFSCILLYIVIQRLSLRLRALRQRQLDQAETVL 1 | |||
2 EPDVVVKRQEALAAARLRMQEDLNAQVEKHKEKLRQ 0 | |||
0 LEEEKRRQKIEMWDSMQEGRSYKRNSGRPQ 0 | |||
0 EEDGPGPSTSSVIKGKSDKKPLRGG 1 | |||
2 GYNPLTGEGGGTCSWRPGRRGPSSGGuG* 0 | |||
>SELS_ratNor Rattus norvegicus (rat) | |||
0 MDRGEEPLSARPALETESLRFLHVT 1 | |||
2 VGSLLASYGWYILFSCVLLYIVIQKLSLRLRALRQRQLDQAEAVL 1 | |||
2 EPDVVVKRQEALAAARLRMQEDLNAQVEKHKEKLRQ 0 | |||
0 LEEEKRRQKIEMWDSMQEGRSYKRNSGRPQ 0 | |||
0 EEDGPGPSTSSVIPKGKSDKKPLRGG 1 | |||
2 GYNPLTGEGGGTCSWRPGRRGPSSGGuS* 0 | |||
>SELS_canFam Canis familiaris (dog) tga confirmed | |||
0 MERDGQQLSARPALETEGLRYLHVT 1 | |||
> | 2 VGSLLATYGWYIVFSCILLYVVFQKLSTRLRALRQRQLDRAEAAV 1 | ||
0 | 2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 | ||
2 | 0 LEEQKRRQKIEMWDSMQEGKSYKGNARKHP 0 | ||
2 | 0 EEDSPGPSTSSVLPKRKPDRKPLRGG 1 | ||
2 | 2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 | ||
>SELS_equCab Equus caballus (horse) | |||
0 | 0 MEPDEERLSARPAVEAEGLRFLHVT 0 | ||
0 | 2 GSLLAAYGWYLVFSCILLYVLFQKLSSRLRALRQRRLDRAAAAV 1 | ||
2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 | |||
2 | 0 LEEEKRRRKIEMWDSMQEGKSYKRNARGPQ 0 | ||
0 | 0 EEDSPGPSTSSVIKRKSDRKPLRGG 1 | ||
2 | 2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 | ||
0 | >SELS_bosTau Bos taurus (cow) tga confirmed | ||
0 | 0 MERDGDQLSARPTLETEGLRFLHVT 1 | ||
2 VGSLLATYGWYIVFSCILLYVVFQKLSTRLRALRQRHLDQAAAAL 1 | |||
2 EPDIVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 | |||
0 LEEEKRRQKIEMWDSMQEGKSYKGNTRKPQ 0 | |||
0 EEDSPGPSTSSVIPKRKSDRKPLRG 1 | |||
2 GGYNPLSGEGGGTCSWRPGRRGPSSGGuG* 0 | |||
>SELS_susScr Sus scrofa (pig) | |||
0 MEQDGDQLSARPALETESLRFLHVT 1 | |||
2 VGSLLATYGWYIVFCCILLYVVFQKLSTRLRALRQRHLDGAAAAL 1 | |||
2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 | |||
0 LEEEKRRQKIERWDSVQEGRSYRGDARKRQ 0 | |||
0 EEDSPGPSTSSVIPKRKSDKKPLRGG 1 | |||
2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 | |||
>SELS_eriEur Erinaceus europaeus (hedgehog) | |||
0 MEADTEPLPLRPALETEGLRLLHAT 1 | |||
2 VGSLLASYGWYIVLCGVVLYVAFQKLSPRLRALRRRQLDRAEAAV 1 | |||
2 EPDVVVKRQEALAAARLKMQEELNAQAEKHKEKLKQ 0 | |||
0 LEEERRRQKIEMWDSMQEGKSYRGNSRRAQ 0 | |||
0 EDDSPGPSTSSAVKRKPDRKPLRGG 1 | |||
2 GYNPLSGEGGGSCSWRPGRRGPSSGGuG* 0 | |||
>SELS_dasNov Dasypus novemcinctus (armadillo) | |||
0 | 0 1 | ||
2 LGSLLATYGWYIVFSCVLLYVVFQKLSTRLRALRQRQLDRAAAAV 1 | |||
2 EPDIVVKRQEAVAAARLKMQEELNAQAEKHKEKLKQ 0 | |||
0 LEEEKRRQKIEMWDSIQEGKYGNAKKP 0 | |||
0 KGDCPGSSTSLVMKCQSAKLRGG 1 | |||
2 GYNPLSGEGGGTCSWRPGRRGPSAGGuG* 0 | |||
>SELS_monDom Monodelphis domestica (opossum) | |||
0 MDLDEEEQQQLLAKPALELEGIRFLQET 1 | |||
2 VGWLLSNYGWYLLFSFIILYVIFQKLFFRVMRQRALESVASSI 1 | |||
0 EPDTVVKQQEALAASRLRMQEELNTQAEKYKEKLKQ 0 | |||
2 LEEEKRKQKIEMWESMQEGKSYKGKAKKPQ 0 | |||
0 QEPEPGPSTSSNIPKPKPDRKSLRGS 1 | |||
2 GYNPLTGEGSGTCSWRPGRRGPSSGGuG* 0 | |||
>SELS_ornAna Ornithorhynchus anatinus (platypus) | |||
0 LFI 1 | |||
2 VGSVLSAYGWYILFGCAVLYLIFQKLSGSLRVMRRRYSDTTGAAI 1 | |||
2 DPEVVVKRQEALAASRLRMQEELNAQAEKYREKQKQ 0 | |||
0 LEEAKRRQKIEIWESMQEGKSYKGNSRLQPQ 0 | |||
0 QETDPGPSTSSVIPKPKPARKPLRG 1 | |||
2 GYNPLSGEGGGTCSWRPGRRGPSSGGuG* 0 | |||
>SELS_tacAcu Tachyglossus aculeatus (echidna) EUEMSW404CHB0V 454 | |||
0 1 | |||
2 LRVMRQRYSDTTGAAI 1 | |||
2 DPEVVVKRQEALAASRLRMQEELNAQAEKYREKQKQ 0 | |||
0 LEEAKRRQKIEIWESMQEGKSYKGNSRLRPK 0 | |||
0 QETDPGPSTSSVIPKPKPARKPLRGG 1 | |||
2 SYNPLSGEGGGTCSWRPGRRGPSSGGuG* 0 | |||
>SELS_galGal Gallus gallus (chicken) | |||
0 MELGDRGGAGPGPGKPALEREGLRFLHVT 1 | |||
2 VGSLLATYGWYIVFSCILLYVVFQKLSTRLRALRQRHLDQAAAAL 1 | |||
2 EPDIVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 | |||
0 LEEEKQRRQKIEMWDSMQEGKSYKGNTRKPQ 0 | |||
0 EEDSPGPSTSSVIPKRKSDRKPLRGG 1 | |||
2 GYNPLSGEGGGTCSWRPGRRGPSSGGuG* 0 | |||
>SELS_xenTro Xenopus tropicalis (frog) tga confirmed | |||
0 MELGNQPGPGNRPEIELEWYQYLQNT 1 | |||
2 VGVVLSSYGWYILLGCILIYLLIQKLPQNFTRAGTSNHSTVT 1 | |||
2 DPDEIVRRQEAVTAARLRMQEELNAQAELYKQKQVQ 0 | |||
0 LQEEKRQRNIETWDRMKEGKSSKVACRLGQEPS 0 | |||
0 PSTSTSAATKPKQEKQERKTLRGS 1 | |||
2 GYNPLTGDGGGTCAWRPGRRGPSSGGuG* 0 | |||
>SELS_danRer Danio rerio (zebrafish) | |||
0 MEAEDGARVRNEDVPPQNQDLSFLQPS 1 | |||
2 VTAFMSEYGWYLLFGCVGVYLLIQHLRKSRSSTQTRSSSGSAEAH 1 | |||
2 DVGSVVRRQEALEASRRRMQEEQDARAAEFREKQRM 0 | |||
0 LEEEKRRQKIEMWDSMQEGKSYKGSAKVAQ 0 | |||
0 QNTEEAASSSSLRPKTEKKPLRSS 1 | |||
2 GYSPLSGDAGGSCSWRPGRRGPSAGGuG* 0 | |||
>SELS_calMil Callorhinchus milii (elephantfish) | |||
0 1 | |||
2 1 | |||
2 EPLLIVKQQEAMEAARRKMQAELDTQAVKYKEKQNL 0 | |||
0 LEEAKRRQKIEAWESIQEGRSYRANTRFNQ 0 | |||
0 QEPGQPSSSNLTKPKSGKKSLAT 1 | |||
2 GFNALTGDGGGSCTWRPNRRGPSSGGuG* 0 | |||
>SELS_squAca Squalus acanthias (spiny_dogfish) ES606337 511 756 tga-sel taa-stop | |||
0 1 | |||
2 ILSEYGWYILFSGVAGYLVLQRISGMLQARSRKWTQASEAQL 1 | |||
2 EPSLIVRRQEAMDAARHRMQQELDAQAAKYREKQKE 0 | |||
0 IEEEKRREKIEAWDNMQEGKSSRKKANLNPE 0 | |||
0 PESSTSSATKPKSDKKSLRG 1 | |||
2 GFNPLTGDGGGSCLWRPGRRGPSAGAuG* 0 | |||
>SELS_petMar Petromyzon marinus (lamprey) cDNA EB082976 39% identity homSap tga-sel taa-stop full | |||
0 MDPRNEEPAVIRGVSDAVSELLARFGWPL 1 | |||
1 | 2 LLFCALLYFTVRRAAPWLRWGRSSSSSTSISNP 1 | ||
2 DLLMNMHSAMERSRIRMQQELDARAAEHQARVKQ 0 | |||
0 LEEERQKQKMESWERGRSLRPRRDPQSQQ 0 | |||
0 EDNSTPSTSLPRTERQRLRDN 1 | |||
2 NYSPLSGGGGPTCLWRPGRRGPASGGGuG* 0 | |||
</pre> | |||
=== SELT: 11 vertebrate sequences === | |||
<pre> | |||
>SELT_homSap Homo sapiens (human) 5 exons recent pseudogenes on chr 5, chr9 | |||
0 MQYATGPLLKFQIC 2 | |||
1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 | |||
1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 | |||
0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 | |||
2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 | |||
>SELT_macMul Macaca mulatta (rhesus) | |||
0 MRLLLLLLVAASAMVRSEASANLGGVPSKRLKMQYATGPLLKFQIC 2 | |||
1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 | |||
1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 | |||
0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 | |||
2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 | |||
>SELT_otoGar Otolemur garnettii (bushbaby) | |||
0 MRLLLLLLVAASAVVRSEASANLGGVPSKRLKMQYATGPLLKFQIC 2 | |||
1 VSuGYRRVFEEYMRVISQRYPDIRIIGKIYSLHHSYR 2 | |||
1 HIASFLSIFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 | |||
0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 | |||
2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 | |||
>SELT_musMus Mus musculus (mouse) | |||
0 MRLLLLLLVAASAVVRSEASANLGGVPSKRLKMQYATGPLLKFQIC 2 | |||
1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 | |||
1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 | |||
0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 | |||
2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 | |||
>SELT_ratNor Rattus norvegicus (rat) | |||
0 MRLLLLLLVAASAVVRSEASANLGGVPSKRLKMQYATGPLLKFQIC 2 | |||
1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 | |||
0 | 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 | ||
1 | 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 | ||
1 | 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 | ||
0 | |||
> | >SELT_bosTau Bos taurus (cow) | ||
0 | 0 MRLLLLLLVAASAVVRSDASANLGGVPGKRLKMQYATGPLLKFQIC 2 | ||
1 | 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 | ||
1 | 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 | ||
0 | 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 | ||
2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 | |||
> | >SELT_canFam Canis familiaris (dog) | ||
0 | 0 MRYLLVLLVAASAVIRSDASANLGGVPSKRLKMQYATGPLLKFQIC 2 | ||
1 | 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 | ||
1 | 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 | ||
0 | 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 | ||
2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 | |||
> | >SELT_monDom Monodelphis domestica (opossum) | ||
1 | 0 MRFLLLLLMAAAAL-QGEASADTGGVPGKRLKMQYATGPLLKFQIC 2 | ||
1 | 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 | ||
1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 | |||
0 IYACMMVFFLSNMIENQCMSTGAFEITLN 1 | |||
2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHH*SQ * 0 | |||
> | >SELT_ornAna Ornithorhynchus anatinus (platypus) | ||
0 | 0 MRLLLLVVVAAAAGGRSEASADLGGLPSKRLKMQYATGPLLKFQIC 2 | ||
1 | 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 | ||
1 | 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 | ||
0 | 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 | ||
2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 | |||
> | >SELT_petMar Petromyzon marinus (lamprey) cDNA EE737658 tga-sel tag-stop full | ||
0 | 0 MRTHLFAGPVLKVEYC 2 | ||
1 | 1 VSuGYRRVFEEYSRVIAERFPDIRVEGDNYLPQPLYR 2 | ||
1 | 1 YIASFFSVFKLVLIGLVLSGKNLFPMLGVDTPGVWTWSQENK 0 | ||
0 | 0 LYACLMIFFVSNMVETQCMSTGAFEVSLN 1 | ||
2 DVPVWSKLQSGRVPSPQEILQILDNHVKLSGGSAGRMQPS* 0 | |||
> | >SELT_eptBur Eptatretus burgeri (hagfish) cdna BJ650136 tga-sel tag-stop full | ||
0 | 0 MKSKMYSGPELLFQYC 2 | ||
1 | 1 ISuGYRRVFEEYSQALRERYPDIRIEGSNYPPPPLYS 2 | ||
1 | 1 TCASVLSVLKVMLIVLVVSGRNPFPLLGLDTPNAWNWSQNNK 0 | ||
0 | 0 IYACLMIFFLTNMIENQCLSTGAFEVVFN 1 | ||
2 DVPIWSKLQSGRVPSLPELAQILDNHLAMGGQASPSNTHGPQ* 0 | |||
</pre> | |||
> | === SEPHS1: 13 chordate sequences === | ||
0 | <pre> | ||
>SEPHS1_homSap Homo sapiens (human) PHYH- SEPHS1- DUF1172- PRFF18+ FRMD4A- | |||
1 | 0 MSTRESFNPESYELDKSFRLTRFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL 1 | ||
0 | 2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 | ||
0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKMTDR 0 | |||
0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 | |||
1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 | |||
2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPGATS* 0 | |||
> | >SEPHS1_musMus Mus musculus (mouse) | ||
0 | 0 MSTRESFNPETYELDKSFRLTRFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL 1 | ||
2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 | |||
1 | 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKMTDR 0 | ||
0 | 0 ERDKVIPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 | ||
1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 | |||
2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPGATS* 0 | |||
> | >SEPHS1_loxAfr Loxodonta africana (elephant) frag | ||
0 | 0 MSVRESFNPESYELDKSFRLTKFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRLGM 1 | ||
2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 | |||
1 | 0 GRIACANVLSDLYAMGVTECDNMLMLLGISNKMTDR 0 | ||
0 | 0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 | ||
1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 | |||
2 1 | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTP 1 | |||
> | >SEPHS1_monDom Monodelphis domestica (opossum) | ||
0 | 0 MSVRESFNPESYELDKSFRLTRFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL 1 | ||
2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 | |||
1 | 0 GRIACANVLSDLYAMGVTECDNMLMLLGISNKMTDR 0 | ||
0 | 0 ERDKVVPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 | ||
1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 | |||
2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPEMAATALYTLDNNVLTLEQRQFYEDNGFLVIGATS* 0 | |||
> | >SEPHS1_ornAna Ornithorhynchus anatinus (platypus) virtual selenocys | ||
0 | 0 MSVRESFNPESYELDKSFRLTRFTELKGTGCKVPQDVLQKLLEALQENHFQEDEQFLGAVMPRL 1 | ||
2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 | |||
1 | 0 GRIACANVLSDLYAMGVTECDNMLMLLGISNKMTDR 0 | ||
0 | 0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 | ||
1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 | |||
2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPGATS* 0 | |||
> | >SEPHS1_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01BJUY7 454 | ||
0 | 0 AEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 | ||
1 | 1 PDNAVPGDVLVLTKPLGTQ 0 | ||
1 | 2 AKQQRSEVSFVIHNLPIIAKMAAITKACGNRFGLLQGTSSETS 1 | ||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPRIIEVTPRGAAAPPPDNNSSA | |||
> | >SEPHS1_galGal Gallus gallus (chicken) | ||
0 | 0 MSvREtFNPESYELDKSFRLTRFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL 1 | ||
2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 | |||
1 | 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKMTDR 0 | ||
0 | 0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 | ||
1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 | |||
2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPGATS* 0 | |||
> | >SEPHS1_xenTro Xenopus tropicalis (frog) frag | ||
0 | 0 MSVRESFNPESYELDKSFRLTRFAELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL | ||
2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 | |||
1 | 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKLTDR 0 | ||
0 | 0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWVVLGGVATTVCQPNEFIM 2 | ||
1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 | |||
2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGSCPETS 1 | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVASQNVNPTPGATS* 0 | |||
> | >SEPHS1_danRer Danio rerio (zebrafish) first occurence of threonine form | ||
0 | 0 MSVRESFNPESYELDKNFRLTRFTELKGTGCKVPQDVLQKLLESLQENHYQEDEQFLGAVMPRL | ||
2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 | |||
1 | 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKLTEK 0 | ||
0 | 0 ERGKVMPLVIQGFKDASEEAGTSVTGGQTVINPWIVLGGVATTVCQPNEFIM 2 | ||
1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMLNMARLNRT 1 | |||
2 AAGLMHTFNAHAATDITGFGILGHAQNLARQQRTEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNTTPGATS* 0 | |||
> | >SEPHS1_calMil Callorhinchus milii (elephantfish) frag | ||
1 | 0 MSVRETFNPENYELDKNFRLTRFAELKGTGCKVPQDVLHKLLEALQENHYQEDEQFLGAVMPRL 1 | ||
1 | 2 GIGMDSCVIPLRHGGLSLVQTTDFFYPLVDDPYMM 0 | ||
0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKMADK 0 | |||
0 ERDKVMPLIIQGFKDAADEAGTAVTGGQTVLNPWIILGGVATTVCQPNEFIM 2 | |||
1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMFNMARLNRT 1 | |||
2 1 | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGYQAWIIGIVEKGNRTARIIDKPRIIEVAPTVATQNVNPTPGATS* | |||
>SEPHS1_petMar Petromyzon marinus (lamprey) cDNA LyEST8076 FD729689 tga-sel tgc-cys tag-stop not so virtual selenocys full | |||
0 MSVHRYFDPEDHDLDKSFRLTKFSELKGuGCKVPQETLLKLLEGLEQDGPYQDEHQQFMGAVMPRL 1 | |||
2 GIGMDACVIPLRHGGLSLVQTTDFFYPLVDDPYMM 1 | |||
2 GKIACANVLSDLYAMGVTECDNMLMLLAISQKLSEK 0 | |||
0 ERDKVIPLMIRGFKDAAEEAGTTVTGGQTVVNPWIVIGGVATTVCQPNEFIM 2 | |||
1 PDNAVPGDVLVLTKPLGTQVAVNAHQWLDM 0 | |||
0 PEKWNKIKLVVTQEDVELAYQEAMFNMARLNRTAAGLMHTFNAH 2 | |||
1 AATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVIAKMAAISKACGNLFGLLQGTSAETSG 1 | |||
2 GLLICLPREQAARFCAEIaKYGEGHQAWIIGIVEKGGRTARIIDKPRIIEVAPRGASPTGVASPDTPTGPPLT* 0 | |||
>SEPHS1_eptBur Eptatretus burgeri (hagfish) cdna BJ649814 BJ654390 tga-sel taa-stop full | |||
0 MSVHRYFDPEDHELDKSFRLTKFSDLKGuGCKVPQESLLKLLEGLEQDSPFQDEHQQFMGAVMPRL 1 | |||
0 | 2 GIGMDSCVIPLRHGGLSLVQTTDFFYPLVDDPYMM 0 | ||
2 | 0 GKIACANVLSDLYAMGVTDCDNMLMLLGISQKLSEK 0 | ||
0 | 0 ERDKVVPLMVRGFKDAAEEAGTTVTGGQTVMNPWIIIGGVASTVCQPNEFIM 2 | ||
0 | 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 | ||
0 | 0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRTAAGLMHTFNAH 1 | ||
2 AATDITGFGILGHAQNLARQQRNEVAFVIHNLPRNEVAFVIHNLPVVAKMAAISKACGNLFGLLQGTSAETSG 1 | |||
2 GLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGARTARIIDKPRIIEVAPRGAPPPTVTTPTTPTAPLS* 0 | |||
> | >SEPHS1_braFlo Branchiostoma floridae (amphioxus) tga confirmed 73% hsa BW738890 BW748096 frag | ||
0 | 0 MAAPEQVQQAVRPGRVFDPVAHGLDKSFRLTRFADLKuCCKVPQEVLLNLLEGLQNQTIPEEQGQFLPPHFAPI 1 | ||
2 | 2 GIGMDSCVVPLRQGGLSLVQTTDFFYPLVDDPYMQ 0 | ||
0 | 0 GKIACANVLSDLYAMGVKTCDNMLMLLGVSNKMSDK 0 | ||
0 | 0 ERDTVVPLIIRGFKDLAEEAGTMVQGGQTVLNPWVTIGGVATTVCPPNEFIE 2 | ||
0 | 1 PGNAVVGDVLVLTKPLGTQVAVNAHQWLDE 0 | ||
0 PERWNRIRLVVSEEDVERAYQESMFLMSKLNRT 1 | |||
2 AAKLMHKYNAHGATDITGFGLLGHAKNLVQHQKNEVSFTIHNLPVIAKMAAVAKACGNMFQLLQGYSAETS 1 | |||
2 * 0 | |||
> | >SEPHS1_strPur Strongylocentrotus purpuratus (sea_urchin) tga confirmed one fused exon, one new | ||
0 | 0 MAGLPQDVVQQQHNQAILVQQQPQFEVTEPVVVQQQQQVVRRFDPEAHELHKDFRLTRYTELKGuGCKVPREALLNLLKGLEPNEGEQGQGDNAEYMQVQSGVPKF 1 | ||
2 | 2 GIGMDCSVTPLRHGGLCLVQTTDFFFPLVDDPYMQ 0 | ||
0 | 0 GQIACANVLSDLCAMGVTECDNMLMLLGVPKAFTDK 0 | ||
0 | 0 ERDAIMPLIMRGFK DKAAEAETSVTGGQTVINPWCLVGGVATCVCLPTDIVS 2 | ||
1 PNNAVVGDVLVLTKPLGTQIAVNAHQWLDQ 12 PEKWKKIQSVVSPEEVERAYQEAMYNMSLLNRT 1 | |||
2 AASLMRKFNAHGSTDVTGFGILGHAKNLVENQKNEVRFVIHNLPVIANMQNVAKTCGNQFQLLQGYSAETS 1 | |||
2 GGLLIAFPREQAAAFCREMEKSTGHTAWIIGIVDRGSRTAQIIEKPRVIAVTTKMN* 0 | |||
</pre> | |||
> | === SEPHS2: 8 vertebrate sequences === | ||
<pre> | |||
>SEPHS2_homSap Homo sapians (human) 1 exon but intronated like SEPHS1 for comparison | |||
MAEASATGACGEAMAAAEGSSGPAGLTLGRSFSNYRPFEPQALGLSPSWRLTGFSGMKGuGCKVPQEALLKLLAGLTRPDVRPPLGRGLVGGQEEASQEAGLPAGAGPSPTFPAL | |||
GIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM | |||
GRIACANVLSDLYAMGITECDNMLMLLSVSQSMSEEE | |||
EKVTPLMVKGFRDAAEEGGTAVTGGQTVVNPWIIIGGVATVVCQPNEFIM | |||
PDSAVVGDVLVLTKPLGTQVAVNAHQWLDN | |||
PERWNKVKMVVSREEVELAYQEAMFNMATLNRT | |||
AAGLMHTFNAHAATDITGFGILGHSQNLAKQQRNEVSFVIHNLPIIAKMAAVSKASGRFGLLQGTSAETS | |||
GGLLICLPREQAARFCSEIKSSKYGEGHQAWIVGIVEKGNRTARIIDKPRVIEVLPRGATAAVLAPDSSNASSEPSS* | |||
> | >SEPHS2_musMus Mus musculus (mouse) 1 exon | ||
MAEAAAAGASGETMAALVAAEGSLGPAGWSAGRSFSNYRPFEPQTLGFSPSWRLTSFSGMKGuGCKVPQETLLKLLEGLTRPALQPPLTSGLVGGQEETVQEGGLSTRPGPGSAFPSL | |||
SIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM | |||
GRIACANVLSDLYAMGITECDNMLMLLSVSQSMSEKER | |||
EKVTPLMIKGFRDAAEEGGTAVTGGQTVVNPWIIIGGVATVVCQQNEFIM | |||
PDSAVVGDVLVLTKPLGTQVAANAHQWLDN | |||
PEKWNKIKMVVSREEVELAYQEAMFNMATLNRT | |||
AAGLMHTFNAHAATDITGFGILGHSQNLAKQQKNEVSFVIHNLPIIAKMAAISKASGRFGLLQGTSAETS | |||
GGLLICLPREQAARFCSEIKSSKYGEGHQAWIVGIVEKGNRTARIIDKPRVIEVLPRGASAAAAAAPDNSNAASEPSS* | |||
> | >SEPHS2_loxAfr Loxodonta africanus (elephant) 1 exon | ||
MAEAAATGASGEAMAVAEGSSGPAGFSLGRGFSSYRPFEPQALGLNPSWRLTGFSGMKGuGCKVPQETLLKLLAGLTRPDMRPPLGRTLVEGHEEAIQEAGLPAGSGPSPTLPSL | |||
GIGLDSCVIPLRHGGLSLIQTTDFFYPLVEDPYMM | |||
GRIACANVLSDLYAMGITECDNMLMLLSVSQNMSEE | |||
EREKVTPLMIKGFRDAAEEGGTAVTGGQTVINPWIIIGGVATVVCQPNEFIM | |||
PDSAVVGDVLVLTKPLGTQVAVNAHQWLDN | |||
PERWNKIKMVVSREEVELAYQEAMFNMATLNRT | |||
AAGLMHTFNAHAATDITGFGILGHSQNLARQQRNEVSFVIHNLPIIAKMAAISKASGRFGLLQGTSAETS | |||
GGLLICLPREQAARFCSEIKSSKYGEGHQAWIVGIVEKGNRTARIIDKPRVIEVLPRGTTATTLVPDNFNASSEPTL* | |||
> | >SEPHS2a_monDom Monodelphis domestica (opossum) processed 1 exon processed | ||
MAAAAAATVGNGACTPATGGSLAGSYRPFEPQALGLSPNWRLTSFSDMKG uGCK VPQETLLTLLAGLTRPEERPPRAPGLNLGFGDVVAEEAAGPAVLEPGPDAGPFLPSAAGPSLPSPTL | |||
GIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM | |||
GRIACANVLSDLYAMGITECDNMLMLLSVSQKMNEE | |||
EREKVMPLMIKGFRDAAEEGGTSVTGGQTVVNPWIIIGGVATVVCQPNEFIM | |||
PDSAVPGDVLVLTKPLGTQVAVNAHQWLDN | |||
PEKWNKIKLVVTREDVELAYQEAMFSMAMLNRT | |||
AAGLMHTFNAHAATDVTGFGILGHAQNLAKQQRSQVSFVIHNLPIIAKMAAISKASGRFGLLQGTSAETS | |||
GGLLICLPREHAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPGIIEVTPRGATVPLHPNNSHASLEPGS* | |||
> | >SEPHS2a_macEug Macropus eugenii (wallaby) 1 exon processed | ||
MRRRQRTVRTEAVTPAVGGSFAGSYSPFEPQSLGLSPNRRLTSFSDMKGuGCKVPQEALLTLLAGLTRPEERPPRAPGLGLGFGDSAAEEAAGPAAPEPGPDAGPFLAPAAGASPSPPTL | |||
GIGMDSCVIPLRHGGLSLVQTTDFFYPLMEDPYMM | |||
GRIACANVLSDLYAMGITECDNMLMLLSVSQKMNEE | |||
EKEKIMPLMIKGFRDAAEEGGTSVTGGQTVINPWIIIGGVATVVCQPNEFIM | |||
PDSAVPGDVLVLTKPLGTQVAVNAHQWLDNPERWNKIKLVVTREDVELAYQEAMFNMAMLNRT | |||
AAGLMHTFNAHAATDVTGFGILGHAQNLAKQQRSQVSFVIHNLPIIAKMAAISKASGRFGLLQGTSAETS | |||
GGLLICLPREHAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPGIIEVTPRGATVPLHPNNSHASLEPGS* 0 | |||
> | >SEPHS2b_monDom Monodelphis domestica (opossum) unprocessed 8 exons no syntenty to 1 frag | ||
0 | 0 MASVPPRAPAAEGAGAASPGAYRPFEPKALGLSPSWRLTGFSELKGuGCKVPPEALLTLLASLTAAPAPAADRGN 1 | ||
2 | 2 GIGMDSCVVPLRHGGLSLVQTTDFFYPLVEDPYMM 0 | ||
0 | 0 GRIACANVLSDLYAMGITECDNMLMLLSVSQKMSEE 0 | ||
0 | 0 EREKIMPLMVRGFRDAAEQGGTSVTGGHTVINPWLIIGGVATSVCQPNEFIM 2 | ||
0 | 1 0 | ||
0 PEKWNKIKLVVSKEDVELAYQEAMFSMAMLNRT 1 | |||
2 AARLMHTFNAHAATDITGFGILGHAQNLAKQQRSEVSFVIHNLPIIAKMAAVSKASGCFGLLQGTSAETS 1 | |||
2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPRIIEVMPRGATLHQDNSNTSPEPAL* 0 | |||
> | >SEPHS2b_macEug Macropus eugenii (wallaby) unprocessed | ||
0 | 0 MAAAPPPPPPPLAAEGASAGAGARSQGAYRPFEPQALGLSPSWRLTGFSELKGuGCKVPQEALLTLLAGLARPPVAPAPDGGD 1 | ||
2 | 2 GIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM 0 | ||
0 | 0 GRIACANVLSDLYAMGITDCDNMLMLLSISQKMNEE 0 | ||
0 | 0 EREKIMPLMVKGFRDAAEEGGTSVTGGHTVINPWIIIGGVATVVCQSNEFIM 2 | ||
0 | 1 0 | ||
0 PERWNKIKLVVSKEDVELAYQEAMFSMAMLNRT 1 | |||
2 AARLMHTFNAHAATDITGFGILGHAQNLAKQQRSEVSFVIHNLPIIAKMAAVSKASGCFGLLQGTSAETS 1 | |||
2 GGLLICLPREQ AARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPRIIEVMPRGAALHHDSSNTSPEPAS* 0 | |||
> | >SEPHS2_ornAna Ornithorhynchus anatinus (platypus) tga confirmed | ||
0 FDPELLGLSPSWRLTGFSELKGuGCKVPQETLLKLLAGLTHPDPRPGAAPPDPQDPPPDPQDRTPHHAGPAPPAL 1 | |||
0 | 2 GIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM 0 | ||
0 | 0 GRIACANVLSDLYALGITECDNMLMLLSVSQKMNEE 0 | ||
0 EREKIMPLMIKGFRDAAEEGGTSVTGGQTVVNPWIIIGGVATVVCQPNEFIM 2 | |||
0 PDNAVPGDVLVLTKPLGTQVAVNAHQWLDN 0 | |||
0 | 0 PEKWNKIKLVVSREDVELAYQEAMFSMAMLNRT 1 | ||
2 | 2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRSEVSFVIHNLPIIAKMAAITKACGNRFGLLQGTSSETS 1 | ||
0 | 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPRIIEVTPRGAAAPPPDNNSSASPETGS* 0 | ||
0 | |||
> | >SEPHS2_tacAcu Tachyglossus aculeatus (echidna) EUPZL4S02I1W5B 454 | ||
0 | 0 KLVVSREDVELAYQEAMFSMAMLNRT 1 | ||
2 | 2 AAGLMHTFNAHAATDI | ||
>SEPHS2_galGal Gallus gallus (chicken) frag | |||
0 MAAPPGPSVPSPTPGPSPGSGCALGPSTSYRPFDPTSLGLDPNWRLTSYSELRGuGCKVPEETLLRLLEGLGGPRSGAAAAAAAAGGGDGEGAEPGGAPTL 1 | |||
2 GIGTDCCVIPLRHGGLSLVQTTDFFYPLVDDPYMM 0 | |||
0 GRIACANVLSDLYAMGITECDNMLMLLSVSQKMTDE 0 | |||
0 | 0 ERDKVMPLIVRGFRDAAEAAGTSVTGGQTVLNPWVIVGGAASVVCQRQEFIV 2 | ||
0 | 1 PDSAVPGDVLVLTKPLGTQVAVTAHQWLDN 0 | ||
0 PERWNKIKLVVTREEVEAAYREAMFSMATLNRT 1 | |||
1 | 2 AAGLMHTFNAHAATDITGFGILGMHKNLAKQQRNEVSLLFTSLPVLAKNGCCQMAAVSKGLWQYCLGLYSLELVPETS 1 | ||
0 | 2 GGLLICLPREQAARFCAE | ||
2 | |||
> | >SEPHS2_xenTro Xenopus tropicalis (frog) 8 exons in genome | ||
0 | 0 maAPGASHLSAACPFFPGYRPFDPVSVGLEASFRLTSFSDLKGuGCKVPRETLLRLLTGLTEEEAAVSGGAIRDPDVPCAGQDGGQNRL 1 | ||
0 | 2 GIGLDSCVIPLRHRGLSLVQTTDFFYPSVEDPYMQ 0 | ||
2 | 0 GRIACANVLSDLYAMGITECDNMLMLLSVSQKMTEE 0 | ||
0 | 0 EREKVTPLMIKGFRDAAEEGGTSVTGGQTVVNPWIIIGGVATVVCQANEFIM 2 | ||
2 | 1 PQNAVAGDVLVLTKPLGTQVAVNAHQWLDN 0 | ||
0 PERWNKIRLVVSREDVELAYQEAHVNMATLNRT 1 | |||
2 AAALMHSFNAHAATDVTGFGILGHAQNLAQEQQNEVSFVIHNLPIIAKAAITKACGNRFgLLQGTSPETS 1 | |||
2 GGLLICLPREQAARFCAEIKSPKHGEGHQAWIIGIVEKGNRSARIIEKPRiiEVTPRGAaTSDNTA* 0 | |||
</pre> | </pre> | ||
=== | === TXNRD1 (TR1): 27 chordate terminal exons === | ||
<pre> | <pre> | ||
> | >TXNRD1_homSap Homo sapiens (human) 13 exons tga-sel taa-stop | ||
0 | 0 MNGPEDLPKSYDYDLIIIGGGSGGLAAAK 0 | ||
0 | 0 EAAQYGKKVMVLDFVTPTPLGTRW 1 | ||
2 GLGGTCVNVGCIPKKLMHQAALLGQALQDSRNYGWKVEET 1 | |||
2 VKHDWDRMIEAVQNHIGSLNWGYRVALREKKVVYENAYGQFIGPHRIK 0 | |||
2 | 0 ATNNKGKEKIYSAERFLIATGERPRYLGIPGDKEYCISS 0 | ||
2 | 0 DDLFSLPYCPGKTLVVGASYVALECAGFLAGIGLDVTVMVRSILLRGFDQDMANKIGEHMEEHGIKFIRQFVPIK 0 | ||
0 VEQIEAGTPGRLRVVAQSTNSEEIIEGEYNT 0 | |||
0 VMLAIGRDACTRKIGLETVGVKINEK 2 | |||
1 TGKIPVTDEEQTNVPYIYAIGDILEDKVELTPVAIQAGRLLAQRLYAGSTVK 0 | |||
2 | 0 CDYENVPTTVFTPLEYGACGLSEEKAVEKFGEENIE 0 | ||
0 VYHSYFWPLEWTIPSRDNNKCYAKIICNTKDN 0 | |||
0 ERVVGFHVLGPNAGEVTQGFAAALKCGLTKKQLDSTIGIHPVCAE 0 | |||
0 VFTTLSVTKRSGASILQAGCuG* 0 | |||
</pre> | </pre> | ||
VFTTLSVTKRSGASILQAGCuG TXNRD1_homSap Homo sapiens (human) | |||
VFTTLSVTKRSGASILQAGCuG TXNRD1_panTro Pan troglodytes (chimp) | |||
VFTTLSVTKRSGASILQAGCuG TXNRD1_ponPyg Pongo pygmaeus (orang_sumatran) | |||
VFTTLSVTKRSGASILQAGCuG TXNRD1_macMul Macaca mulatta (rhesus) | |||
VFTTLSVTKRSGGSVLQAGCuG TXNRD1_otoGar Otolemur garnettii (bushbaby) | |||
VFTTLSVTKRSGGSILQAGCuG TXNRD1_tupBel Tupaia belangeri (tree_shrew) | |||
IFTTLSVTKRSGGDILQSGCuG TXNRD1_musMus Mus musculus (mouse) | |||
IFTTLSVTKRSGGDILQSGCuG TXNRD1_ratNor Rattus norvegicus (rat) | |||
VFTTLSVTKRSGASVLQSGCuG TXNRD1_cavPor Cavia porcellus (guinea_pig) | |||
VFTTLSVTKRSGGDILQAGCuG TXNRD1_oryCun Oryctolagus cuniculus (rabbit) | |||
VFTTLSVTKRSGASILQAGCuG TXNRD1_canFam Canis familiaris (dog) | |||
IFTTLSVTKRSGGNILQTGCuG TXNRD1_equCab Equus caballus (horse) | |||
VFTTLSVTKRSGGNILQTGCuG TXNRD1_bosTau Bos taurus (cow) | |||
VFTTLSVTKRSGASILQAGCuG TXNRD1_susScr Sus scrofa (pig) | |||
VFTTLSVTKRSGGSILQAGCuG TXNRD1_eriEur Erinaceus europaeus (hedgehog) | |||
VFTTLSVTKRSGGSILQTGCuG TXNRD1_echTel Echinops telfairi (tenrec) | |||
VFTTLSVTKRSGGSILQAGCuG TXNRD1_monDom Monodelphis domestica (opossum) | |||
IFTTLTVTKRSGGNVIQAGCuG TXNRD1_ornAna Ornithorhynchus anatinus (platypus) | |||
VFTTLSITKRSGENTLQSGCuG TXNRD1_galGal Gallus gallus (chicken) | |||
IFTTLSVTKRSGENILQSGCuG TXNRD1_anoCar Anolis carolinensis (lizard) | |||
IFTTLTVTKRSGGNILQSGCuG TXNRD1_xenTro Xenopus tropicalis (frog) | |||
IFTTMEVTKSSGGDITQSGCuG TXNRD1_danRer Danio rerio (zebrafish) | |||
IFTTLEVTKSSGKSIIQTGCuG TXNRD1_tetNig Tetraodon nigroviridis (pufferfish) | |||
IFTTMEVTKSSGGNINQSGCuG TXNRD1_takRub Takifugu rubripes (fugu) | |||
VFTTLDVTKRSGGNITQSGCuG TXNRD1_gasAcu Gasterosteus aculeatus (stickleback) | |||
IFTTLQVTKSSGGDIKQAGCuG TXNRD1_oryLap Oryzias latipes (medaka) | |||
IFTMLAVTKRSGADIKQTGCuG TXNRD1_petMar Petromyzon marinus (lamprey) | |||
IFTTMDITKGSGEDPTKTGCuG TXNRD1_braFlo Branchiostoma floridae (amphioxus) | |||
=== | === TXNRD3 (TGR): 29 chordate terminal exons === | ||
<PRE> | |||
< | >TXNRD3_homSap Homo sapiens (human) 12 exons tga-sel tga-stop | ||
> | 0 MVLDFVVPSPQGTSW 1 | ||
0 | 2 GLGGTCVNVGCIPKKLMHQAALLGQALCDSRKFGWEYNQQ 1 | ||
2 VRHNWETMTKAIQNHISSLNWGYRLSLREKAVAYVNSYGEFVEHIK 0 | |||
0 ATNKKGQETYYTAAQFVIATGERPRYLGIQGDKEYCITS 2 | |||
1 DDLFSLPYCPGKTLVVGASYVALECAGFLAGFGLDVTVMVRSILLRGFDQEMAEKVGSYMEQHGVKFLRKFIPVM 0 | |||
0 VQQLEKGSPGKLKVLAKSTEGTETIEGVYNT 0 | |||
0 | 0 VLLAIGRDSCTRKIGLEKIGVKINEK 2 | ||
0 | 1 SGKIPVNDVEQTNVPYVYAVGDILEDKPELTPVAIQSGKLLAQRLFGASLEK 0 | ||
0 CDYINVPTTVFTPLEYGCCGLSEEKAIEVYKKENLE 0 | |||
0 IYHTLFWPLEWTVAGRENNTCYAKIICNKFDH 0 | |||
0 DRVIGFHILGPNAGEVTQGFAAAMKCGLTKQLLDDTIGIHPTCGE 0 | |||
0 VFTTLEITKSSGLDITQKGCuG* 0 | |||
</PRE> | |||
1 | |||
0 | |||
0 | |||
0 | |||
VFTTLEITKSSGLDITQKGCuG TXNRD3_homSap Homo sapiens (human) | |||
VFTTLEITKSSGLDITQKGCuG TXNRD3_panTro Pan troglodytes (chimp) | |||
VFTTLEITKSSGLDITQKGCuG TXNRD3_macMul Macaca mulatta (rhesus) | |||
VFTTLEITKSSGLDITQKGCuG TXNRD3_musMus Mus musculus (mouse) | |||
VFTTMEITKSSGLDITQKGCuG TXNRD3_ratNor Rattus norvegicus (rat) | |||
VFTTLDITKSSGLDITQRGCuG TXNRD3_cavPor Cavia porcellus (guinea_pig) | |||
VFTTLEITKSSGLAVTQQGCuG TXNRD3_oryCun Oryctolagus cuniculus (rabbit) | |||
VFTTLEITKSSGLDITQKGCuG TXNRD3_canFam Canis familiaris (dog) | |||
VFTTLEITKSSGLDITQKGCuG TXNRD3_felCat Felis catus (cat) | |||
VFTTLEITKASGLDITQKGCuG TXNRD3_equCab Equus caballus (horse) | |||
VFTTLEITKSSGLDITQKGCuG TXNRD3_myoLuc Myotis lucifugus (microbat) | |||
VFTTLEITKASGLDITQKGCuG TXNRD3_bosTau Bos taurus (cow) | |||
IFTTLEITKASGLDILQRGCuG TXNRD3_sorAra Sorex araneus (shrew) | |||
VFTTLEITKSSGLDITQKGCuG TXNRD3_dasNov Dasypus novemcinctus (armadillo) | |||
VFTTLEITKSSGLDITQKGCuA TXNRD3_loxAfr Loxodonta africana (elephant) A=single trace | |||
VFTTLEITKSSGLDITQKGCuG TXNRD3_proCap Procavia capensis (hyrax) | |||
VFTSLEITKSSGLDISQKGCuG TXNRD3_echTel Echinops telfairi (tenrec) | |||
VFTTLEITKSSRTVISPRGCuG TXNRD3_monDom Monodelphis domestica (opossum) | |||
IFTTLTVTKRSGGNVIQAGCuG TXNRD3_ornAna Ornithorhynchus anatinus (platypus) | |||
VFTTMDITKSSGQDITQRGCuG TXNRD3_galGal Gallus gallus (chicken) | |||
VFTTMDITRASGKDIAQSGCuG TXNRD3_anoCar Anolis carolinensis (lizard) | |||
VFTTMDVSKSSGGDITQKGCuG TXNRD3_xenTro Xenopus tropicalis (frog) | |||
IFTTMEVTKSSGGDITQSGCuG TXNRD3_danRer Danio rerio (zebrafish) | |||
IFTTLEVTKSSGKSIIQTGCuG TXNRD3_tetNig Tetraodon nigroviridis (pufferfish) | |||
IFTTMEVTKSSGGNINQSGCuG TXNRD3_takRub Takifugu rubripes (fugu) | |||
VFTTLDVTKRSGGNITQSGCuG TXNRD3_gasAcu Gasterosteus aculeatus (stickleback) | |||
IFTTLQVTKSSGGDIKQAGCuG TXNRD3_oryLap Oryzias latipes (medaka) | |||
VFTTLSVSKRSGGNIQQSGCuG TXNRD3_calMil Callorhinchus milii (elephantfish) | |||
IFTMLAVTKRSGADIKQTGCuG TXNRD3_petMar Petromyzon marinus (lamprey) | |||
IFTTMDITKGSGEDPTKTGCuG TXNRD3_braFlo Branchiostoma floridae (amphioxus) | |||
> | === TXNRD2 (TR3): 33 vertebrate terminal GCUG* exons === | ||
0 | <pre> | ||
>TXNRD2_homSap Homo sapiens (human) 17 exons tga-sel taa-stop | |||
0 MAAMAVALRGLGGRFRWRTQAVAGGVRGAARGAA 1 | |||
1 | 2 AGQRDYDLLVVGGGSGGLACAKE 1 | ||
0 | 2 AAQLGRKVAVVDYVEPSPQ 1 | ||
0 | 2 GTRWGLGGTCVNVGCIPKKLMHQAALLGGLIQDAPNYGWEVAQPVPHDW 2 | ||
1 RKMAEAVQNHVKSLNWGHRVQLQDR 2 | |||
1 KVKYFNIKASFVDEHTVCGVAKGGKE 0 | |||
0 ILLSADHIIIATGGRPRYPTH 0 | |||
0 IEGALEYGITSDDIFWLKESPGKT 2 | |||
1 LVVGAS 1 | |||
2 YVALECAGFLTGIGLDTTIMMRSIPLRGFDQ 0 | |||
0 QMSSMVIEHMASHGTRFLRGCAPSRVRRLPDGQLQVTWEDSTTGKEDTGTFDTVLWAI 1 | |||
2 GRVPDTRSLNLEKAGVDTSPDTQKILVDSREATSVPHIYAIGDVVE 0 | |||
0 GRPELTPIAIMAGRLLVQRLFGGSSDLMDYDN 0 | |||
0 VPTTVFTPLEYGCVGLSEEEAVARHGQEHVE 0 | |||
0 VYHAHYKPLEFTVAGRDASQCYVK 0 | |||
0 MVCLREPPQLVLGLHFLGPNAGEVTQGFALGIK 2 | |||
1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 | |||
</pre> | |||
1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_homSap Homo sapiens (human) | |||
0 | 1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_ponPyg Pongo pygmaeus (orang_sumatran) | ||
1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_macMul Macaca mulatta (rhesus) | |||
1 CGASYVQVMQTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_calJac Callithrix jacchus (marmoset) | |||
1 | 1 CGASYAQVMRTVGIHPTCSEEVVRLRISKRSGLDPTVTGCuG* 0 TXNRD2_otoGar Otolemur garnettii (bushbaby) | ||
0 | 1 CGASYAQVTRTVGIHPTCAEEVVRLRISKRSGLDPTVTGCuG* 0 TXNRD2_micMur Microcebus murinus (mouse_lemur) | ||
0 | 1 CGASYAQVMQTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_tupBel Tupaia belangeri (tree_shrew) | ||
0 | 1 CGASYAQVMQTVGIHPTCSEEVVKLHISKRSGLEPTVTGCuG* 0 TXNRD2_musMus Mus musculus (mouse) | ||
1 CGASYAQVMQTVGIHPTCSEEVVKLHISKRSGLDPTVTGCuG* 0 TXNRD2_ratNor Rattus norvegicus (rat) | |||
1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_cavPor Cavia porcellus (guinea_pig) | |||
0 | 1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_ochPri Ochotona princeps (pika) | ||
1 | 1 CGASYAQVMRTVGIHPTCAEEVAKLRITKRSGLDPTVTGCuG* 0 TXNRD2_canFam Canis familiaris (dog) | ||
0 | 1 CGASYAQVMRTLGIHPTCAEEVAKLRITKRSGLDPTVTGCuG* 0 TXNRD2_equCab Equus caballus (horse) | ||
0 | 1 CGASYAQVMRTVGIHPTCAEEVTKLRISKRSGLDPTVTGCuG* 0 TXNRD2_myoLuc Myotis lucifugus (microbat) | ||
0 | 1 CGASYQQLMRTVGIHPTCAEEVAKLRISKRSGLDPTVTGCuG* 0 TXNRD2_bosTau Bos taurus (cow) | ||
1 CGASFQQVIRTVGIHPTCAEEVAKLRISKRSGLDPTVTGCuG* 0 TXNRD2_oviAri Ovis aries (sheep) | |||
1 CGASYEQVMRTVGIHPTCAEEVAKLRISKRSGLDPTVTGCuG* 0 TXNRD2_susScr Sus scrofa (pig) | |||
1 CGASYTQVMQTVGIHPTCAEEVAKLRISKRSGLDPTVTGCuG* 0 TXNRD2_eriEur Erinaceus europaeus (hedgehog) | |||
1 CGASYAQVMQTVGIHPTCAEEVAKLHISKRSGLDPTVTGCuG* 0 TXNRD2_sorAra Sorex araneus (shrew) | |||
1 CGASYAQLMQTVGIHPTCAEEVAKLHISKRSGQDPTVTGCuG* 0 TXNRD2_dasNov Dasypus novemcinctus (armadillo) | |||
1 CGASYAQMMRTVGIHPTCAEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_loxAfr Loxodonta africana (elephant) | |||
1 CGATYSDLMKTVGIHPTCAEEVTKLKITKRSGLSAVITGCuG* 0 TXNRD2_monDom Monodelphis domestica (opossum) | |||
1 CGATYPQMKKTVGIHPTSAEEVTKLWITKRSGLPATVTSCuG* 0 TXNRD2_macEug Macropus eugenii (wallaby) | |||
1 CGATYSQMMKTVGIHPTCAEEVTKLHITKRSGLDPTVTGCuG* 0 TXNRD2_galGal Gallus gallus (chicken) | |||
1 CGATYSQMMKTVGIHPTCAEEVTKLHITKRSGLDPTVTGCuG* 0 TXNRD2_anoCar Anolis carolinensis (lizard) | |||
1 CGATYHQLMRTVGIHPTCAEEVTKLHITKRSGLDPTVTGCuG* 0 TXNRD2_xenTro Xenopus tropicalis (frog) | |||
1 CGLTYEHLRNTVGIHPTCAEELTKLNITKRSGLDATVTGCuG* 0 TXNRD2_danRer Danio rerio (zebrafish) | |||
1 CGATYSHLLQTVGIHPTCAEEVIKVNITKRSGLDATVTGCuG* 0 TXNRD2_tetNig Tetraodon nigroviridis (pufferfish) | |||
1 CGATYSHLLQTVGIHPTCAEEVIKVNITKRSGLDATVTGCuG* 0 TXNRD2_takRub Takifugu rubripes (fugu) | |||
1 CGATYSHLLQTVGIHPTCAEEVVKVNITKRSGLDPAVTGCuG* 0 TXNRD2_gasAcu Gasterosteus aculeatus (stickleback) | |||
1 CGATYRQLMQTVGIHPTSAEELVKINITKRSGLDPTVTGCuG* 0 TXNRD2_oryLap Oryzias latipes (medaka) | |||
1 CEATYSQLVATVGIHPTCAEDVNKLNITKRSRLDPTVTGCuG* 0 TXNRD2_calMil Callorhinchus milii (elephantfish) | |||
1 CNATYSQLASTVGIHPTCAEDVNKLNITKRSGQDATVTGCuG* 0 TXNRD2_squAca Squalus acanthias (spiny_dogfish) | |||
CLRVQIFTMLAVTKRSGADIKQTGCLAVTKRSGADIKQTGCuG* 0 TXNRD2_petMar Petromyzon marinus (lamprey)* | |||
SGLTYQQLASSVGIHPTCAEEVVKMGITKRSGLDPTVTGCuG* 0 TXNRD2_braFlo Branchiostoma floridae (amphioxus)* | |||
*may not be orthologs: TXNRD2_braFlo has last 3 exons fused though correct phase 0 | |||
0 IYHAFYKPLEYYIPERDASQCYIKAVCKRVGDQEIVGLHFLGPNAGEVTQGFAVALRSGLTYQQLASSVGIHPTCAEEVVKMGITKRSGLDPTVTGCuG* 0 | |||
not available TXNRD2_ornAna Ornithorhynchus anatinus (platypus) | |||
=== SEPP1 (SELP): 15 vertebrate sequences === | |||
> | >SEPP1_homSap Homo sapiens (human) chr5 4 exons 10 selenocysteines | ||
0 MWRSLGLALALCLLPSGGTESQDQSSLCKQPPAWSIRDQDPMLNSNGSVTVVALLQAS<span style="color: #FF0000;">u</span>YLCILQASK 2 | |||
1 LEDLRVKLKKEGYSNISYIVVNHQGISSRLKYTHLKNKVSEHIPVYQQEENQTDVWTLLNGSKDDFLIYDR 2 | |||
1 CGRLVYHLGLPFSFLTFPYVEEAIKIAYCEKKCGNCSLT 0 | |||
1 | 0 TLKDEDFCKRVSLATVDKTVETPSPHYHHEHHHNHGHQHLGSSELSENQQPGAPNAPTHPAPPGLHHHHKHKGQHRQGH | ||
0 | PENRDMPASEDLQDLQKKLCRKRCINQLLCKLPTDSELAPRS<span style="color: #FF0000;">u</span>CCHCRHLIFEKTGSAIT<span style="color: #FF0000;">u</span>QCKENLPSLCS<span style="color: #FF0000;">u</span>QGLRAEEN | ||
ITESCQ<span style="color: #FF0000;">u</span>RLPPAA<span style="color: #FF0000;">u</span>QISQQLIPTEASAS<span style="color: #FF0000;">u</span>R<span style="color: #FF0000;">u</span>KNQAKK<span style="color: #FF0000;">u</span>E<span style="color: #FF0000;">u</span>PSN* 0 | |||
>SEPP1_panTro Pan troglodytes (chimp) | |||
> | 0 MWRSLGLALALCLLPSGGTESQDQSSLCKQPPAWSIRDQDPMLNSSGSVTVVALLQAS<span style="color: #FF0000;">u</span>YLCILQASK 2 | ||
1 | 1 LEDLRVKLKKEGYSNISYIVVNHQGISSRLKYTHLKNKVSEHIPVYQQEENQTDVWTLLNGSKDDFLIYDR 2 | ||
0 | 1 CGRLVYHLGLPFSFLTFPYVEEAIKIAYCEKKCGNCSLT 0 | ||
0 TLKDEDFCKRVSLATVDKTVETPSPHYHHEHHHNHGHQHLGSSELSENQQPGAPNAPTHPAPPGLHHHHKHKGQH | |||
RQGHPENQDMPGSEDLQDLQKKLCRKRCINQLLCKLPKDSELAPRSCCCHCRHLIFEKTGSAIT<span style="color: #FF0000;">u</span>QCKENLPSLCS<span style="color: #FF0000;">u</span> | |||
QGLRAEENITESCQ<span style="color: #FF0000;">u</span>RLPPAA<span style="color: #FF0000;">u</span>QISQQLIPTEASTS<span style="color: #FF0000;">u</span>C<span style="color: #FF0000;">u</span>KNQAKK<span style="color: #FF0000;">u</span>E<span style="color: #FF0000;">u</span>PSN* 0 | |||
> | |||
0 | >SEPP1_ponPyg Pongo pygmaeus (orang_sumatran) | ||
1 | 0 MWRSLGLALALCLLPLGGTESQDQSSLCKQPPAWSIRDQGPMLNSNGSVTVVALLQAS<span style="color: #FF0000;">u</span>YLCILQASK 2 | ||
0 | 1 LEDLRVKLKKEGYSNISHIVVNHQGISSRLKYTHLKNKVSEHIPVYQQEENQTDVWTLLNGSKDDFLIYDR 2 | ||
1 CGRLVYHLGLPFSFLTFPYVEEAVKIAYCEKKCGYCSLT 0 | |||
0 TLKDEDFCKSVSLATVDKTVEAPSPHYHHEHHHNHRHQHLGSSELSENQQPGAPDAPTHPAPPGLHHHHKHKGQH | |||
RQGHPENRDMPGSEDIQDLQKKLCRKRCINQLLCKLPKDSELAPRSCCCHCRHLIFEKTGSAIT<span style="color: #FF0000;">u</span>QCKENLPSLCS<span style="color: #FF0000;">u</span> | |||
> | QGLRAEENITESCQ<span style="color: #FF0000;">u</span>RLPPPA<span style="color: #FF0000;">u</span>QISQQLIPTEASTS<span style="color: #FF0000;">u</span>R<span style="color: #FF0000;">u</span>KNQAKK<span style="color: #FF0000;">u</span>E<span style="color: #FF0000;">u</span>PSN* 0 | ||
0 | |||
>SEPP1_macMul Macaca mulatta (rhesus) | |||
0 MWRSLGLALALCLLPSGGTESQDQSSFCKQPPAWSIRDQDPMLDSNGSVTVVALLQAS<span style="color: #FF0000;">u</span>YLCILQASK 2 | |||
1 | 1 LEELRVKLEKEGYSNISYIVVNHQGISSRLKYTHLKNKVSEHIPVYQQEENQTDVWTLLNGSKDDFLIYDR 2 | ||
0 | 1 CGLLVYHLGLPFSFLTLPYVEEAIKIVYCEKKCGNCSLT 0 | ||
0 TLKDEDFCKSVSLATVDETVEAPQAHYHHEHHHNQGHQSSELSENQQPGAPGAPTHPAPPGLHHHHKHKGQHRQGH | |||
PES<span style="color: #FF0000;">u</span>DMPGSEGLQHLQKKL<span style="color: #FF0000;">u</span>RKRCINQLLCKLPKDSELAPRS<span style="color: #FF0000;">u</span>CCHCRHLIFEKTGSAIT<span style="color: #FF0000;">u</span>QCKENLPSLCS<span style="color: #FF0000;">u</span>QGLLAEET* 0 | |||
> | >SEPP1_musMus Mus musculus (mouse) | ||
0 | 0 MWRSLGLALALCLLPYGGAESQGQSSACYKAPEWYIGDQNPMLNSEGKVTVVALLQAS<span style="color: #FF0000;">u</span>YLCLLQASR 2 | ||
1 LEDLRIKLESQGYFNISYIVVNHQGSPSQLKHSHLKKQVSEHIAVYRQEEDGIDVWTLLNGNKDDFLIYDR 2 | |||
1 | 1 CGRLVYHLGLPYSFLTFPYVEEAIKIAYCEERCGNCNLT 0 | ||
0 | 0 SLEDEDFCKTVTSATANKTAEPSEAHSHHKHHNKHGQEHLGSSKPSENQQPGPSETTLPPSGLHHHHRHRGQHRQGH | ||
LES<span style="color: #FF0000;">u</span>DTTASEGLHLSLAQRKL<span style="color: #FF0000;">u</span>RRGCINQLLCKLSKESEAAPSSCCCHCRHLIFEKSGSAIA<span style="color: #FF0000;">u</span>QC<span style="color: #FF0000;">u</span>ENLPSLCS<span style="color: #FF0000;">u</span>QGLF | |||
AEEKVTESCQCRSPPAA<span style="color: #FF0000;">u</span>QNQPMNPMEANPN<span style="color: #FF0000;">u</span>S<span style="color: #FF0000;">u</span>DNQTRK<span style="color: #FF0000;">u</span>K<span style="color: #FF0000;">u</span>HSN* 0 | |||
> | >SEPP1_ratNor Rattus norvegicus (rat) | ||
0 | 0 MWRSLGLALALCLLPYGGAESQGQSPACKQAPPWNIGDQNPMLNSEGTVTVVALLQAS<span style="color: #FF0000;">u</span>YLCLLQASR 2 | ||
1 LEDLRIKLENQGYFNISYIVVNHQGSPSQLKHAHLKKQVSDHIAVYRQDEHQTDVWTLLNGNKDDFLIYDR 2 | |||
1 | 1 CGRLVYHLGLPYSFLTFPYVEEAIKIAYCEKRCGNCSFT 0 | ||
0 | 0 SLEDEAFCKNVSSATASKTTEPSEEHNHHKHHDKHGHEHLGSSKPSENQQPGALDVETSLPPSGLHHHHHHHKHKGQH | ||
RQGHLES<span style="color: #FF0000;">u</span>DMGASEGLQLSLAQRKL<span style="color: #FF0000;">u</span>RRGCINQLLCKLSEESGAATSSCCCHCRHLIFEKSGSAIT<span style="color: #FF0000;">u</span>QCAENLPSLCS<span style="color: #FF0000;">u</span>Q | |||
GLFAEEKVIESCQCRSPPAA<span style="color: #FF0000;">u</span>HSQHVSPTEASPN<span style="color: #FF0000;">u</span>S<span style="color: #FF0000;">u</span>NNKTKK<span style="color: #FF0000;">u</span>K<span style="color: #FF0000;">u</span>NLN* 0 | |||
> | >SEPP1_canFam Canis familiaris (dog) | ||
0 | 0 MWRSLGLALALCLLPWGGAESQGQSSFCKQPPAWSIRDQNPMLNSSGSVTVVALLQAS<span style="color: #FF0000;">u</span>YLCILQASR 2 | ||
1 LEDLRVKLEKEGFLNISYVVVNHQGLSSQLKYMYLKNKVSEHIPVYQQEENQTDVWTLLNGKKDDFLIYDR 2 | |||
1 | 1 CGRLVYHLGLPYSFLTFPYVEEAIKRAYCEEKCGNCSLT 0 | ||
0 | 0 VLEDEEVCKMVSSGTVESTTEAPQPHPHDHHLHHHHHHHHKHWHRLMPHGNDELSENQQPEEPDVSEHPAPQGLHRHHKHKDHQRQGH | ||
PDN<span style="color: #FF0000;">u</span>DMPAGSESLQLSVPQNQL<span style="color: #FF0000;">u</span>RKGCRNQLLCKLPRDSGLAPSS<span style="color: #FF0000;">u</span>C<span style="color: #FF0000;">u</span>H<span style="color: #FF0000;">u</span>RHLIFEKTGSAIT<span style="color: #FF0000;">u</span>QCKETLPSLCS<span style="color: #FF0000;">u</span>QGLWAEENVIES<span style="color: #FF0000;">u</span>Q | |||
<span style="color: #FF0000;">u</span>RWPPAA<span style="color: #FF0000;">u</span>QASQQLRPTEASTN<span style="color: #FF0000;">u</span>S<span style="color: #FF0000;">u</span>KYKTKM<span style="color: #FF0000;">u</span>K<span style="color: #FF0000;">u</span>LTY* 0 | |||
> | >SEPP1_equCab Equus caballus (horse) | ||
1 | 0 MWRSLGLALALCLLPWGGTESQGQSSFCKQPPAWSIRDQDPMLNSYGSVTVVALLQAS<span style="color: #FF0000;">u</span>YLCLLQASR 2 | ||
0 | 1 LEDLRVKLEKEGYSNISYVVVNHQEISARLKYIHLKNKVSEYIAVYQQEENQTDIWTLLNGSKDDFLIYDR 2 | ||
1 CGRLVYHLGLPYSFLTFPYVEEAIKIAYCEKKCGNCSLM 0 | |||
0 TLEDEDFCKTLSLATVEKTTEDSQPHHHHHHQHHHKHGHQHVGNSQLSENQQPEATGAPEHPPPPGLHHHHKHKGQQ | |||
RRGHPEN<span style="color: #FF0000;">u</span>DMPGGESLQLSLPQKKL<span style="color: #FF0000;">u</span>QKGCINQLLCKMPKDSKLAPSS<span style="color: #FF0000;">u</span>C<span style="color: #FF0000;">u</span>HCRHLIFENTRSAIT<span style="color: #FF0000;">u</span>QCTENLPSLCS<span style="color: #FF0000;">u</span>Q | |||
> | GLWAEENVIESCQ<span style="color: #FF0000;">u</span>RLPPAA<span style="color: #FF0000;">u</span>QPS-QQLKPTEASTN<span style="color: #FF0000;">u</span>S<span style="color: #FF0000;">u</span>KYKAAM<span style="color: #FF0000;">u</span>K<span style="color: #FF0000;">u</span>PSN* 0 | ||
0 | |||
>SEPP1_bosTau Bos taurus (cow) 12 sel | |||
1 | 0 MWRGLGLALALCLLLTGGTESQGQSSYCKQPPPWSIKDQDPMLNSYGSVTVVALLQAS<span style="color: #FF0000;">u</span>YLCILQASR 2 | ||
0 | 1 LEDLRVKLEKEGYSNISYVVVNHQGISSRLKYVHLKNKVSEHIPVYQQEENQPDVWTLLNGNKDDFLIYDR 2 | ||
0 | 1 CGRLVYHLGLPYSFLTFTYVEDSIKTVYCEDKCGNCSLS 0 | ||
0 RPQDEDVCKNVFLATKEKTAEASQRHHHPHPHSHPHPHPHPHPHPHPHPHHGHQLHENAHLSESPKPDTPDTPENPPTSGLHHHHHRHKGPQRQGH | |||
SDNCDTPLGSESLQPSLPQKKL<span style="color: #FF0000;">u</span>RKRCINQLL<span style="color: #FF0000;">u</span>QFPKYSESALSSCCCHCRHLVFEKTGSAIT<span style="color: #FF0000;">u</span>QCTEKLPSLCS<span style="color: #FF0000;">u</span>QGLLAEENVIES<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>RLPPAA<span style="color: #FF0000;">u</span> | |||
> | QAAGQQLNPTEASTK<span style="color: #FF0000;">u</span>S<span style="color: #FF0000;">u</span>KNKAKM<span style="color: #FF0000;">u</span>K<span style="color: #FF0000;">u</span>PSN* 0 | ||
0 | |||
>SEPP1_choHof Choloepus hoffmanni (sloth) | |||
1 | 0 MWKSLGLALALCLLPWGGTESQEQSPFCKRPPAWTIRGQDPMLNSNGSVTVVALLQAS<span style="color: #FF0000;">u</span>YLCILQASR 2 | ||
0 | 1 LEDLRVKLEKEGYSNISYIIVNHHEITSQLKYIHLKNKVSEHISVYQQKENERDVWALLNGKKDDFFIYDR 2 | ||
1 CGHLVYHFRLPYSFLTFPYVEEGIKIAYCENKCGNCPPT 0 | |||
0 FINLFQALKDEDVCENVSLAITEKTTEVSQPHPHHNHHHKHPGNSQLPENQQTGAPDAPQQPPPPGLHHHHKHQGQQREGHPDNCDMPESEGLQLSPHQKKL<span style="color: #FF0000;">u</span>RKRCINQLFCKVSKDSE | |||
LAPRSCC<span style="color: #FF0000;">u</span>HCRHLVFEKTGSAITCQCKENLPSLCS<span style="color: #FF0000;">u</span>QGLWAEENVIES<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>RLPPAA<span style="color: #FF0000;">u</span>QISQQLLKPTEVSTN<span style="color: #FF0000;">u</span>E<span style="color: #FF0000;">u</span>NNKADM<span style="color: #FF0000;">u</span>K<span style="color: #FF0000;">u</span>PAN* 0 | |||
> | |||
1 | >SEPP1_ornAna Ornithorhynchus anatinus (platypus) | ||
0 | 0 MWQGLGLALALCLLPGGGAESQSASSHCKEAPRWQIRDQDPMLNSLGTVTVVALLQAS<span style="color: #FF0000;">u</span>YLCILQASR 2 | ||
1 LEDLRVKLENEGYSNISYIIVNHQGMPSQLNHKTLKEKVSEHIPVYQQDEKQTDVWSTLKGNKDDFLIYDR 2 | |||
1 CGRLVYHLSLPYTFLSFSYVEDSIKTTYCEQNCGNCSYT 0 | |||
0 MPEAEEFCTNTSSAAKEKATEAPLPHNDRPHHHHHHHHHHGHKPHPSGTEQAPADPDGPLRSPAPQGLHKRLRPAGQPRQGQ | |||
> | GGSREAAEGRGEELPSPRKKA<span style="color: #FF0000;">u</span>RKGNASCQNQLL<span style="color: #FF0000;">u</span>DWHKRSGPAPSS<span style="color: #FF0000;">u</span>C<span style="color: #FF0000;">u</span>HCRHLLFGSKATATAL<span style="color: #FF0000;">u</span>QCRDALPALCS<span style="color: #FF0000;">u</span>QGRQS | ||
0 2 | GEDVIES<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>RSPLPA<span style="color: #FF0000;">u</span>PPAAQLPSPSPTDPNAA<span style="color: #FF0000;">u</span>K<span style="color: #FF0000;">u</span>ENTAGM<span style="color: #FF0000;">u</span>K<span style="color: #FF0000;">u</span>PTR* 0 | ||
1 | |||
0 | >SEPP1_galGal Gallus gallus (chicken) reqd traces tga-sel taa-stop | ||
0 MWAGLGLALVLCLLPGGGTESQRCQEPPEWHIGEESPMLNARGSVAVVALLQAS<span style="color: #FF0000;">u</span>YLCLLQASR 2 | |||
1 LEDLRVKLENEGLVNISYVVVNHQSPHSQKKFHLLQESVSDHITVYQQDDHQADVWTTLNGNKDDFLIYDR 2 | |||
1 CGRLVYHLGLPYSFLSFQYVEEAIKIAYCENNCGNCSYT 0 | |||
> | 0 EPDIDNICENITKKEDENLAGIEPEPEPSGQHSHHHHQLHRHRHHHHHREGGRHSKNQNHQAPSESQRRHPHNGRRHRVFNHNRHDQ | ||
0 2 | IGSHEQVETLPPGEGVENLPRVTKL<span style="color: #FF0000;">u</span>KKGKTICKNQLT<span style="color: #FF0000;">u</span>NWQTASDSTTSS<span style="color: #FF0000;">u</span>CCHCRHLLFEELGNSIT<span style="color: #FF0000;">u</span>Q | ||
1 | CRGALPNSCR<span style="color: #FF0000;">u</span>HGQLLAEDITES<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>RLLTAA<span style="color: #FF0000;">u</span>ESAAGGGSETSDT<span style="color: #FF0000;">u</span>L<span style="color: #FF0000;">u</span>ERAGN<span style="color: #FF0000;">u</span>A<span style="color: #FF0000;">u</span>KTN* 0 | ||
0 | |||
0 | >SEPP1_xenTro Xenopus tropicalis (frog) frag | ||
0 | 0 MWKGFALALALCLLPWGGAESQGHRKRCKEPPEWSIGDQNPMMQSAGKVTVVALLQAS<span style="color: #FF0000;">u</span>SLCLLQASR 2 | ||
</ | 1 LEDLRLKLEKEKLVGISYVVVNHQGRQSRAKYDLLKSKVSEHIPVYQQEEYQPDVWSLLKGDKDDFFVYDR 2 | ||
1 CGRLVQHLELPYSLLHFPYVEEAIRIAYCEDKCGECEHK 0 | |||
0 ILDADVCKKPEEQQEQDKPVEEKVERPRHHRNHHRHHRPKHSGHRHRHHHSEDGQVAELDVLRSSAQANNRAGSQNGQGQ | |||
SVVQVVPQSEVLFVPQREADIPVLARQP<span style="color: #FF0000;">u</span>KKAKS<span style="color: #FF0000;">u</span>KKQYL<span style="color: #FF0000;">u</span>DWREEGGKAFNS<span style="color: #FF0000;">u</span>C<span style="color: #FF0000;">u</span>H<span style="color: #FF0000;">u</span>RQLSFEVTENEVA<span style="color: #FF0000;">u</span>H<span style="color: #FF0000;">u</span>QEALPSSC | |||
S<span style="color: #FF0000;">u</span>QELLSDSLPES<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>RLSAAA<span style="color: #FF0000;">u</span>HSESTGLPETKLESEPNAP<span style="color: #FF0000;">u</span>A<span style="color: #FF0000;">u</span>PQEAEN<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>KEL<span style="color: #FF0000;">u</span>RFLM* 0 | |||
>SEPP1_xenTro Xenopus laevis (frog) | |||
0 MWKGFALALALCLLPWGGAESQGHRSRCKQPPDWSIGDQNPMIQSAGKVTVVALLQAS<span style="color: #FF0000;">u</span>YLCLLQASR 2 | |||
1 LEDLRLKLEKEKLVGISYVVVNHQGRHSRAKYDLLKSKVSEHIPVYQQEENQPDVWSLLKGDKDDFFIYDR 2 | |||
1 CGRLVQHLELPYSLLHFPYVEEAVRIAYCGDKCGECEHK 0 | |||
0 IPDADVCKKPEEQPELEKPVEEKVERPRPHRNHHRHHRPKHSGHRHRHHNNEGGQAAEVDAFQTNNRAGSHNGQGQSVVPQSEVVFVPQ | |||
READVPVLALQP<span style="color: #FF0000;">u</span>KKAKS<span style="color: #FF0000;">u</span>KKQYL<span style="color: #FF0000;">u</span>EWREDAGKAFNS<span style="color: #FF0000;">u</span>C<span style="color: #FF0000;">u</span>H<span style="color: #FF0000;">u</span>RQLSFEIAQNEVT<span style="color: #FF0000;">u</span>RCQEALPAS<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>QELLSDSLSES<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>RLSAAA<span style="color: #FF0000;">u</span>HSHS | |||
TGLPELDTESETNAP<span style="color: #FF0000;">u</span>A<span style="color: #FF0000;">u</span>PQEAEN<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>KEL<span style="color: #FF0000;">u</span>RFLM* 0 | |||
>SEPP1_danRer Danio rerio (zebrafish) 17 sel | |||
0 MWKALSLTLALCLLVGCSAESETEGARCKLPPEWKVGDVEPMKNALGQVTVVAYLQAS<span style="color: #FF0000;">u</span>LFCLEQASK 2 | |||
1 LNDLLLKLENQGYPNIAYMVVNNREERSQRLHHLLQERLLNITLYAQDLSQPDAWQAVNAEKDDILVYDR 2 | |||
1 CGRLTYHLSLPYTILSHPHVEEAIKHTYCDRICGECSLES 0 | |||
0 SAQLEECKKATEEVNKPVEEEPRQDHGHHEHGHHEHQGEAERHRHGHHHPHHHHHHHRGQQQVDVDQQVLSQVDFGQVAVETPMMKRP<span style="color: #FF0000;">u</span>AKHSR<span style="color: #FF0000;">u</span>KVQYS<span style="color: #FF0000;">u</span>QQGADSPVAS<span style="color: #FF0000;">u</span> | |||
C<span style="color: #FF0000;">u</span>H<span style="color: #FF0000;">u</span>RQLFGGEGNGRVAGL<span style="color: #FF0000;">u</span>HCDEPLPAS<span style="color: #FF0000;">u</span>P<span style="color: #FF0000;">u</span>QGLKEQDNHIKET<span style="color: #FF0000;">u</span>Q<span style="color: #FF0000;">u</span>RPAPPAE<span style="color: #FF0000;">u</span>ELSQPT<span style="color: #FF0000;">u</span>V<span style="color: #FF0000;">u</span>PAGDAT<span style="color: #FF0000;">u</span>G<span style="color: #FF0000;">u</span>RKK* 0 | |||
>SEPP1_torCal Torpedo californica (electric_ray) 1 tga-sel taa-stop | |||
0 MQQGLGVALALCLLLVGQAESQGQCDKPSEWTIGDKNPMKDSLGKVTVLALFQAS<span style="color: #FF0000;">u</span>HFCLEQAGS 2 | |||
1 LKNLKRKLDQDGLEHISYMVVNHQGNDSRKRYQELKRNVQGKLPVYQQEIDQPDVWNLLQGQKDDFFIYDR 2 | |||
1 CGQQTFHLELPFTILSQMYVEHAIKQTYCQEICTNCS 0 | |||
0 LAELPFGMQQHKMQKSQRRWEEDGSPVPKHRGSTMGHKTTPPQSPR* | |||
=== | === SEPP2 (SELPB): 16 vertebrate sequences === | ||
<pre> | <pre> | ||
> | >SEPP2_monDom Monodelphis domestica (opossum) chr2:9,451,774-9,455,977 1 selenocys tga stop taa | ||
0 | 0 MPPPGLSLAVLLGLLGASLAFENRTRFCQPAPPWQVGGGRAPMEEALGNVTVVALLKASuHFCLKQAAS 2 | ||
1 | 1 MGSLQERLARMGAPDVRFIIVNEKSPQSRALHGELELHAPPGVPVYGQPELGPDIWSILGGAKDDFLIYDR 2 | ||
1 CGRLTFHIRLPFSFLHFPYVESAIRFTHRQDSCGNCSFYPAQ 0 | |||
0 VNSTDERKGESKHSPGLEGEGQEPLGEKPDSRGLTLGSSAPTAHAHDPVHGGMEKPSPSLPSPALEPSLHEGEAPG* 0 | |||
0 | |||
> | >SEPP2_macEug Macropus eugenii (wallaby) trace archive tga stop taa | ||
0 | 0 MGLPGLAAALLLGLVGATLASENGTRICQPAPKWQVDRGASPMEEALGQVTVVALLKASuHFCLKQAAS 2 | ||
1 | 1 IGNLQERLARVGATGVRFIIINEKSPLSQALYGELELHAPSGVLVYNQQGPGPDIWSILGGAKDDFLIYDR 2 | ||
1 CGRLTFHLPLPLSFLYFPYVESAIRFTHQQDHCGNCSFYPAQ 0 | |||
0 VNNTDKGKAESLTQSPRLEGEGQESLAEGPSTHGPTLWPSTPHPAHARGPVHSPGSKRLSPSLPPPSLEWPPHEEANK* 0 | |||
0 | |||
> | >SEPP2_ornAna Ornithorhynchus anatinus (platypus) synteny last exon uncertain | ||
0 | 0 MAGSGLLGPALTLATLLAAAGALPDLENGTRICQPAPRWTVNGVAPMEGTEGQVIVVALLKASuHFCLKQAAR 2 | ||
1 | 1 LAGLRERLAGHGAGNVSFLIVNQRDPTAQLLHTELERHAPPGVPVYAQDGPDPDVWSVLGGDKDDFFVYDR 2 | ||
1 CGRLTFHIQLPFSFLHFPYVEAAVRFTHRRDFCGNCSYYFPQVGT 0 | |||
0 VNDTTTQESELEKSPGAPGEEPEGSPVREPDRPQSQDPTGPFSGVLLQGKENKIIPWKTPLQAAPRKPSHPPGAHD* 0 | |||
0 | |||
> | >SEPP1_tacAcu Tachyglossus aculeatus (echidna) EUEMSW408ERBZB length=227 run=R_2007_08_22_12_11_10_ 59% | ||
1 VPVYAQDGPDPDVWSILGGGKDDFLVYDR 2 | |||
1 | 1 CGRLTFHIRLPFSFLHFPYVEAAVLFSHRHDFCGNCS | ||
> | >SEPP2_galGal Gallus gallus (chicken) taa stop synteny Zswim5 HPDL MUTYH | ||
0 | 0 MGSLLLALASCLGLAVASEGATNGSRLCHEAPAWRINGSSPMEGAAGQVTVVALLKASuHFCLLQARS 2 | ||
1 | 1 LGALRERLGQQGVSDVRYVIVNEQAPLSRAMFGELQRHAPPGVPVLQQQPHEPDVWQLLGGDKDDFLVYDR 2 | ||
1 CGRLAFHIQLPYSFLHLPYVESAIRFTHRKDFCGNCSLYPNSTQE 0 | |||
0 ANSTMEVPATLTPLPKQEEKESETPAHHQPNHLHPHHRAVGNGTAPEPSGDHRPAHAHHHHGAHGKLHPKGQTPEGRDP* 0 | |||
0 | |||
> | >SEPP2_taeGut Taeniopygia guttata (finch) | ||
0 | 0 MGLLVLALATWLGLGLASASEETANSSRICQEAPAWTINGSSPMEGAAGQVTVVALLKASuQFCLRQAHS 2 | ||
1 | 1 LGGLRERLARQGMADVRYMIVNEKAPLSRAMLPELQRHAPPGVPVFQPEREDPDVWQVLGGDKDDFLVYDR 2 | ||
1 CGRLAFHIQLPFSFLHFPYVEAAIRSSHIKDFCGNCSLYPNTTQE 0 | |||
0 ANSTMEGPATPSSLPEHEGMESETPVHQHKPLHPHRHHEVNSERDTNPSEDHKPATHAHHHHGDHGQLHHKGRKQKEGDEH* 0 | |||
0 | |||
> | >SEPP2_anoCar Anolis carolinensis (lizard) scaffold_4:5,993,109-5,994,846 | ||
0 | 0 MDYSLATRILLLGLVVISATQAEVTENKTRICQPAPLWKINGTAPMAGMEGQVTVVALLKASuPFCLKQAAK 2 | ||
1 | 1 IGGLQKKLSNEGVANVSFLIVNEKTPLSRAMYWELKRNAPQGIPVYQQQILEPDVWQILDGDKDDFLIYDR 2 | ||
1 CSRLTFHIQLPYSFLHLPYVEAAVHYTHRKDYCGNCSRYYSE 0 | |||
0 * 0 | |||
0 | |||
> | >SEPP2_xenTro Xenopus tropicalis (frog) NM_001006907 misannotated, no selenocysteine | ||
1 | 0 MHNLALTVSILMGLLGQVSSSEQTNSSICKPPPKWSIEGEVPMAEALGKVTVVALLQASCGFCLVQAAR 2 | ||
1 MGPLRYKLSLQGMTDIKYMIVNDQSLHSANMFPELKRWAPEGIPVYQQTPGQDDVWELLDGNKDDFLIYDR 2 | |||
0 | 1 CGRLTFHVRLPLSFLHFPYVEAAILSTYNESFCGNCSFTSNSTLIPM 0 | ||
0 | 0 NGTTVSPSGDDSSSPLQNKDEPVNKEPSPTLEKHNDQRKLDSELRLHDHSQHHPINSHKRQENQNNHPRNLIKNGKQN* 0 | ||
> | >SEPP2_danRer Danio rerio (zebrafish) 3 possible tga-sel taa-stop | ||
0 | 0 MQALWPLLLSALPALLGASSLFVEKESNGSRICKPAPQWEIDGKTPMKELLGNVVVVALLKASuHFCLTQAAR 2 | ||
1 | 1 LGDLRDKLANGGLTNISFMVVNEQDSQSRAMYWELKRRTAQDIPVYQQSPLQNDVWEILEGDKDDFLVYDR 2 | ||
1 CGYLTFHIVLPFSFLHYPYIEAAIRATYHKNMCNCS 0 | |||
0 | 0 LNANFSISESSDSTKNEPAGENNQRPNSTEPVTAAHHHHHQQHEPHHHHNPYPNNHKKSGDSDVTGKPKEPTHHSHQEHVHNHRFYTVFSFIILFYIMFFFFVNIWSCSCVRTSYLPQRG | ||
VFGCLQCIFVLNEVYTLNLRQGSVCGCuTQESFIYTKLLISPLFGKMuLCuTKWLLGLLC* 0 | |||
> | >SEPP2_tetNig Tetraodon nigroviridis (pufferfish) from cDNA, genome misassembled | ||
0 | 0 MSSPWLLWLQVALTGLLWASQGQSATSRICKAAPRWEVGGQAPMEALVGRVVVVALLKASuQFCHIQASK 2 | ||
1 | 1 IGPLREKLSRRNVTEVSFVIVNEQEPVSRALYWQLRRRAPPGVPVYQQAPLQDDVWEALDGDKDDFLIYDR 2 | ||
1 CGQLTFHIGLPYSSLRYTYVEAAVIATHQGNICNCS 0 | |||
0 ANFTSLSISNSSGSGGMPSQTNQTVTAETDGPHTTHHHPHPHRHHHHHQHLSPEQPTPTAMPGQATPTPA* | |||
0 | |||
> | >SEPP2_gasAcu Gasterosteus aculeatus (stickleback) tga-sel taa-stop | ||
0 | 0 MQALWPLLYAALPGLLWASHVSLLVEGDNDASRICKPAPHWTIKERAPMQELLGNVVVVALLKASuQFCLTQASK 2 | ||
1 | 1 IGGLRDKLNRSNLTDVSFMIVNEREPHSRAMYWELKRRAPPGVPVYQQAALQSDVWEALDGDKDDFLVYDR 2 | ||
1 CGLLTFHIVLPYSFLHNVYVEAAIRATYLKNICNCT 0 | |||
0 VDSVVSSLNNSVMNNETDFNVSQTNATPRIQPDTNDPEGAGTPPPPTHHHSHHHHHHHQHHHPHHPHLNQPLHDTSHPLSVHHHHHPHHHHHQQQQPGNHSGTAVDH* 0 | |||
0 | |||
> | >SEPP2_oryLat Oryzias latipes (medaka) | ||
0 | 0 MLRLCSALPALLWASLSVLSAEGDSNASKICKPAPYWDIEGHVPMQEHLGNVVVVALLKATuEFCLTQASK 2 | ||
1 | 1 IGNLRDKLNRNNITEVSFMIVNELEALSQTMHWKLKKKAPTGVPVYQQSSLQKDVWEILDGDKDDFLIYDR 2 | ||
1 CGLLTFHIVLPNSFLQNADVENAITATYTQDICNCS 0 | |||
0 GNSTLSGGGNNFTRNSSQSHSGVQRPADEPENVTTTLEASPNDSRHEHAHHHHPHHQHLHHHHPHHQHLHHHHPHHQHLHHHHPHLQQHSKHQNDDSY* 0 | |||
0 | |||
> | >SEPP2_cypCar Cyprinus carpio (carp) | ||
0 | 0 MQALWALWLSAFPALLWASPLFLERESNASRICKPAPKWEIDGEIPMKNLLGNVVVVALLKASuQFCLTQAAR 2 | ||
1 | 1 LGKLRDKLALGKLTDISFLVVNEQDAQSRAMYWELKRRTTEGIPVYQQSPLQNDVWDILEGDKDDFLVYDR 2 | ||
1 CGHLTFHIVLPFSFLHYPYIEAAIRATYHKNICNCS 0 | |||
0 LHANSSISEGFKNNTNNANKQTEVKNQKTNNTEPVTVEHHHHRHEHHHQHVHYHYHQHVHHHHQNKNNSTGDSDVTSVPKEPIQHSHQEHS | |||
0 | |||
> | >SEPP2_oncMyk Oncorhynchus mykiss (trout) | ||
0 | 0 MQGLLTLWLCAALPGLLCASPLLVEGDNDASKICKPAPHWEIKGHEAPMKELLGNVVVLALLKASuHFCRTQASK 2 | ||
1 | 1 LGGLRDKLLRSNLTDVSFLIVNEREAQSRAMYWELKRRAPPGIPVYQQAPLQDDVWEALDGDKDDFLVYDR 2 | ||
1 CGRLTFHIVLPYSFLHYPYIEAAVRATYHKDICGNCT 0 | |||
0 VDSNTTSSAGWNSTRRNETLSSSQMRVNETDSTVRSIDVDTVSNPVPSDGPQMSAEGGW* 0 | |||
0 | |||
> | >SEPP2_squAca Squalus acanthias (spiny_dogfish) | ||
0 2 | 0 MGLEMPLLMILLGLTITVAVAKNETRICQVAPHWGIANQSPIEEQFGHVVVVALLKASuQFCLTQAAK 2 | ||
1 | 1 LGSLRDKLARQGLKDVRYMIVNEKTPRSRAMLWELKRHAPKDVPVYQQSPFQPDVWRILQGNKDDFLVYDR 2 | ||
1 CGKLTFHIVLPYSYLHFRYIEAAIRATYNKDICGNCS 0 | |||
0 VSDITLERERNITTTYKVTQSNTTSL | |||
0 | |||
> | >SEPP2_leuEri Leucoraja erinacea (skate) potential 2 terminal sel uRYFKIGEuWKKFPSQIRAKCQY* | ||
0 | 0 MGLQRSLLIILVGAAITLAAANNDTRICQVAPHWEIGNQSPMEQLSGQLVVVALLKASuQFCLTQAAK 2 | ||
1 | 1 LGILRDKLSLQGLKNIHYIIVNEKTLESRAMFWKLKLKTPKNITVYQQSAFQPDVWRILRGNKDDFLIYDR 2 | ||
1 CGKLTFHITSPYSYLNFRYVEAAIMATYNTDYCGNCM 0 | |||
0 GSSTTLEATSYISTGPNWEYSQHHSRSLK* 0 | |||
0 | |||
> | >SEPP2_calMil Callorhinchus milii (elephantfish) | ||
1 LGSLQDKLARQGLKDVHYMIVNEKAPESRAMLWELKRHVPNNVSVYQQSPIQPDVWHSLQGGKDDFLIYDR 2 | |||
1 | 1 CGRLTFHVVLPYSSLQYPYIEAAIRATHKRDICGECT 0 | ||
</pre> | |||
=== SELU1: 13 vertebrate sequences === | |||
> | <pre> | ||
0 | >SELU1_homSap Homo sapiens (human) chr10 processed pseudogenes chr8 and chr12 | ||
0 MSFLQDPSFFTMGMWSIGAGALGAAALALLLANTDVFLSKPQKAALEYLEDIDLKTLEK 1 | |||
0 | 2 EPRTFKAKELWEKNGAVIMAVRRPGcFLCREE 0 | ||
0 | 0 AADLSSLKSMLDQLGVPLYAVVKEHIRTEVKDFQPYFKGEIFLDEK 0 | ||
0 | 0 KKFYGPQRRKMMFMGFIRLGVWYNFFRAWNGGFSGNLEGEGFILGGVFVVGSGKQ 0 | ||
0 GILLEHRENEFGDKVNLLSVLEAAKMIKPQTLASEKK* 0 | |||
> | >SELU1_monDom Monodelphis domestica (opossum) tgt-cys | ||
0 2 | 0 MSFLDLNFFSMSMWSLGAGALGAAALSLILANTDLFLTKSVDATLEFLEEIQLKTLDN 1 | ||
2 ESPKTFKARELWEHRGAVIMAVRRPGCFLCREV 0 | |||
0 | 0 AADLSALKPQLDLLGVPLYAVVKEKIGSEVENFQPYFKGKIFLDER 0 | ||
0 | 0 KKFYGPQKRKMMFMGFVRLGVWQNFFRARSKGFSGNLEGEGFVLGGVYVIGPGKQ 0 | ||
0 | 0 GILLEHREKEFGDKVNPASVLEAAKKIKPHTSTSEGK* 0 | ||
> | >SELU1_macEug Macropus eugenii (wallaby) EX196548 full | ||
0 | 0 MSFLDLSFLSMGMWSLGAGALGAAVLSLILANTDVFLTKSVTATLEFLEDIELKTLDN 1 | ||
2 KTFKARELWEHRGAVIMAVRRPGCFLCREE 0 | |||
0 | 0 AADLSALKPQLDQLGIPLYAVVKEKIGSEVEDFQPYFKGKIFLDER 0 | ||
0 | 0 KKFYGPQKRKMMFMGFVRLGVWQNFFRARSKGFSGNLEGEGFILGGVYVIGPRKQ 0 | ||
0 | 0 GILLDHREKELGDKVNPASVLEACKKIKLHA* 0 | ||
> | >SELU1_triVul Trichosurus vulpecula (opossum) EC360881 | ||
0 | 0 MSFLDLSFFSMGMWSLGAGALGAAVLSLILANTNLFLTKSVTATLEFLEEIELKTLDN 1 | ||
2 ESPKTFKARELWEHRGAVIMAVRRPGCFLCREE 0 | |||
0 | 0 AAELSALKPQLDQLGIPLYAVVKEKIGSEVENFQPYFKGKIFLDER 0 | ||
0 | 0 KKFYGPQKRKMMFMGFVRLGVWQNFFRARSKGFSGNLEGEGFILGGVYVIGPGKQ 0 | ||
0 | 0 GILLEHREKEFGDKVDPASVLEAA * 0 | ||
> | >SELU1_ornAna Ornithorhynchus anatinus (platypus) taa early stop full | ||
0 | 0 MPLPPDLGLFNLGMWSVGVGALGAAAVGLLLANTDLLLTKPEKATLEYLEDTELKTLGK 1 | ||
2 EPRTFKARELWQRNGAVIMAVRRPGuFLCREE 0 | |||
0 | 0 AAELSSLKPQLDRLGVPLYAVVKEKIGTEVEDFQPYFKGEIFLDER 0 | ||
0 | 0 KKFYGPHKRKMLFLGFIRLGVWQNFLRARNRGFSGNLEGEGLILGGVYVLGAGKQ 0 | ||
0 | 0 GILLEHREREFGDKVSPASVLEAAQRIKPQPL* 0 | ||
> | >SELU1_tacAcu Tachyglossus aculeatus (echidna) 454:EUEMSW405C31QQ (74%) tSASEKK terminus? frag | ||
0 | 0 1 | ||
2 EPRTFKARELWQRNGAVIMAVRRPGuFLCREE 0 | |||
0 | 0 AAELSSLKPQLDQLGVPLYAVVKENIGTEVEDFQPYFKGEIFLDER 0 | ||
0 | 0 KRFYGPHKRKMLFLGLIRLGVWQNFIRARNKGFPPVTWEGEG 0 | ||
0 | 0 GVLLEHREREFGDKVSPASVLEAAQKIKPQ* 0 | ||
> | >SELU1_galGal Gallus gallus (chicken) | ||
0 2 | 0 MSFLPDFGIFTMGMWSVGLGAVGAAITGIVLANTDLFLSKPEKATLEFLEAIELKTLGS 1 | ||
2 EPRTFKASELWKKNGAVIMAVRRPGuFLCREE 0 | |||
0 | 0 ASELSSLKPQLSKLGVPLYAVVKEKIGTEVEDFQHYFQGEIFLDEK 0 | ||
0 | 0 RSFYGPRKRKMMLSGFFRXGVWQNFFRAWKNGYSGNLEGEGFTLGGVYVIGAGRQ 0 | ||
0 | 0 GVLLEHREKEFGDKVSLPSVLEAAEKIKPQAS* 0 | ||
> | >SELU1_taeGut Taeniopygia guttata (finch) | ||
0 | 0 msflpdfgiFTMGMWSVGLGAIGAAVTGIVLANTDLFLSKPEKATLEFLEEIELKTLGS 1 | ||
2 EKRTFKAGELWKQNGAVIMAVRRPGuFLCREE 0 | |||
0 | 0 ASELSSLKPQLSKLGVPLYAVVKENIGTEVEDFQHYFKGEIFLDEK 0 | ||
0 | 0 KGFYGPRRRKMMLSGFFRLGVWQNFVRAWRSGYSGNLEGEGFTLGGVYVIGAGRQ 0 | ||
0 | 0 GVLLEHREKEFGDKVSLPSVLEAAEKIKPQAS* 0 | ||
> | >SELU1_anoCar Anolis carolinensis (lizard) | ||
0 | 0 MWTIGLGAIGAAVTGIILANTDLFLSKAEQASLDFLEAIDLKTLGE 1 | ||
2 NQRTFKAEELWKKNGAVIMAVRRPGuFLCREV 0 | |||
0 | 0 AAELSSLKPQLDKLGVPLYAVVKENLGTEVMDFQPYFKGEIFLDEK 0. | ||
0 | 0 KQFYGPQKRKMLFMGFIRCSVWRNFFRAWKSGYTGNIDGEGFVLGGVFVVGPGKQ 0 | ||
0 | 0 GVLLEHREKEFGDKVSLDAVLEAVKNIQPQPSEKDK* 0 | ||
> | >SELU1_takRub Takifugu rubripes (fugu) | ||
0 | 0 MGLLAKLLAAVGGFVTAVMNSVTDAFLTPPLRATLEHLEETDLKTLSG 1 | ||
2 ALVIRLIPTRTETKTFKAKSLWENSGAVVMAVRRPGuFLCRE 0 | |||
0 | 0 EAAELSSLKPRLDQLGVPLYAVVKEDVGTEIQNFRPYFQGEIFLDEK 0 | ||
0 RRFYGPRERKMGLLGFLRVGVWMNGLRAFRSGFMGNVLGEGFVLGGVFVIGREQQ 0 | |||
0 GILLEHREREFGDKVNIEDVIQAVDRIAQELMPVTQN* 0 | |||
> | >SELU1_calMil Callorhinchus milii (elephantfish) frag | ||
1 GAuGYEPRYQKLAIVIKDEFPDADVSGKVGRT 1 | 2 ENRTFRASELWAGRGAVIMAVRRPGuFLCRE 0 | ||
2 GSFEIEINGQLIFSKLETGGFPYEND 0 | 0 AAALSSLRPSLAQLGVPL | ||
0 ISEAVQKANNGEELQKIENSRPPCVIL* 0 | 0 GHLLEHREKEFGDAVNLTAVMEAAGKISPRQSAE* 0 | ||
> | >SELU1_squAca Squalus acanthias (spiny_dogfish) also selenocysteine | ||
0 MPLKIHIVYC 2 | 0 MVVVVEDFHMGLWTLGLGALGAAITGVILANTDLLLPKAETASLAYLSGAELRTLDR 1 | ||
1 GAuGYRSR 0 | 2 EERTLKAGDLWSRSGAVIMVVRRPGuFLCREE 0 | ||
0 FHRLKDELETEFPGELEI 0 | 0 AAEISSLRPQLDELGVPLYGVIKENINNELKNFQPFFKGEIFLDVE 0 | ||
0 TGEGTPTQTGFLEVQIVGGKLLHSKA 0 | 0 MRFYGPKPRTMGLMGFMRLGVWKNFVRAWQKGFSGNTDGEGFILgGVFVIGAGQQ 0 | ||
0 NGDGFVDSDEKLQKIFSGVEKALKK* 0 | 0 GVLLEHREKEFGDVVNISSVLEARRKIETQRTEP* 0 | ||
> | >SELU1_braFlo Branchiostoma floridae (amphioxus) 8.9 m traces somewhat different intronation | ||
0 MPLKIHVVYC 2 | 0 MGWRLLVA 00 TMGWVVNAVGVAVVGMVAVNLNFWLPKHIPAQLVHIA 1 | ||
1 GSuGYASK 0 | 2 DAALEELDGNKKKLQAQELWKNHGAVIMAVRRPGuQLCREE 0 | ||
0 FRALKVKLDHEFPGKLEI 0 | 0 ALGLSSLKPQLDRAGVPLYAVVHERKGVPEFQPYFQGKVFLDLE 0 | ||
0 TSEGTPGLTGKFEVQVGEKLVHSKK 0 | 0 RRFYGPHERWMSLAGLLRVNFWLNIRRVEKKVEGNYEGEGRLLGGVFVVGSGNQGILMQHAEQEFGHHANLTDIMKAVGEIQHINAKSKI* 0 | ||
0 NGDGFVDSPEKLQKIFKAVENALKGQ* 0 | </pre> | ||
> | === SELU2: 1 vertebrate sequence === | ||
0 MVVIVKVIYC 2 | |||
1 GGuGYGPR 0 | <pre> | ||
0 YRRLEQELKDEFGDDVDM 0 | >SELU2_homSap Homo sapiens (human) 7 exons chr1p36.32 36% id NM_152371 | ||
0 VGESTPGVTGWLEVEVNGKLIHSKK 0 | 0 MSTVDLARVGACILKHAVTGE 0 | ||
0 NGDGYIDSEGKLKKIVNAVSAAM* 0 | 0 AVELRSLWREHACVVAGLRRFGCVVCRWIAQDLSSLAGLLDQHGVRLVGVGPEALGLQEFLDGDYFAG 1 | ||
2 ELYLDESKQLYKELGFKR 2 | |||
</pre> | 1 YNSLSILPAALGKPVRDVAAK 0 | ||
0 AKAVGIQGNLSGDLLQSGGLLVVSK 1 | |||
=== DIO1: 50 vertebrate iodothyronine deiodinases === | 2 GGDKVLLHFVQKSPGDYVPKEHILQVLGISAEVCASDPPQ 0 | ||
<pre> | 0 CDREV* 0 | ||
>DIO1_homSap Homo sapiens (human) iodothyronine deiodinase type I 4 exons chr1 uc001cwb.1 | |||
0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWEFMQ 1 | >SELU2_strPur Strongylocentrotus purpuratus (sea_urchin) | ||
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLVIYIEEAHAS 1 | 0 MAFDLSKIASNLVKNFATGE 0 | ||
2 DGWAFKNNMDIRNHQNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYIIQEGRILYK 0 | 0 MVAISSLWEKQACVIHFMRRFGuSACRLGASELDSLRPQLDEADVRLVGIGLEDLGAQEFLDGGFWKG 1 | ||
0 GKSGPWNYNPEEVRAVLEKLHS* 0 | 2 DLFIDQQQKSYQGLGFKKYSLMSIMKAVFSAKTRAALSR 0 | ||
0 ASDKKIKANLKGDKLQIGGTLIISK 1 | |||
>DIO1_panTro Pan troglodytes (chimp) | 2 GGEKVLVDFKQEAPGDHIPLQTVLDAFGIKGETPKTEGAEGGTVVCNDDVCQMK* 0 | ||
0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVHLSGQRCNIWEFMQ 1 | </pre> | ||
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLVVYIEEAHAS 1 | |||
2 DGWAFKNNMDIRNHQNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYVIQEGRILYK 0 | === SELU3: 3 eukaryote sequences === | ||
0 GKSGPWNYNPEEVRAVLEKLHS* 0 | |||
<pre> | |||
>DIO1_ponPyg Pongo pygmaeus (orang_sumatran) | >SELU3_homSap Homo sapiens (human) 6 exons chr9q22.32 25% id processed pseudogene chrX | ||
0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTETGGLAPNCPVVRLSGQRCNIWEFMQ 1 | 0 MAAPAPVTRQVSGAAALVPAPSGPDSGQPLAAAVAELPVLDARGQRVPFGALFRERRAVVVFVR 0 | ||
0 HFLCYICKEYVEDLAKIPRSFLQ 0 | |||
>DIO1_macMul Macaca mulatta (rhesus) | 0 EANVTLIVIGQSSYHHIE 0 | ||
0 MGLPQSGLWVKKLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWDFMQ 1 | 0 PFCKLTGYSHEIYVDPEREIYKRLGMKRGEEIASS 1 | ||
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLIIYIEEAHAS 1 | 2 GQSPHIKSNLLSGSLQSLWRAVTGPLFDFQGDPAQQGGTLILGP 1 | ||
2 DGWAFKNNMDIRNHRNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYVIQEGRILYK 0 | 2 GNNIHFIHRDRNRLDHKPINSVLQLVGVQHVNFTNRPSVIHV* 0 | ||
0 GKSGPWNYNPEEVRAVLEKLYS* 0 | |||
>SELU3_petMar Petromyzon marinus (lamprey) cDNA EE741479 ti|1430987375 tga-sel taa-stop frag | |||
>DIO1_macFas Macaca fascicularis (crab_macaque) | VRPDPYIYKALELKMGEDVDKIY 1 | ||
0 MGLPQSGLWVKKLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWDFMQ 1 | 2 KSPHVHCGLWWGVARGLWRAMWSESFDFQGDPKQQGGALVLGP 1 | ||
2 GGRVLFSHRDEAVLDHTPINRLLAVAGIPPVDFTHKHMVKHVuPRPLTPTG* 0 | |||
>DIO1_macNem Macaca nemestrina (pigtailed_macaque) | |||
0 MVLPQSGLWVKKLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGPSCNIWDFMQ 1 | >SELU3_monBre Monosiga brevicollis (choanoflagellate) tga-sel | ||
MATGAGVGASWEDVAKMTVYDHQQEPCSLGSLCSPGQMTAIGTLARDNSARVPPPLITHHATLPPRVCRLVFVR | |||
HFLuFVCKDYVTDLARVPDEHWA | |||
GARVVVIGCANPVFID | |||
RFAADTGFPASNVYCDPKREVYQALQLPSVLAAINA | |||
SKSIHIHSSALGGTMNSIYRGLRAMRAQGNVKQQGGAFVVNG | |||
QGQLIYAHRDQRPEDHCPINTLLEQVGGRPFF* | |||
</pre> | |||
=== SELV: 19 placental mammal sequences === | |||
<pre> | |||
>SELV_homSap Homo sapiens (human) chr 19 uc002oly.1 66 pro + 32 thr +21 ser first exon | |||
0 MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTLTPVLTPSPAGTSPLVLTPAPAQIPTLVPTPALARIPRLVPPPAPAWIPTPVPT | |||
PVPVRNPTPVPTPARTLTPPVRVPAPAPAQLLAGIRAALPVLDSYLAPALPLDPPPEPAPELPLLPEEDPEPAPSLKLIPSVSSEAGPAPGPLPTRTPL | |||
AANSPGPTLDFTFRADPSAIGLADPPIPSPVPSPILGTIPSAISLQNCTETFPSSSENFALDKRVLIRVTYC 2 | |||
1 GLuSYSLR 0 | |||
0 YILLKKSLEQQFPNHLLF 0 | |||
0 EEDRAAQATGEFEVFVNGRLVHSKK 0 | |||
0 RGDGFVNESRLQKIVSVIDEEIKKR* 0 | |||
>SELV_panTro Pan troglodytes (chimp) essentially identical to humna | |||
MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTLTPVLTPSPAGTPPLVLTPAPAQIPTLVPTPALARIPRLVPPPAPAWIPTPVPTPV | |||
PVRNPTPVPTPARTLTPPVRVPAPAPAQLLAGIRAALPVLDSYLAPALPLDPPPEPAPELPLLPEEDPEPAPSLKLIPSVSSEAGPAPGPLPTRTPLAANS | |||
PGPTLDFTFRADPSATGLADPPIPSPVPSPILGTIPSAISLQNSTETFPSSSENFALDKRVLIRVTYC 2 | |||
1 GLuSYSLR 0 | |||
0 YILLKKSLEQQFPNHLLF 0 | |||
0 EEDRAAQATGEFEVFVNGRLVHSKK 0 | |||
0 RGDGFVNESRLQKIVSVIDEEIKKR* 0 | |||
>SELV_ponPyg Pongo abelii (orang_borneo) | |||
0 MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTLTPVLTPSPAGTPPLVLTPAPAQIPTLVPTPALARIPRLVPPPAPAWIPTPVPTPVPVRNPTPVPTPARTLTTPV | |||
RVPAPAPAPPPAQVLAGIRAALPILDSYLAPALRLHPPPEPAPELPLSPEEDPEPAPSLKLIPSVSSEAGPAPGPLPARTPQAANSPGPTLDFTFRADLSATGLADPPIPSPVPSPILGT | |||
TSSAISLQNSTENFASSSENFALDKRVLIRVTYc 2 | |||
1 GLuSYSLR 0 | |||
0 YILLKKSLEQQFPNHLLF 0 | |||
0 EEDRAAQATGEFEVFVNGRLVHSKK 0 | |||
0 RGDGFVNESTLQKIVSVIDEEIKKR* 0 | |||
>SELV_macMul Macaca mulatta (rhesus) distal seq from M. nemestrina transcript | |||
0 MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTQTPVLTPSPARLPALVQPPAPAHIPTQVPTPALARIPGLVPPPARAWIPTPVPTPARVRNPTPVPTPARTLTPPV | |||
RVPAPAPDPAQVLAGIRTALPALDSYPAPALSLDSPPEPAQEPPLSPEEDPEPAPSLKLIPSVSSEAGPALGPLPAHTPLAAKSSGPTLDFTFRADPSATGLADPHIPSPVPAPILGTIP | |||
SALSLQNFTETLVSTSENFALDKRVLIRVTYC 2 | |||
1 GLuSYSLR 0 | |||
0 YILLKNSLEQQFPSHLLF 0 | |||
0 EEDRAAQATGEFEVFVNGRLVHSKK 0 | |||
>SELV_calJac Callithrix jacchus (marmoset) | |||
0 MNNQARTPAPSSARTSTSVRASTPTRTPTPVRTPTAVRIRTPIRTLTPSLAGTPALVPTPTPARISRLVPTPAPARTPTPIPTLVRTLTPVPLPAPARIPAPAPAPAPAPAPAPSPALVP | |||
AGIRATLPVLDSYPALALPWDPPPEPVPEPLVSVSSEEDPEPAPSLKLVPSVSGETGPAPGPLPACTPLATNPPEPTLDFTADSSATELAVPTIPGSVPAPILGTIPLAASLLNSTESFL | |||
SASENFALDKRVPIRVTYc 2 | |||
1 GLuSYSLR 0 | |||
0 YILLKKSLEQRFPDCLLF 0 | |||
0 EEDRAAQATGDFEVFVDGRLVHSKK 0 | |||
0 RGDGFVDEASLQKIVSVIDEEIKKR* 0 | |||
>SELV_otoGar Otolemur garnettii (bushbaby) | |||
0 MNSQARASVHPSRTSTAVRASIPARVHPRARTTPVQPRTLITQDRIPAPVRVPP | |||
1 GLuSYGLQ 0 | |||
0 YILLRQNLEHHFPNRLLF 0 | |||
0 EEGRAAQATGEFEVFVDGKLVHSKK 0 | |||
0 NGDGFVDEIRLQKIVNIIDEEIKKRQ 0 | |||
>SELV_musMus Mus musculus (mouse) AV279316 58 Pro, 25 Thr, 25 Ser syntenic chr 7 46% hsa! | |||
MNNKARVPAPSSVRANTPARTPAPIRTATPVRAPNPAHNSTPVRTSIRVRAPAQVPNPVPIRFPTPAPVPAPTLTPAPTPAPVRHAAPVRTPAPVRAPNLGRV | |||
FPKISPGRFFPSLASPTAQPLSSRAASALLKDPTLAQNQKPSIHSLAEAIQGPLPVLTPSSSKTQGSIPDTASPIDSLASTAMASSTLGPIPGPNPTLEFLAS | |||
PLKETPGLGKLSTISPAPSFGSTKEIPSTSEDVPTPNRILIRVMYC 2 | |||
1 GLuSYGLR 0 | |||
0 YIILKRTLEHQFPNLLEF 0 | |||
0 EEERATQVTGEFEVFVDGKLIHSKK 0 | |||
0 KGDGFVDESGLKKLVGAIDEEIKKR* 0 | |||
>SELV_ratNor Rattus norvegicus (rat) chr 1 synteny 83% mmu | |||
MNNKARNPAPSSVRANTPSRTPTPVRTATPVRASTPAHNRSPVRTSIRVRTPANPVPIRFPTPAPAPAPTPTPAPTPAPVPAAAPVRTPAPVRASIQGRSFPTIS | |||
PVRFLRNLALPAAQPLSSGGAGSLSKDLTLAQKQKPSIHSLAEAIQGPFPVLTPSASSETHGSIPDPAPPTDSLASTAMASSTLDPIPGPKPTLEFLASPLKETP | |||
DLGKLSTISPAQNFVSTKEVPSTSEDVPTANRILIRVMYC 2 | |||
1 GLuSYGLR 0 | |||
0 YILLKKTLEHQFPNLLEF 0 | |||
0 EEERATQVTGEFEVFVDGKLIHSKK 0 | |||
0 KGDGFVDETSLKKLVGAIDEEIKKR* 0 | |||
>SELV_cavPor Cavia porcellus (guinea_pig) | |||
1 GLuSYGFQ 0 | |||
0 YTLLKMSLKQQFPNLLRF 0 | |||
0 EDERATQVTGEFEVFVEGKLVHSKK 0 | |||
0 QGDGFVDDNRMQTIVNAINEEIKRR* 0 | |||
>SELV_oryCun Oryctolagus cuniculus (rabbit) | |||
0 MNNQARTPAPAQARTSSVVRASAPTRVSTQIRTPATGWTPTPVQASTPVRTQTPVRTPTLVQASTPVRTPTLGQASTPVRTPTPVQALTPVRTPTPVSGPDSGRTPTPVGTPVGTL | |||
1 GLuSYGLR 0 | |||
0 YILLKKNLEELFPDCLLF 0 | |||
0 EEERATQASGEFEVFVNGKLVHSKK 0 | |||
0 KGDGFVDEVKLRKIVTAINEEIK... 0 | |||
>SELV_canFam Canis familiaris (dog) CO599097 | |||
0 MNNQARAPPRTSARVLAWVRASTPVRTSIPVRTPTPARIPTSSRAPTSVQTPAPARTPNPVQIPTPVQTSTSARIPNPARTLTPVQTPASAWTPNPVQMLTPARTPTPVPTP | |||
VPTPIPARTPTPARTPTPVEAAAAAPASESFGSSALPLEPPPEPASEPTTSPHQDLSPTPSVKPLPSVTNGFGSTQEPLPDLTPPATDFLGPTLGSTSRADSSATKLTDSSESVR | |||
VPIPGTPSATALATSTNTFAPVGESCSVKIAVRVIYC | |||
1 GLuSYGLR 0 | |||
0 YILLKKSLEQQFPNCLLF 0 | |||
0 EEERAAQATGEFEVFVDGKLVHSKK 0 | |||
0 KGDGFVDEARLQKIMNVIEEEVRKR* 0 | |||
>SELV_felCat Felis catus (cat) | |||
0 MNNQARAPNPSPARVLALVRASTPVRSSIPVRIPTPARIPARTRNPQTPVRAPNPVQIPNPVQISARASNPARAPRPVQIPARTPNPPQTLTPGRAPNPVPTPVQTPTLVQTP | |||
TPVQTPTPVWTETPVQTPTAVGTPTRVQTLTPVIPRVRTPTPIRPLWPDPSPNLIPSGPDHPAPESSLRNSAPSFWINSPDSLPAPILETPSTAFASTFENLPEDSKILIRVIYC | |||
1 GLuSYSLR 0 | |||
0 YILLRKSLELQFPNCLRF 0 | |||
0 EEERSAQATGEFEVFVDGKLVHSKK 0 | |||
0 KGDGFVDEARLQKIVSTIDEEIRKR* 0 | |||
>SELV_bosTau Bos taurus (cow) DT829759 | |||
0 MNNQPRTPAPTPARASTPVRGSTLHRTSIQVRTPTPGPDSGPTRITTLLRTPALIRTLTPIRTPTPVRTPTPVPPSTPVRSPIPVRTPTPVPPSTPVRTPTPVPLSTPVRPPTPVRPPTPVRPPTPVRPPTP | |||
IGTPTLIRSPTPVQIPIPEPIPTPIPSRVLIPPLESFPDSALPSGPPLELEPTLTVSPAKNLEPSPAKNLEPSPRVKQVSSAANGFPPIQEPLPALTPLATDLRSPSLGSPLRTDTSTTNLIASSSGHVPGTPILGAIQAILPVPATALASISGNLKEENKIMIRVVYC | |||
1 GLuSYSLR 0 | |||
0 YILLRKSLEQQFPNSLIF 0 | |||
0 EEEISAQATGEFEVFVDGKLIHSKK 0 | |||
0 NGDGFVDEVKLQTIVNLLNEEFKKR* 0 | |||
>SELV_susScr Sus scrofa (pig) ti|851198642 CX061656 | |||
0 MNNPARAPAPSPVRPSASLRPLASVRASTPARGSTLARTSLLVRTPRTPNLVPASGPGPIRTPTPVRTPTPVRTPTPVRTLTPVRTPTPVRTPTPVRTPTPVRTPTPVRTPTPVQVPTPVRVPTTVGIPTP | |||
TLSQVLVPALESLPNPALPSLDPPLEPDPELTLSPDEDPAPTPRAKHLPLVANGFVPVQEPLPALSPLATNLLESTPGAGTDSSTTKLTDSTSGHVPGTPILATIPLAVALPVSTNALASTSENIQVEQQILIRVVYC 2 | |||
1 GL*SYGLR 0 | |||
0 YILLKKSLEQQFPNCLVF 0 | |||
0 EEDISAQATGAFEVFVDGKLVHSKK 0 | |||
0 KGDGFVDETKLQKIVSHINEEIKTKVAGSC* 0 | |||
>SELV_equCab Equus caballus (horse) | |||
1 GLuSYGLR 0 | |||
0 YILLRKSLDQQFPNRLVF 0 | |||
0 EEDVGAQASGEFEVFVEGKLVHSKK 0 | |||
0 KGDGFVDEARLRKIVSAINEEIKRR* 0 | |||
>SELV_myoLuc Myotis lucifugus (microbat) | |||
0 MNHQARASHPFPARTPASVRASIPVRPSNQTPAPSSAPTRTSIPVPASTAVRTPTLVRTPIPFQAVSPVRTSTRFSASVPVQFPTPVSASTPVQTPTRVPAPVRTSTRFPASV | |||
PVQFPTPVPASTPVRTSTPVPASTPVQTPTRVPAPAQVRTSIPVPAPAPARTSTPVPAPAPVRTSTTVPAPVPFRTPTPVPAPAPVRTSTPVPAPAPVRTSTPVPAPGPNPDSSPCPGPG | |||
1 GLuSYNLR 0 | |||
0 YILLKKSLEQQFPNCLYF 0 | |||
0 EEDISEQATGEFEVFVDGKLVHSKK 0 | |||
0 KGDGFVDEVKMQKIVNIIDEEIRKN* 0 | |||
>SELV_pteVam Pteropus vampyrus (macrobat) | |||
0 MNNKARASAPSLPGTSALVRASAPVRPSTPVRAPTLIRTPTPVRTPNPVPASTPARVPTPVRTLTPVRTPTPIRTPTPVRTQIPFRAANPVRAANPVRTPTPLRTPTPVRTSIPVRTL | |||
APVSTPTPIPPPTPPAVPAVVPAAVPGLEFFPSPPPEPVPEPALSPDKDPAPTQSVKHVPSVANEFGLTQEPLSALAPLATDLLGPTRASIRRADPEATKLTGSTAEPSPASILELISSAHIGIASEYLQVDTGSVIRVILL | |||
1 GLuSYNFR 0 | |||
0 YILLRKNLEQQFPHGLLF 0 | |||
0 EEEVSAEGTGEFEVLVDGKLIHSKK 0 | |||
0 RGDGFVDEARLQKIVNVINE IRKR* 0 | |||
>SELV_dasNov Dasypus novemcinctus (armadillo) ti|593909023 | |||
1 GLuSYALR 0 | |||
0 YILLKKSLEMQFPNRLIF 0 | |||
0 EEARSSQAVGEFEVFVDGALVHSKK 0 | |||
0 RGDGFVDDSKMEKIVSSIEEAVKKT* 0 | |||
>SELV_loxAfr Loxodonta africana (elephant) | |||
0 2 | |||
1 GLuSYGLR 0 | |||
0 YILLRKSLEQQFPNHLIF 0 | |||
0 EEDISSQATGEFEVFVDGKLVHSKK 0 | |||
0 KGDGFVDDTRLQKIVNSINETIKKE* 0 | |||
>SELV_proCap Procavia capensis (hyrax) | |||
0 2 | |||
1 GLuSYSLR 0 | |||
0 YILLRKNLEQQFPNHLLF 0 | |||
0 EVDISSQATGEFEVFVDGKLVHSKK 0 | |||
0 KGDGFVDDAQLQKIVNSINETIQRR* 0 | |||
</pre> | |||
=== SEPW1: 29 vertebrate sequences === | |||
<pre> | |||
>SEPW1_homSap Homo sapiens (human) Selenoprotein W chr19 87 aa uc002phn.1 has retroprocessed pseudogene | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGRLDI 0 | |||
0 CGEGTPQATGFFEVMVAGKLIHSKK 0 | |||
0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_panTro Pan troglodytes (chimp) | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGRLDI 0 | |||
0 RGEGTPQATGFFEVMVAGKLIHSKK 0 | |||
0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_ponPyg Pongo pygmaeus (orang_sumatran) CR926472 | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGRLDI 0 | |||
0 CGEGTPQATGFFEVMVAGKLIHSKK 0 | |||
0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_macMul Macaca mulatta (rhesus) | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGRLDI 0 | |||
0 CGEGTPQATGFFEVMVAGKLIHSKK 0 | |||
0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_macFas Macaca fascicularis (cynomolgus_monkey) AB169486 | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGRLDI 0 | |||
0 CGEGTPQATGFFEVMVAGKLIHSKK 0 | |||
0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_papAnu Papio anubis (baboon) EY285690 | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGRLDI 0 | |||
0 CGEGTPQATGFFEVMVAGKLIHSKK 0 | |||
0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_calJac Callithrix jacchus (marmoset) | |||
0 MALTVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGRLDI 0 | |||
0 SGEGTPQATGFFEVTVAGKLIHSKK 0 | |||
0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_micMur Microcebus murinus (mouse_lemur) | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGCLDI 0 | |||
0 CGEGTPQATGFFEVMVAGKLVHSKK 0 | |||
0 GDGYVDTESKFLKLV | |||
>SEPW1_musMus Mus musculus (mouse) | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKPK 0 | |||
0 YLQLKEKLEHEFPGCLDI 0 | |||
0 CGEGTPQVTGFFEVTVAGKLVHSKK 0 | |||
0 RGDGYVDTESKFRKLVTAIKAALAQCQ* 0 | |||
>SEPW1_ratNor Rattus norvegicus (rat) BC087625 | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKPK 0 | |||
0 YLQLKEKLEHEFPGCLDI 0 | |||
0 CGEGTPQVTGFFEVTVAGKLVHSKK 0 | |||
0 RGDGYVDTESKFRKLVTAIKAALAQCQ* 0 | |||
>SEPW1_cavPor Cavia porcellus (guinea_pig) | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKPK 0 | |||
0 YLQLKEKLEDEFPGCLDI 0 | |||
0 CGEGTPQTTGFFEVTVAGKLVHSKK 0 | |||
0 GGDGFVDTEGKFRKLVAAIKAALAQG* 0 | |||
>SEPW1_oryCun Oryctolagus cuniculus (rabbit) | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKPK 0 | |||
0 YLQLKKKLEDEFPGCLDI 0 | |||
0 CGEGTPQVTGFFEVTVAGKLVHSKK 0 | |||
0 RGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_ochPri Ochotona princeps (pika) | |||
0 MALSVRVVYW 2 | |||
1 GAuGYKPK 0 | |||
0 YLQLKKRLEDEFPGCLDI 0 | |||
0 GEGTPQVTGFFEVMVAGKLVHSKK 0 | |||
0 SGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_canFam Canis familiaris (dog) | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGCLDI 0 | |||
0 RGEGTPQATGFFEVTVAGKLVHSKK 0 | |||
0 RGDGYVDTESKFLRLVAAIKTALAQG* 0 | |||
>SEPW1_felCat Felis catus (cat) | |||
0 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGCLDI 0 | |||
0 RGEGTPQATGFFEVMVGGKLVHSKK 0 | |||
0 RGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_bosTau Bos taurus (cow) | |||
0 MAVVVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPSRLDI 0 | |||
0 RGEGTPQVTGFFEVFVAGKLVHSKK 0 | |||
0 GGDGYVDTESKFLKLVAAIKAALAQA* 0 | |||
>SEPW1_oviAri Ovis aries (sheep) | |||
0 MAVVVRVVYC 2 | |||
1 GAuGYKPK 0 | |||
0 YLQLKKKLEDEFPSRLDI 0 | |||
0 CGEGTPQVTGFFEVFVAGKLVHSKK 0 | |||
0 GGDGYVDTESKFLKLVAAIKAALAQA* 0 | |||
>SEPW1_susScr Sus scrofa (pig) AF380118 | |||
0 MGVAVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGRLDI 0 | |||
0 CGEGTPQVTGFFEVLVAGKLVHSKK 0 | |||
0 GGDGYVDTESKFLKLVAAIKAALAQG* 0 | |||
>SEPW1_eriEur Erinaceus europaeus (hedgehog) | |||
0 MALAVRVVYC 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGCLDI 0 | |||
0 RGEGTPQGTGFFEVLVAGKLVHSKK 0 | |||
0 KGDGYVDTETKFLKLVTAIKAALAQG* 0 | |||
>SEPW1_sorAra Sorex araneus (shrew) | |||
0 2 | |||
1 GAuGYKSK 0 | |||
0 YLQLKKKLEDEFPGCVDV 0 | |||
0 CGEGTPQVTGFFEVMVAGKLVHSKK 0 | |||
0 RGDGYVDSESKYVRLVTAIKTALAQA* 0 | |||
>SEPW1_choHof Choloepus hoffmanni (sloth) | |||
0 MALAVRVVYW 2 | |||
1 GAuGYKPK 0 | |||
0 YVQLKKKLEDEFPGCLDI 0 | |||
0 SGEGTPQTTGFFEVMVAGKLVHSKK 0 | |||
0 QKGDGFVDTESKFLRLVAAIKAALAQG* 0 | |||
>SEPW1_monDom Monodelphis domestica (opossum) diverged | |||
0 MAIQVRVVYW 2 | |||
1 GAuGYKPK 0 | |||
0 YLLLKKKLEDEYPGLLRH 0 | |||
0 NGEGTPEVTGFFEVTVAGKLVHSKK 0 | |||
0 AGHGFVDTADKYLQIVAEIKAALA* 0 | |||
>SEPW1_ornAna Ornithorhynchus anatinus (platypus) | |||
0 MASLEAFPRGVVPVHVVYC 2 | |||
1 GAuGYKPK 0 | |||
0 FLQLKKKLENEFPGQVEI 0 | |||
0 SGEGTPQVTGWFEVTVAGKLVHSKK 0 | |||
0 EGDGFVDSESKFAKIRMAIKAALVPGY* 0 | |||
>SEPW1_galGal Gallus gallus (chicken) tga confirmed | |||
0 MPLRVTVLYC 2 | |||
1 GAuGYKPK 0 | |||
0 YERLRAELEKRFPGALEM 0 | |||
0 RGQGTQEVTGWFEVTVGSRLVHSKK 0 | |||
0 NGDGFVDTNAKLQRIVAAIQAALP* 0 | |||
>SEPW1_anoCar Anolis carolinensis (lizard) | |||
0 2 | |||
1 GAuGYSPK 0 | |||
0 YQQLKRGLEKEFPGKLEI 0 | |||
0 TGEGTPQVTGWFEVTVAGKLVHSKK 0 | |||
0 NGDGFVDNDTKLHKILMAIKAALA* 0 | |||
>SEPW1_xenTro Xenopus tropicalis (frog) tga confirmed | |||
0 MPDTMVKVNVVYC 2 | |||
1 GAuGYLSK 0 | |||
0 FRRLKKELEQRFPGKLSI 0 | |||
0 DGEGTERMTGWFEVSINGKLVHSKK 0 | |||
0 NGDGYVDNDAKLQKIILAIEAALKQ* 0 | |||
>SEPW1_danRer Danio rerio (zebrafish) tga confirmed | |||
0 MTVKVHVVYC 2 | |||
1 GGuGYRPK 0 | |||
0 FIKLKTLLEDEFPNELEI 0 | |||
0 TGEGTPSTTGWLEVEVNGKLVHSKK 0 | |||
0 NGDGFVDSDSKMQKIVTAIEQAMGK* 0 | |||
>SEPW1_takRub Takifugu rubripes (fugu) | |||
0 MGVTIRVEYC 2 | |||
1 GGuGYGPR 0 | |||
0 YEELARVVRAEFPDADVSGFVGRM 1 | |||
2 GSFEIQINEQLIFSKLETGGFPYEDD 0 | |||
>SEPW1_calMil Callorhinchus milii (elephantfish) | |||
1 GAuGYEPRYQKLAIVIKDEFPDADVSGKVGRT 1 | |||
2 GSFEIEINGQLIFSKLETGGFPYEND 0 | |||
0 ISEAVQKANNGEELQKIENSRPPCVIL* 0 | |||
>SEPW1_squAca Squalus acanthias (spiny_dogfish) EE722256 | |||
0 MGVKIHFEYC 2 | |||
1 GAuGYKPR 0 | |||
0 YQELANTIMGTFPDAAISGDVGRT 1 | |||
2 GSFEIEINGQLVFSKLETSGFPYEDD 0 | |||
0 IMDAVQKASAGDDVQKIVKSRPPCVIL* 0 | |||
>SEPW1_torCal Torpedo californica (electric_ray) | |||
0 MSVKVHVEYC 2 | |||
1 GAuGYGNR 0 | |||
0 YQDLANNILKTFPDADISGDVGRK 1 | |||
2 GSFEVEINGQLVFSKLETKGFPFEND 0 | |||
0 IITAVQNASNGVEMQQITNSRASCVIL* 0 | |||
>SEPW1_petMar Petromyzon marinus (lamprey) cDNA FD700531 tga-sel tag-stop full | |||
0 MPLKIHIVYC 2 | |||
1 GAuGYRSR 0 | |||
0 FHRLKDELETEFPGELEI 0 | |||
0 TGEGTPTQTGFLEVQIVGGKLLHSKA 0 | |||
0 NGDGFVDSDEKLQKIFSGVEKALKK* 0 | |||
>SEPW1_eptBur Eptatretus burgeri (hagfish) cdna BJ662449 tgn-sel tag-stop | |||
0 MPLKIHVVYC 2 | |||
1 GSuGYASK 0 | |||
0 FRALKVKLDHEFPGKLEI 0 | |||
0 TSEGTPGLTGKFEVQVGEKLVHSKK 0 | |||
0 NGDGFVDSPEKLQKIFKAVENALKGQ* 0 | |||
>SEPW1_strPur Strongylocentrotus purpuratus (sea_urchin) tga confirmed | |||
0 MVVIVKVIYC 2 | |||
1 GGuGYGPR 0 | |||
0 YRRLEQELKDEFGDDVDM 0 | |||
0 VGESTPGVTGWLEVEVNGKLIHSKK 0 | |||
0 NGDGYIDSEGKLKKIVNAVSAAM* 0 | |||
</pre> | |||
=== SEPWb: 2 vertebrate sequences === | |||
<pre> | |||
>SEPWb_squAca Squalus acanthias (spiny_dogfish) | |||
0 MPVEVAIEYC 2 | |||
1 SGuGFLTR 00 YLSLANEIRERVPGTNVTGFIGQA 1 | |||
2 GSFEVTINNKLVFSKLESNGFPYQED 0 | |||
0 VIEAVKTAKEGSAVERIISEEEIITQKRGCIII* 0 | |||
>SEPWb_danRer AAO86697 | |||
0 MVVKVKIEYC 2 | |||
1 GAuGYEPR 00 FQELKREICGNCPDAEVSGFVGRR 1 | |||
2 GCFEIQINDFLVFSKLESGGFPYSED 0 | |||
0 IIEAVVKAKDGKPEKIIRNRKECIIL* 0 | |||
</pre> | |||
=== RDX12 (C17orf37, SEPW2A): 49 deuterostome sequences === | |||
<pre> | |||
>RDX12_homSap C17orf37 NM_032339 | |||
0 MSGEPGQTSVAPPPEEVEPGSGVRIVVEYC 2 | |||
1 EPCGFEATYLELASAVKEQYPGIEIESRLGGT 1 | |||
2 GAFEIEINGQLVFSKLENGGFPYEKD 0 | |||
0 LIEAIRRASNGETLEKITNSRPPCVIL* 0 | |||
>RDX12_panTro | |||
MSGEPGQTSVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_gorGor | |||
MSGEPGQTSVAPPPEEVEPGSGVRMVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_ponAbe | |||
MSGESGQTFVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_rheMac | |||
MSGEPGQTSVAPPPEEVEPGSGVRIMVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASKGEPLEKITNSRPPCVIL* | |||
>RDX12_calJac | |||
MSGEPGQTSVAPLPGEVEPGSGVHIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASTGEPLEKITNSRPPCIIL* | |||
>RDX12_otoGar | |||
MSGEPGHSSVAPPPGEVEPGSGVRIMVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCIIL* | |||
>RDX12_tupBel | |||
MSGESGETSVAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGT-------------------------------------------TNSRPPCVIL* | |||
>RDX12_musMus | |||
MSGEPAPVSVVPPPGEVEAGSGVHIVVEYCKPCGFEATYLELASAVKEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASNGEPVEKITNSRPPCVIL* | |||
>RDX12_ratNor | |||
MSGEPGQVSVVPPPGEVEAGSGVHIVVEYCKPCGFEATYLELASSLEEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_dipOrd | |||
MSGEPGEVSLAPPPGEAEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_cavPor | |||
MSGEPGELSVARPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIQIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_oryCun | |||
MSGEPGPTSAAPSPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCTIL* | |||
>RDX12_ochPri | |||
MSGEPGPTSAAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELANAVKEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_vicPac | |||
MSGETGPTSVAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_turTru | |||
MSGETGPASSAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_bosTau | |||
MSGDTGTTSVAPPPGETEPGHGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_equCab | |||
MSGEPGLTSVAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_canFam | |||
MSGEPGPTSEAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_myoLuc | |||
MSGDAGPVSAAPHPGELEPGSGVRIVVEYCEPCGFEATYLELASAVKEQFPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_pteVam | |||
MSGDLGPTSAAPHPREIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCIIL* | |||
>RDX12_eriEur | |||
MSGEPGPTAVAPPPGEVEPGSGVRIVVEYCEPCGFEATYQELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_sorAra | |||
MSG----LAVAPPPGEGEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_loxAfr | |||
MNGEPGPISLAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_proCap | |||
MNGEPGPVSLAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* | |||
>RDX12_echTel | |||
MSGEPGPVSLAPPPGEAEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITSSRPPCAIL* | |||
>RDX12_monDom | |||
MSGESGAELVAPLPGEALPGSGVRIVVEYCEPCGFESTYLELASAVKEEYPGIKIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASSGEPLEKITNSCPPCVIL* | |||
>RDX12_macEug | |||
MSGEPGADLEEPLP.....GSGVRIVVEYCEPCGFESTYLELASAVKEEYPGIQIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASSGEPLQKITNSRPPCVIL* | |||
>RDX12_ornAna | |||
---------------------------------------------------------------AFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASLGEPLEKITNSRPPCSIL* | |||
>RDX12_galGal | |||
MSSSGGNGAAAVGTESEAGDGDGFGSDSGSERRVHIMVEYCEPCGFGATYEELASAVREEYPDIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCTIL* | |||
>RDX12_melGal | |||
MSSSGGGNGAAAAGTETEAGNGADGFVSGSGSERRVHIMVEYCEPCGFGATYEELASAVREEYPDIEIESRLGGTGAFEIEINGQLIFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCVIL* | |||
>RDX12_taeGut | |||
MSGGTGDGTGDGNGAERRVRIVVEYCEPCGFEATYQELASAVRDEYPDIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCVIL* | |||
>RDX12_anoCar | |||
MSDGSGEPAAEAPPATEGGVRIVVEYCKPCGFESAYLELANAVKEEYPDVEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRKAINGEPLEKITKSRPPCVIL* | |||
>RDX12_xenTro | |||
MSVSIVVEYCEPCGFKSHYEELASAVLEEFPDVTIDSRPGGTGAFEIEINGQLVFSKLELGGFPYAKDLIEAIRKASNGEPVEKITNSQAPCVIL* | |||
>RDX12_ambMex | |||
MAAVTIVVEYCKHCGFESHYLELESAVKEEFPDVVIESRCGETGTFEIKINGQIVFSKLELGGFPYEKDLMEAIKRASNGEPVERITNSRAPCVIL* | |||
>RDX12_tetNig | |||
MAVKIIVEFCGGuGYGPRYEELARVVKAEFLDADVSGFVGRLGSFEIVINEQLVFSKLETGGFPYEDDVLQVIQCAYDGKPVEKLTKSRPPCVIM* | |||
>RDX12_fugRub | |||
MGVTIRVEYCGGuGYGPRYEELARVVRAEFPDADVSGFVGRMGSFEIQINEQLIFSKLETGGFPYEDDVMHAIQCVSDGKPVEKITKSRPPCVIM* | |||
>RDX12_gasAcu | |||
MGVTMRVEYCGNuGYEPRYQELRTAVKQDFPDADVTGFVGRRGSFEIVLNGQLIFSRLESWGFPHVEDVLDAVKKAADGKPVDKITISRAPCVIM* | |||
>RDX12_oryLat | |||
MGVKIDVEYCGRuGYEPRYQDLASTVKDEFPEAEVSGFVGRSGSFEIQINGQLVFSKLELGGFPYEDDVLNAVQNAHDGKPVQKITKSRAPCVIM* | |||
>RDX12_danRer | |||
MGVQIKVEYCGGuGYEPRYQELKRVVTAEFTDADVSGFVGRQGSFEIEINGQLIFSKLETSGFPYEDDIMGVIQRAYDGQPVEKITKSQPPCVIL* | |||
>RDX12_oreMas | |||
MGVKVRVEYCGGuGYEPRYRELARVVKGEFSDADVTGVVGRTGSFEIEINGQLVFSKLETGGFPYEDDVMDAIHNAYDGKPLQKITKSRAPCVIM* | |||
>RDX12_calMil | |||
..........GAuGYEPRYQKLAIVIKDEFPDADVSGKVGRTGSFEIEINGQLIFSKLETGGFPYENDISEAVQKANNGEELQKIENSRPPCVIL* | |||
>RDX12_squAca | |||
MGVKIHFEYCGAuGYKPRYQELANTIMGTFPDAAISGDVGRTGSFEIEINGQLVFSKLETSGFPYEDDIMDAVQKASAGDDVQKIVKSRPPCVIL* | |||
>RDX12_torCal | |||
MSVKVHVEYCGAuGYGNRYQDLANNILKTFPDADISGDVGRKGSFEVEINGQLVFSKLETKGFPFENDIITAVQNASNGVEMQQITNSRASCVIL* | |||
>RDX12_petMar | |||
MAVNVNVEYCGGuGYEPRYQELAANILKQAPGVEVIGQVGRSGSFEVTINGELIFSKLECGGFPFAEDIIAEVKKVQGGEKVGKVTKSQAPCVIL* | |||
>RDX12_cioInt | |||
MADSNKVKVEIEYCGSuGYYGRFMDLKNDLESGCPNALVSGFVGREGSFEVSINGKQIFSKLETFGFPYPNDLIEAMKTAQNGEEVAAINNSQSPCTIL* | |||
>RDX12_braFlo | |||
MNYSRSKMAPKVEVEYCGGuGYAPRYWELANQIKTAVPDAEVTGVVGRSSSFEIIVDGQLLFSKLESGGFPQEQEILEALSNYKEGEKVEQVTNIQFPWCTIL* | |||
>RDX12_strPur | |||
MAQKKVTIEYCGSuGYYPRYRELHEMIESAIKGVDVSGKRGRPSSFEVKLNGQVLYSKLKNGGFPDLDAIVEAIEDYKGEGTVTPVEKKAASCIIL* | |||
>RDX12_astPec | |||
MATRKVEIEYCGGuGFYPRFMDLHNDIKSGVPGVAVDGRVGRSTSFEVTLNGKLLFSKLKTGGFPINKEIVTAIKEYKGEGDVTPVTNTASNCVLL* | |||
</pre> | |||
=== DIO1: 50 vertebrate iodothyronine deiodinases === | |||
<pre> | |||
>DIO1_homSap Homo sapiens (human) iodothyronine deiodinase type I 4 exons chr1 uc001cwb.1 | |||
0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWEFMQ 1 | |||
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLVIYIEEAHAS 1 | |||
2 DGWAFKNNMDIRNHQNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYIIQEGRILYK 0 | |||
0 GKSGPWNYNPEEVRAVLEKLHS* 0 | |||
>DIO1_panTro Pan troglodytes (chimp) | |||
0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVHLSGQRCNIWEFMQ 1 | |||
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLVVYIEEAHAS 1 | |||
2 DGWAFKNNMDIRNHQNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYVIQEGRILYK 0 | |||
0 GKSGPWNYNPEEVRAVLEKLHS* 0 | |||
>DIO1_ponPyg Pongo pygmaeus (orang_sumatran) | |||
0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTETGGLAPNCPVVRLSGQRCNIWEFMQ 1 | |||
>DIO1_macMul Macaca mulatta (rhesus) | |||
0 MGLPQSGLWVKKLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWDFMQ 1 | |||
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLIIYIEEAHAS 1 | |||
2 DGWAFKNNMDIRNHRNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYVIQEGRILYK 0 | |||
0 GKSGPWNYNPEEVRAVLEKLYS* 0 | |||
>DIO1_macFas Macaca fascicularis (crab_macaque) | |||
0 MGLPQSGLWVKKLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWDFMQ 1 | |||
>DIO1_macNem Macaca nemestrina (pigtailed_macaque) | |||
0 MVLPQSGLWVKKLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGPSCNIWDFMQ 1 | |||
>DIO1_calJac Callithrix jacchus (marmoset) | >DIO1_calJac Callithrix jacchus (marmoset) | ||
| Line 2,774: | Line 3,480: | ||
</pre> | </pre> | ||
=== DIO3: | === DIO3: 37 vertebrate iodothyronine deiodinases === | ||
<pre> | <pre> | ||
>DIO3_homSap Homo sapiens (human) 1 exon chr14 | >DIO3_homSap Homo sapiens (human) 1 exon chr14 | ||
| Line 2,906: | Line 3,612: | ||
0 MNTMKALKNAIVCLLLLPRFLLAAVVFWLLDFSCIRKRVFLRMKEQGGDATDPPLCISDSNRLFSVESLKAVWHGHKLDFLKAAHLGRGAPNTEVVHLEDQRCSRILDYAKDKRPLILNFGS | 0 MNTMKALKNAIVCLLLLPRFLLAAVVFWLLDFSCIRKRVFLRMKEQGGDATDPPLCISDSNRLFSVESLKAVWHGHKLDFLKAAHLGRGAPNTEVVHLEDQRCSRILDYAKDKRPLILNFGS | ||
CTuPPFMARLKAFQGVVQQNADIADSLVVYIEEAHPSDGWMSTDAPYQIPKHRRLEDRLNAAQLMHLEVPGCLVVVDSMENSSNAAYGAFFDRLYILQEGKVVYQGGRGPEGYRISELRDWLDQYRKGMEKSNNLVINM* 0 | CTuPPFMARLKAFQGVVQQNADIADSLVVYIEEAHPSDGWMSTDAPYQIPKHRRLEDRLNAAQLMHLEVPGCLVVVDSMENSSNAAYGAFFDRLYILQEGKVVYQGGRGPEGYRISELRDWLDQYRKGMEKSNNLVINM* 0 | ||
>DIO3_calMil Callorhinchus milii (elephantfish) note ATU not CTU | |||
0 MKLLKEAMAVLVLLPRFIVTALTLWLLDILCIRKRLLCKLRARYEE AAELLGPGAGPPAPD HRMLTAEGMQVVWQSFLLDALKQVKVGLEAPNSAVARLDGGAPCRLLDFASRDRPLVVNFGS | |||
ATuPPFVSRLPAFRQMVERYAEVADFLLVYVDEAHPSDGWALRSRFQLRRHRSQEERCSAAGLLAREFGLPAACGVVADLMDNNANRAYGVAFERLCVVQSQKIAYLGGKGPFFYNLNGVREWLERHSGQRWG* 0 | |||
>DIO3_squAca Squalus acanthias (spiny_dogfish) note CTU | >DIO3_squAca Squalus acanthias (spiny_dogfish) note CTU | ||
| Line 2,911: | Line 3,621: | ||
CTuPPFMARLKAFQRVAIQYADIADFLLVYIEEAHPSDGWVSTDAPYDIPRHRCLEDRLKAARLMHKENPSCLVVADTM | CTuPPFMARLKAFQRVAIQYADIADFLLVYIEEAHPSDGWVSTDAPYDIPRHRCLEDRLKAARLMHKENPSCLVVADTM | ||
> | >DIO3_chiPun Chiloscyllium punctatum (bambooshark) EU275162 835-1324 | ||
0 | 0 MDLLKEIVAILILLPRFFITAFMLWLLDILCIRKRLLPKRREQTEGSADDPPLCVSDTNRMFTLESVKAIWYGQKLDFFKSAHVGSPAPNTEVVQLQDQRKVRLLDYSRGARPLVLNFGS | ||
CTuPPFMARLKSFQRVATQYADIADFLLIYIEEAHPSDGWVSTDAPYNIPRHRSLEDRLKAASLIDKESPGCLVVADTMDNSSNSAYGAYFERLYVLRDQKVVYQGGRGPEGYKISELRLWLEQYKSQSHNSSTVLIEV* 0 | |||
>DIO3_braFlo Branchiostoma floridae (amphioxus) taa 1 exon closer match to DIO3 note YTU not CTU 10x trace coverage | >DIO3_braFlo Branchiostoma floridae (amphioxus) taa 1 exon closer match to DIO3 note YTU not CTU 10x trace coverage | ||
| Line 4,682: | Line 5,392: | ||
2 TSKMEPRWNFWKYLVNPKGQVIKVWRTEEPMASIKQEVSELVANIILKKRDEF* 0 | 2 TSKMEPRWNFWKYLVNPKGQVIKVWRTEEPMASIKQEVSELVANIILKKRDEF* 0 | ||
</pre> | </pre> | ||
== Basal selenoproteins: 28 platypus 18 echidna 19 elephantshark 18 lamprey 8 hagfish 14 amphioxus 6 tunicate 4 urchin == | == Basal selenoproteins: 28 platypus 18 echidna 19 elephantshark 18 lamprey 8 hagfish 14 amphioxus 6 tunicate 4 urchin == | ||
| Line 4,821: | Line 5,529: | ||
>SELU1_strPur Strongylocentrotus purpuratus (sea_urchin) | >SELU1_strPur Strongylocentrotus purpuratus (sea_urchin) | ||
>SELU2_strPur Strongylocentrotus purpuratus (sea_urchin) 2nd copy | >SELU2_strPur Strongylocentrotus purpuratus (sea_urchin) 2nd copy | ||
<pre> | |||
Temporary fragments of sea urchin selenoproteins needing more work... | |||
>SELU1_strPur Strongylocentrotus purpuratus (sea_urchin) tga confirmed | |||
0 MALLWGAAAAAVGGLLMANMDFWLPRGGAATTEAYLAGAVLQLLGETKLQAQLSLVDEAVRTFKAKDLWKDTGAVVLAVRRPGUSLCREEAKE | |||
LSSLKPELDALGIPLYAVLLEPGGYKEFLPFFSGEVFLDTETRFYGEEKRNISLVGLLRI | |||
STLLQVRENKKQGIQGNYIGHGLTLXGIFVIGPGDRGYSWNIAXRTLAITHQMR | |||
>SELU2_strPur Strongylocentrotus purpuratus (sea_urchin) 2nd copy CD317360 84% id also Sec | |||
PRVRRDGGDLFLWEKQACVIHFMRRFGuSACRLGASELDSLRPQLDEADVRLVGIGLEDL | |||
GAQEFLDGGFWKGDLFIDQQQKSYQGLGFKKYSLMSIMKAVFSAKTRAALSRASDKKIKA | |||
NLKGDKLQIGGTLIISKGGEKVLVDFKQEAPGDHIPLQTVLDAFXIKGETPKTEGAEGGT | |||
VVCXDDVCQMK* 0 | |||
>SELO_strPur Strongylocentrotus purpuratus (sea_urchin) tga confirmed CD336539 fragments 35-end | |||
NSIIGNIASRSLLAVQRGHHRFSDSRSFSKSPHKMARIDKLNFDNRVLRTLPIDPETENYARQVPGACFSRVKPTPVANPKTVAYSVPAMKLLDLSEEELKREDFPAYFSGSKPLPGSDPAAHCYCGHQFGNFAGQLGDGAAMLVCILFESYFRNL...MNATNPKFILRNYIAENAIKEAENGDFSEVHRVLKLLESPFSDDVSLPFIEHTSELAGDEDAGASKPVEPTISPSNTSTVNSCTGVAYNSRPPEWAMELRVSUSS- | |||
>SELN1_strPur Strongylocentrotus purpuratus (sea_urchin) | |||
HAEFQLNEPPSHPFWFTPAQFIGHLIIRKDASHVKYF | |||
NMHVPSNRSLNVDMEWMNGPNEVENMEVDIGFMPKMELHAPNPSSQVTIYSESGEVLYEPSNQQEMSSGTIHWDEEISMEEALVLLEKEMYPFKKIEYMPLNAAFKRTHA | |||
</pre> | |||
Latest revision as of 23:24, 28 December 2008
Introduction to selenoprotein evolution
(more selenoprotein classes shortly)
SEPP2: lost in placentals
SEPP2 is a newly discovered paralog of SEPP1 with a UxxC motif (plus a later conserved cysteine and distal CxxC motif) and high-scoring putative SECIS element in 3' UTR that is quite conserved in vertebrates but only through marsupials. No corresponding gene -- or even decayed debris -- can be found in syntentic position in any placental mammal, including Atlantogenata. Also, cysteine has displaced the selenocysteine in Xenopus, a species much depleted in selenoproteins. SEPP2 does not appear at GenBank non-redundant other than mis-annotated SEPP1 chicken mRNAs. SEPP2 transcripts are available only in skate, zebrafish, and echidna (a short 454 read) .
The two paralogs have 4 coding exons with identical intron phases so clearly represent a segmental gene duplication. The gene duplication can be tracked back to before chondricthyes divergence but only one copy can be located in earlier diverging deuterostomes (sea urchin and acorn worm). SEPP2 retains the selenocysteine TGA only in its first exon and lacks the cluster of them in exon 4 of SEPP1. Exon 4 in SEPP2 is diverging very rapidlly, about neutrally, and is difficult to recover from blast searches without transcripts.
It can be surmised the first seleoncysteine in both SEPP1 and SEPP2 has a different functional role from terminal selenocysteines in SEPP1, possibly a conserved redox role distinct from selenostorage function of exon 4 in SEPP1. SEPP2 is very likely functional in all the species in which it occurs because of its conservation. There is no indication it is "on its way out" in marsupials -- the SECIS would be obliterated very quickly if the gene were non-functional.
SELU: 3 paralogs, variable timing losses
SELU: This family consists of three deeply diverged (distinct exon patterns) paralogs. The encoding gene has several average-length exons with anomalously short introns like many selenoproteins. In the SELU1 group, selenocysteine occurs in a UxxC motif already in the earliest deuterostomes but drops out in mammals after monotremes, being replaced by CxxC in marsupials and placentals. Amphibia separately lost selenocysteine.
The second paralog SELU2 has selenocysteine in bilaterans only to the node of sea urchin, suggesting it was lost early in the deuterostome ancestor (note though the early eukaryote Monosigna has cysteine). It is the closer paralog of SelU1, 36% vs 27% percent identity. No vestigal SECIS element persists in living species that encode cysteine. (The decayed SECIS elements still identifiable in 3' UTR of cysteine-containing GPX6 genes in rodents and human GPX5 represent much more recent loss of selenocysteine.)
The third paralog SELU3 has cysteine in all metazoans for which a sequence is available with the exception of the immediate outgroup choanflagellate Monosigna. It might be called virtual selenoprotein in eumetazoans. The pattern suggests a scenario in which selenocysteine was present in an ancestral gene prior to gene duplications followed by conversion to cysteine in different phylogenetic patterns within each gene subfamily.
This family exhibits the "selenocysteine rachet": if selenocysteine happens to be replaced by ordinary cysteine (despite putative catalytic inferiority) in some stem lineage, the unselected 3' UTR SECIS element then deteriorates over a few million years from accrued mutations, for the same reason (lack of purifying selection) the crayfish in the cave loses its imaging opsins. Consequently the whole following clade will contain cysteine -- a reversion to TGA at the cystein codon might occur but it would simultaneously require a multi-step reversion or de novo evolution of a SECIS element, ie all SECIS elements are ancient and selenocysteines cannot wink back on paraphyletically. (However the overall selenoproteome can still increase over time because of gene duplications elsewhere.)
A phylogenetic overview of the occurence of selenocysteine in SELU:
SELU1 SELU2 SELU3 genus species (common) CFLC CVVC CYIC Homo sapiens (human) CFLC CVVC CYIC Macaca mulatta (rhesus) CFLC CMVC CVYC Rattus norvegicus (rat) CFLC CMVC CVYC Mus musculus (mouse) CFLC .... CVYC Oryctolagus cuniculus (rabbit) CFLC CSVC CYIC Canis familiaris (dogs) CFLC CMVC CYIC Bos taurus (cow) CFLC .... CYIC Dasypus novemcinctus (armadillo) CFLC .... .... Echinops telfairi (tenrec) CFLC CMVC CYIC Monodelphis domesitca (opossum) CFLC .... .... Macropus eugenii (wallaby) CFLC CMVC .... Trichosurus vulpecula (opossum) UFLC .... .... Ornithorhynchus anatinus (platypus) UFLC .... .... Tachyglossus aculeatus (echidna) UFLC .... CYTC Gallus gallus (chicken) UFLC .... .... Taeniopygia guttata (finch) UFLC .... .... Anolis carolinensis (lizard) CFLC CQIC CYTC Xenopus tropicalis (frog) CFLC CQIC CYTC Xenopus laevis (frog) UFLC CQVC CYTC Danio rerio (zebrafish) UFLC CQIC CYAC Takifugu rubripes (fugu) UFLC CQVC CSYC Gasterosteus aculeatus (stickleback) UFLC .... CHTC Oncorhynchus mykiss (trout) UFLC .... .... Ictalurus punctatus (catfish) UFLC CQVC CSYC Oryzias latipes (medaka) UFLC .... .... Callorhinchus milii (elephantfish) UFLC .... .... Squalus acanthias (spiny_dogfish) .... CVVC DYVA Ciona intestinalis (tunicae) UQLC CQVC .... Branchiostoma floridae (amphioxus) USLC USAC .... Strongylocentrotus purpuratus (purple .... .... CYTA Saccoglossus kowalevskii (acornworm) .... UPFC CFVC Mytilus californianus (mussel) .... UPFC CFIC Crassostrea gigas (bivalve) .... UAFC CFMC Lottia gigantea (limpet) .... UQLC CFMG Capitella capitatus (annelid) .... UKLC .... Hydra magnipapillata (hydra) .... UQLC .... Nematostella vectensis (anemone) .... UQLC .... Oscarella carmela (sponge) .... CRLC UFCK Monosiga brevicollis (choanoflagellate)
SEPW: odd paralog SEPV gained in placentals
Selenoprotein SEPW is one of the shortest known mammalian proteins at 87 aa. With its CxxU motif, it is likely limited to simple redox reactions. Curiously, despite its small size, the protein still has 5 coding exons. One of these is of relatively recent origin because chondrichtyes and telost fish have the second and third exons fused (which, given the phylogenetic tree and rarity of intron gain/loss and assuming parsimony, must be the ancestral condition.
Like many genes on the densely packed human chromosome 19, SEPW seems to have given rise to a segmental duplication, here during the placental stem (ie, is absent marsupial and earlier diverging vertebrates). The second copy, called SELV, retains the same intron placements and phases but with greatly expanded and compositionally anomalous exon 1 which is quite prone to large indels (from replication slippage).
This long exon retains some conservation at its distal end to the last 5 residues of SEPW (unsurprising because the cysteine of the CxxU motif is at the end). There is observable conservation also at the amino terminus but otherwise the exon is evolving unconstrained. Possibly the initial methionine of SEPW was lost at the time of duplication but the terminal residues and splice donor were retained. Transcription far upstream lead to a remote methionine serving as replacement.
This is a very odd gene but transcripts in a half dozen species and moderate conservation (75% within placental mammals) in exons 2-5 over hundreds of millions of years of branch length witin placentals proves that it is evolving under selective constraint and so is functional. Distal exons however retain only 50% conservation to those of SEPW, far too little to support ongoing gene conversion.
RDX12: selenocysteine from echinoderm to teleost fish
This gene has also been denoted C17orf37 (human) and SEPW2A (zebrafish). It is a weak paralog of SEPW and SELV with a related thioredoxin-like fold.
It shares but a single intron with SEPW1, the phase 21 intron that splits the cysteine codon in the CxxC or CxxU, suggesting gene duplication took place in early eukaryotes. As in so many selenoproteins, an ancestral selenocysteine codon that persists to living fish, sharks, lamprey, cephalochordates, urochordates, and echinoderms is lost to cysteine in tetrapods, with the timing still unknown in the understudied lobefinned fish.
The length of the first exon is highly variable, causing considerable difficulty in gene recovery. This variability seems to result from an wandering start codon rather than indels or splice donor read-through. In mammals however, both length and sequence are well conserved. Smaller than the smallest known enzyme, RDX12 probably functions as an auxillary redox protein, though where exactly in metabolism is not known. There are no known disease alleles.
>RDX12_homSap C17orf37 NM_032339 >SEPW1_homSap
0 MSGEPGQTSVAPPPEEVEPGSGVRIVVEYC 2 0 MALAVRVVYC 2
1 EPCGFEATYLELASAVKEQYPGIEIESRLGGT 1 1 GAuGYKSK 0
2 GAFEIEINGQLVFSKLENGGFPYEKD 0 0 YLQLKKKLEDEFPGRLDI 0
0 LIEAIRRASNGETLEKITNSRPPCVIL* 0 0 CGEGTPQATGFFEVMVAGKLIHSKK
0 KGDGYVDTESKFLKLVAAIKAALAQG* 0
Alignment of ~34% identity:
RDX12 VRIVVEYCEPCGFEATYLELASAVKEQYPG-IEI--ESRLGGTGAFEIEINGQLVFSKLENGGF 83
||+| || |+++ ||+| +++++|| ++| | || ||+ + |+|+ || + |+
SEPW1 VRVV--YCGAuGYKSKYLQLKKKLEDEFPGRLDICGEGTPQATGFFEVMVAGKLIHSKKKGDGY 66
SELV VLIRVTYCGLuSYSLRYILLKKSLEQQFPNHLLFEEDRAAQATGEFEVFVNGRLVHSKKRGDGF
* *
>RDX12_homSap MSGEPGQTSVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGETLEKITNSRPPCVIL*
>RDX12_panTro MSGEPGQTSVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_gorGor MSGEPGQTSVAPPPEEVEPGSGVRMVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_ponAbe MSGESGQTFVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_rheMac MSGEPGQTSVAPPPEEVEPGSGVRIMVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASKGEPLEKITNSRPPCVIL*
>RDX12_calJac MSGEPGQTSVAPLPGEVEPGSGVHIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASTGEPLEKITNSRPPCIIL*
>RDX12_otoGar MSGEPGHSSVAPPPGEVEPGSGVRIMVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCIIL*
>RDX12_tupBel MSGESGETSVAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGT-------------------------------------------TNSRPPCVIL*
>RDX12_musMus MSGEPAPVSVVPPPGEVEAGSGVHIVVEYCKPCGFEATYLELASAVKEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASNGEPVEKITNSRPPCVIL*
>RDX12_ratNor MSGEPGQVSVVPPPGEVEAGSGVHIVVEYCKPCGFEATYLELASSLEEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_dipOrd MSGEPGEVSLAPPPGEAEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_cavPor MSGEPGELSVARPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIQIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_oryCun MSGEPGPTSAAPSPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCTIL*
>RDX12_ochPri MSGEPGPTSAAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELANAVKEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_vicPac MSGETGPTSVAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_turTru MSGETGPASSAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_bosTau MSGDTGTTSVAPPPGETEPGHGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_equCab MSGEPGLTSVAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_canFam MSGEPGPTSEAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_myoLuc MSGDAGPVSAAPHPGELEPGSGVRIVVEYCEPCGFEATYLELASAVKEQFPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_pteVam MSGDLGPTSAAPHPREIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCIIL*
>RDX12_eriEur MSGEPGPTAVAPPPGEVEPGSGVRIVVEYCEPCGFEATYQELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_sorAra MSG----LAVAPPPGEGEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_loxAfr MNGEPGPISLAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_proCap MNGEPGPVSLAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL*
>RDX12_echTel MSGEPGPVSLAPPPGEAEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITSSRPPCAIL*
>RDX12_monDom MSGESGAELVAPLPGEALPGSGVRIVVEYCEPCGFESTYLELASAVKEEYPGIKIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASSGEPLEKITNSCPPCVIL*
>RDX12_macEug MSGEPGADLEEPLP.....GSGVRIVVEYCEPCGFESTYLELASAVKEEYPGIQIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASSGEPLQKITNSRPPCVIL*
>RDX12_ornAna ---------------------------------------------------------------AFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASLGEPLEKITNSRPPCSIL*
>RDX12_galGal AVGTESEAGDGDGFGSDSGSERVHIMVEYCEPCGFGATYEELASAVREEYPDIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCTIL*
>RDX12_melGal AAAGTETEAGDGFVSGSGSERRVHIMVEYCEPCGFGATYEELASAVREEYPDIEIESRLGGTGAFEIEINGQLIFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCVIL*
>RDX12_taeGut MSGGTGDGTGDGNGAERRVRIVVEYCEPCGFEATYQELASAVRDEYPDIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCVIL*
>RDX12_anoCar MSDGSGEPAAEAPPATEGGVRIVVEYCKPCGFESAYLELANAVKEEYPDVEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRKAINGEPLEKITKSRPPCVIL*
>RDX12_xenTro MSVSIVVEYCEPCGFKSHYEELASAVLEEFPDVTIDSRPGGTGAFEIEINGQLVFSKLELGGFPYAKDLIEAIRKASNGEPVEKITNSQAPCVIL*
>RDX12_ambMex MAAVTIVVEYCKHCGFESHYLELESAVKEEFPDVVIESRCGETGTFEIKINGQIVFSKLELGGFPYEKDLMEAIKRASNGEPVERITNSRAPCVIL*
>RDX12_tetNig MAVKIIVEFCGGuGYGPRYEELARVVKAEFLDADVSGFVGRLGSFEIVINEQLVFSKLETGGFPYEDDVLQVIQCAYDGKPVEKLTKSRPPCVIM*
>RDX12_fugRub MGVTIRVEYCGGuGYGPRYEELARVVRAEFPDADVSGFVGRMGSFEIQINEQLIFSKLETGGFPYEDDVMHAIQCVSDGKPVEKITKSRPPCVIM*
>RDX12_gasAcu MGVTMRVEYCGNuGYEPRYQELRTAVKQDFPDADVTGFVGRRGSFEIVLNGQLIFSRLESWGFPHVEDVLDAVKKAADGKPVDKITISRAPCVIM*
>RDX12_oryLat MGVKIDVEYCGRuGYEPRYQDLASTVKDEFPEAEVSGFVGRSGSFEIQINGQLVFSKLELGGFPYEDDVLNAVQNAHDGKPVQKITKSRAPCVIM*
>RDX12_danRer MGVQIKVEYCGGuGYEPRYQELKRVVTAEFTDADVSGFVGRQGSFEIEINGQLIFSKLETSGFPYEDDIMGVIQRAYDGQPVEKITKSQPPCVIL*
>RDX12_oreMas MGVKVRVEYCGGuGYEPRYRELARVVKGEFSDADVTGVVGRTGSFEIEINGQLVFSKLETGGFPYEDDVMDAIHNAYDGKPLQKITKSRAPCVIM*
>RDX12_calMil ..........GAuGYEPRYQKLAIVIKDEFPDADVSGKVGRTGSFEIEINGQLIFSKLETGGFPYENDISEAVQKANNGEELQKIENSRPPCVIL*
>RDX12_squAca MGVKIHFEYCGAuGYKPRYQELANTIMGTFPDAAISGDVGRTGSFEIEINGQLVFSKLETSGFPYEDDIMDAVQKASAGDDVQKIVKSRPPCVIL*
>RDX12_torCal MSVKVHVEYCGAuGYGNRYQDLANNILKTFPDADISGDVGRKGSFEVEINGQLVFSKLETKGFPFENDIITAVQNASNGVEMQQITNSRASCVIL*
>RDX12_petMar MAVNVNVEYCGGuGYEPRYQELAANILKQAPGVEVIGQVGRSGSFEVTINGELIFSKLECGGFPFAEDIIAEVKKVQGGEKVGKVTKSQAPCVIL*
>RDX12_cioInt MADSNKVKVEIEYCGSuGYYGRFMDLKNDLESGCPNALVSGFVGREGSFEVSINGKQIFSKLETFGFPYPNDLIEAMKTAQNGEEVAAINNSQSPCTIL*
>RDX12_braFlo MNYSRSKMAPKVEVEYCGGuGYAPRYWELANQIKTAVPDAEVTGVVGRSSSFEIIVDGQLLFSKLESGGFPQEQEILEALSNYKEGEKVEQVTNIQFPWCTIL*
>RDX12_strPur MAQKKVTIEYCGSuGYYPRYRELHEMIESAIKGVDVSGKRGRPSSFEVKLNGQVLYSKLKNGGFPDLDAIVEAIEDYKGEGTVTPVEKKAASCIIL*
>RDX12_astPec MATRKVEIEYCGGuGFYPRFMDLHNDIKSGVPGVAVDGRVGRSTSFEVTLNGKLLFSKLKTGGFPINKEIVTAIKEYKGEGDVTPVTNTASNCVLL*
DIO1, DIO2, and DIO3: a curious history
The iodothyronine deiodinases of selenoproteins have been thoroughly studied because of their role in thyroid hormone metabolism. The three paralogs seen in mammals diverged long ago -- the percent identity today is about 45% and exon breaks and phases are completely unrelated. One scenario is that DIO2 and DIO3 in mammals arose as an initial retroprocessed gene that then duplicated again, with DIO2 subsequently acquiring an intron and DIO3 remaining a single coding exon. The critical era for exon acquisition and loss is poorly represented in database species.
DIO2 and DIO3 are found on the same chromosome arm of chr 14 in human but separated by 22 Mbp, rather far for a tandom duplication. This structure is conserved in the chicken genome with only 8.7 Mbp intervening. Again, it is hard to sort out coincidence from correlation. Note the SECIS element might well accompany both retroprocessing and tandem duplications.
No 3D structures are available for this family of proteins (which have a single N-terminal transmembrane segment) but deiodinases have thioredoxin-like fold (which includes glutathione peroxidase selenoproteins) according to SuperFamily. This domain might serve to define trimming for purposes of better protein trees as well as serve as a partial template for folding (noting too a short insert with strong similarities to the active site of iduronidase, a GH-A-fold of glycoside hydrolase).
DIO1 catalyzes the outer ring deiodination of T4 to the biologically active hormone T3, as well as the degradation of T3 and sulfated iodothyronines. DIO2 is also involved in outer ring deiodination. DIO3 converts T(4) to T(3) and T(3) to 3, 3'-diiodothyronine (T2) by inner-ring deiodination. This gene is imprinted and has an anti-sense transcript. These genes are of great interest in the evolution of the thyroid (resp. endostyle) and its hormones.
The putative redox motif varies significantly across the three paralogs, with the CxU motif of DIO1 and DIO3 becoming AxU in DIO2 (meaning it cannot form a local seleno-disulfide). A similar observation holds in some basal DIO3: calMil ATU not CTU, braFlo YTU not CTU. All three proteins contain a distal conserved cysteine; DIO2 has an additional conserved cysteine still further downstream.
DIO1 GSCTuPSF...PQCPV DIO2 GSATuPPF...PQCRV...RVCIV...KRuKK DIO3 GSCTuPPF...PGCAL
Remarkably, a second selenocysteine may have arisen in DIO2 near-terminal position post-lungfish divergence (ie after teleost fish divergence). This TGA codon is conserved over billions of branch length years and is presumably supported by the same SECIS element that supports the first selenocysteine. It may have arisen from read-through of a TGA stop codon; no cysteine counterpart is seen in this position in earlier diverging species DIO2 (fish, shark, lamprey) or in DIO1 or DIO3 paralogs. Since ancestral CTU in the proximal selenocysteine had already morphed into ATU by lamprey and chondrichthyes divergence, possibly the second selenocysteine now contributes the second half of a redox pair (as alanine has a chemically inert sidechain). Bioinformatics alone cannot be definitive here -- the possibility remains that this TGA is the actual stop codon and became conserved for some other reason, or that multiple losses of selenocysteine occured in the earlier diverging species.
>DIO2_homSap PFSYNLQEVRHWLEKNFSKRuKKTRLAG* >DIO2_panTro PFSYNLQEVRHWLEKNFSKRuKKTRLAG* >DIO2_ponPyg PFSYNLHEVRHWLEKNFSKRuKKTRLAG* >DIO2_macMul PFSYNLQEVRHWLEKNFSKRuKKTRLAG* >DIO2_calJac PFSYNLQEVRHWLEKNVSKRuKKTRLAG* >DIO2_otoGar PFFYNLQEVRRWLEKNFSKRuNRLAG* >DIO2_micMur PFFYNLQEVRHWLEKNFSKRuNLD* >DIO2_tupBel PFYYNLQEVRRWLEKNFSKRuNSLAG* >DIO2_musMus PFSYNLQEVRSWLEKNFSKRuILD* >DIO2_ratNor PFSYNLQEVRSWLEKNFSKRuILD* >DIO2_speTri PFCYNLQEVRHWLEKNFSKRuILD* >DIO2_cavPor PFCYNLQEVRRWLEKNFHKRuNRLDG* >DIO2_oryCun PFSYNLQEVRHWLEKNFSKRuKKTRLAG* >DIO2_ochPri PFCYNLQEVRRWLEKNFNKRuI* >DIO2_canFam PFYYNLQEVRRWLEKNFSKRuNRLAG* >DIO2_equCab PFCYNLQEVRHWLEKSFSQRuNRLC* >DIO2_myoLuc PFFYNLQEVRHWLEKNFNKRuNLD* >DIO2_pteVam PFFYNLQEVRRWLEKNFNKRuNLD* >DIO2_bosTau PFFYNLQEVRRWLEKNFSKRuKRLAG* >DIO2_turTru PFYYNLQEVRRWLEKNFSKRuKLD* >DIO2_susScr PFYYNLQEVRRWLEKNFSKRuKLD* >DIO2_vicVic PFHYNLQEVRRWLEKNFSKRuNLD* >DIO2_eriEur PFCYNLQEVRHWLEKSFSQRuNLD* >DIO2_sorAra PFCYNLQEVRDWLEKNFSKRuNSL* >DIO2_dasNov PFCYNLQEVQHWLEKNFSKRuNRLAG* >DIO2_choHof PFYYSLQEVRQWLEKNFSKRuNLD* >DIO2_echTel PFCYNLQEVRCWLEKNFSKRuTRVAGQ* >DIO2_monDom PFFYNLQEVRLWLEKNFSKRuNPG* >DIO2_macEug PFFYNLQEVRLWLE-NFSKRuNPG* >DIO2_ornAna PFFYNLQEVRLWLEQNFSKRuNPD* >DIO2_galGal PFFYNLQEVRLWLEQNFSKRuNPLSTEDLSTDVSL* >DIO2_taeGut PFFYNLQEVRLWLEQNFSKRuNPFSTGVTSIDVSL* >DIO2_anoCar PFFYNLQEVRLWLEQNFRKRuNPNLPQEDMLNISPLK** >DIO2_phiOlf PFFYNLQEVRLWLEQNFRKRuNPNLPDKKSRGLSF* >DIO2_xenTro PFFYNIQEIRRWLELSFGKRuT* >DIO2_neoFor PFFYNLKEVRHWLEQTYRKRuVPTCELIM* >DIO2_danRer PFFYNLKDVRRWLEKCYGK** >DIO2_tetNig PFFYNLKEVRQWLEHSYGKR* >DIO2_takRub PFFYNLKEVRQWLEQSYGKR* >DIO2_gasAcu PFFYNLKDVQHWLEQNFGKRFSRTSAEKDMSHISKKGIIHQ* >DIO2_oryLap PFFYNLKEVRQWLEQLRQTVGPNTEE** >DIO2_funHet PFFYSLKDVRQWLELSYGRR* >DIO2_oncMyk PFFYNLKDVRQYLEQSYGKR* >DIO2_parOli PFFYNLKDVQKWLEQSYGKR* >DIO2_calMil PFFYNLNGVREWLERHSGQRWG* >DIO2_petMar PFFYRVREVKSFLESVKASR*
The collection of DIO1 below from 49 species illustrates the importance of rare genomic events in defining the topology of the mammalian species tree. Here we see a 5 residue insert in the first exon restricted to Pegasoferea (ie excluding artiodactyls) with no hint of homoplasy. Additionally a longer deletion is restricted to Afrotheres.
DIO1_homSa MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWEFMQ DIO1_panTr ..................................................................................................H............. DIO1_macMu .....S...V.K................................................................................................D... DIO1_calJa ....G..................A......T.......K......D........N...........................................H.........D... DIO1_otoGa ....R..........F.......A...M..........SQ.....QQ.V.AK.............LQHPV.LVCPEGPL.....M..R.........SAS...K....D... DIO1_musMu .....LW......VIF.Q..LE.A.....MT...G...QS.....Q....A...R.AP...V.....I................RA.F.......T..C....K....D.I. DIO1_ratNo ...S.LW......VIF.Q..LE.AT....MT...E...Q......Q........R.AP...V.....I................RA.Y.......T.......K..V.D.I. DIO1_oryCu ....R...........VQ...E.A.....MT...E...Q......Q...IAQ..N.AQ.S........................A..P.......S.......Q.S..D..R DIO1_cavPo ...TW...........VQ...E.AM....MT...E.I.KS..............Q..................I.........E.A.......D.S..C...EKRT..D..H DIO1_canFa ....R.V...R......Q...Q.A....F.K...A...QH.V..NGN-----K....Y...A..LY.M.........Q......R..P....................D... DIO1_ursAr ....R.V...R......Q..M..A..........E...QQV...NK.-----.....Y...L...Y.M................R..P....................D... DIO1_felCa ...S.L....R.....FQ..LQ.A....F.....S...QH.V..NR.-----.....Y...A..LY.V................R..P.................S..D..K DIO1_equCa ....RA...........Q..LQ.A......T.......QH.V..NQ.-----.....Y...V..LY...........H........KR......S.............D... DIO1_myoLu ..............I..Q..L..TL...Q.K...R...QH....NR.-----.....Y...A..L...P....I..........K..E.S...............H..D... DIO1_pteVa .E..W..R.........Q..L..A....Q.T...R...Q..V..NR.-----.....F...L..L...............Q.....KE..........C.........D... DIO1_sunMu ....GL..L...FG..VR..LK.A......T.W.SAIRPHL...S.....AK..R.TYED.A...............N..Q...R.KQ.DI..DS...H.....ARL.D... DIO1_bosTa ....S...........FQ..L..AI.....T...R...Q...................E..............I..........M..Q..............E..S..D... DIO1_turTr ....L...........FQ.GL..AM.....T...R...Q.....S.....AK.....YE........A.....I..........M..Q..R.................D... DIO1_susSc .E..L...........FQ..L..AM....MT...G...QD....SQ....AK......E........A................K..E..........S......H..D... DIO1_vicVi ...SL...........FQ.VL..AL.....T...G...QD....SQR...AQ.....YE..............I.............Q.....D....C..-D.VH..D... DIO1_eriEu ....S...........FQ..L..AI.....T...R...Q...................E..............I..........M..Q..............E..S..D... DIO1_dasNo ...S..........I.FQ..L..A...T..T...G...Q....KSQ.SHKAE....PY...G....N......L..IG......K..Q..........H.........D... DIO1_choHo ...SW............Q..L..AM..I..T...G...Q.....SRRANN.KD.Q.PY...G....N.................K..Q..........H..R......D... DIO1_loxAf ..............IF.K..L..AM.........G...K....Q--------....AY.M.GS.L..IP....I...Y......K..E..P..D....C........SD... DIO1_proCa ......V..........R..L..AM.....A...G...K....Q--------....AY.M.CS.L..VP........Y......K..E..........H.....R...D... DIO1_monDo .LRLWLW.........Q.VG..LM..LMKM.S...M.QH..G..Q.SSIFQ..N.KYE..G....TLP..L...R........QALQ..P..D....S.R..PRRL.D..HA DIO1_triVu . AG.L..VR.F.A..Q..F......L.KT...NMM.KH..SL.QRSSISQ.TQ.AYE..G.....I...F............QALQ......P...T.K.ESRH..D..H DIO1_anoCa . FKA.RLVLKT.L..Q.CLSTA...LFM....ATA..Y..KQS.RSS.G...N.VYE..G.....F..LL.....K.K....KALQ.CP...T...DFD.KIHH.LD...
SELH: rapid evolution
SELH has been given the highly unsatisfactory official name C11orf31 by HGNC. It is another small selenoprotein with a conserved CxxU redox motif split by a phase 21 intron. The introns in mammalian SELH are exceedingly short (eg, 93 and 162 bp in human with gene coding span very short at 619 bp) but the level of transcription is not remarkable so provides no explanation for the lack of retroposons. Some species like orangutan have processed pseudogenes, implying transcription in germ-line tissues. Zebrafish has a diverged duplicate gene with tryptophan at the seleocysteine site in addition to an intronless transcribed gene with TGA selenocysteine -- it appears that this has displaced the normal three-intron gene (seen in other fish). That can work in selenoproteins since retropositioning begins at the 3' end of transcripts, meaning the SECIS likely accompanies the coding region.
Protein conservation is below average -- SELH percent identities (to human) drop to the low 80's within Laurasiatheres, to 72% with marsupial, and 57% with chicken. Further, rodents exhibit significant residue loss upstream of the CxxU motif, very unusual in such a short protein but indicative of an inessential structural region. Indeed observed conservation is primarily centered in the middle of the protein. No phylogenetic conversion of selenocysteine to cysteine is observed within vertbrates
SELM2: perinuclear but ER retention signal
Another selenoprotein of unknown function, SELM2 with CxxU motif, surfaces during Blastp searches using selenoprotein SELM2 (aka SEP15 which oddly has a CxU motif) as query. A 3' UTR SECIS element cannot be located with SECISearch 2.19 yet it seemingly must be located in the comparative genomic peaks of conservation lying between SELM and converently transcribed nearby neighboring gene SMTN. Indeed, that has been validated by direct experiment.
The SECIS element has CC in place of supposedly critical apical loop AA. The evolutionary origin of this divergence has not been dated and would be difficult to do. This SECIS element might be recognized by KIAA0256 rather than SECISBP2.
Since the protein is quite short with 5 introns, complete SELM sequences are best recovered from cDNAs and genomic alignments in the UCSC 28way. This is an ancient protein recoverable from vertebrates, amphioxus, sea urchin, shrimp, mite, moths, and hydra, and plants. Moths (silkworm and hawkmoth) -- but not their outgroups -- encode cysteine in a CxxC motif in place of CxxU. No loss of selenocysteine is seen in 22 species of phylogenetically dispersed vertebrates.
SELM begins with an experimentally established signal peptide. There are no glycosylation sites nor additional conserved cysteines beyond the CxxU motif. SuperFamily finds no similarity to proteins of known 3D structure. The terminal residues appear to be a phylogenetically conserved KDAL-class endoplasmic retention signal though it is reported perinuclear.
In whole-mount zebrafish embryos, SELM is expressed within the notochord and anterior somites, axial fin fold, dorsal spinal chord neurons, then in lateral line neuromasts. SelM is located in the ER/Golgi as is its distant homolog Sep15 (found associated with UDP-glucose glycoprotein glucosyltransferase, an ER-resident protein involved in quality control of protein folding).
MSRB123: methionine sulfoxidases
MSRB1 is a short odd protein with HGNC gene name SEPX1, rich in cysteines, with two CxxC motifs, a near-amino terminal cysteine and a more distal selenocysteine in a motif with serine. MSRB1 is now known to be a stereospecific methionine-R-sulfoxide reductase repairing oxidative damage to methionine in native proteins. The two pairs of cysteines bind a zinc atom. The all-beta structure has been determined in a bacterial homolog with a internal structural duplication so weak it needs an xray determination to be revealed. Humans have 9 such domains in 9 proteins according to SuperFamily. These could potentially encode other selenoproteins at least in some species.
The fold exists as a small family of paralogs, three in mammals, with specialization to cell compartment -- cytosol, mitochondria and endoplasmic reticulum (via KAEL* signal), respectively.). All contain catalytic zinc. This multiplicity of MSRB contrasts with a single non-homologous methionine-S-sulfoxide reductase (not a selenoprotein in any species).
MSRB1 has selenocysteine in its active site in conserved motif UxxS, whereas MSRB2 and MSRB3 contain Cys in the well-conserved motif CINS. The cysteine in the Ux53xC motif of prokaryotes is replaced by serine or threonine in all eukaryotic MSRBs as U/CxxS/T. While serine and threonine have polar hydroxyl groups reminiscent of a cysteine, whether they form a covalent bond with the selenocysteine or otherwise contribute to catalysis remains unresolved. Thus this protein has a defunct CxxU motif that was unusual among selenoproteins in having 53 intervening residues.
The exon structures, relative to reliably locatable CxxC anchors, show MSRB2 and MSRB3 more closely related (having a distinctive break within the second CxxC motif), whereas MSRB1 introns are placed altogether differently. This suggests two rounds of gene duplication with intronation of MSRB2/3 prior to the second round. A tree based on the alignable conserved core of these proteins indicates the same result.
The phylogenetic distribution of orthologs of MSRB1 is orderly, with selenocysteine back through fish, with no counterpart currently locatable in amphioxus or tunicates (which otherwise have 3 MSRB genes). Further, there is no sporadic appearance of selenocysein in MSRB2/3 -- cysteine is always found, even prior to bilatera, suggesting though selenocysteine is ancestral (viz prokaryotes) but became cysteine prior to the second duplication giving rise to MSRB2/3.
Care must be taken at greater depth because of possible confusion among paralog members (and lineage-specific expansions and contractions). Synteny is lost so intron phasing and siting must be used along with blast clustering. The selenoprotein rachet predicts as more species are sequenced, some clades may exhibit lost selenocysteine in MSRB1 but none will acquire it in MSRB2 or MSRB3 lineages.
This raises the question that if selenocysteine is not necessary, why is it retained in MSRB1? Possibly that has some connection to localization in the reduced cytosol whereas more oxidizing mitochondrial and ER compartments utilize cysteines. In this view, selenocysteine will not be lost in any vertebrate clade no matter how many species are sequenced for MSRB1; whereas in MSRB23 cysteine was an adaptive change rather than mere rift, allowing wider intracellular distribution. Evidently methionine sulfoxide forms in all three compartments.
32 vertebrate MSRB1 aligned in exon 3: VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK homSap Homo sapiens (human) VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK panTro Pan troglodytes (chimp) VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK ponPyg Pongo pygmaeus (orang_sumatran) VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK macMul Macaca mulatta (rhesus) VSCGRCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFIPK otoGar Otolemur garnettii (bushbaby) VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK micMur Microcebus murinus (mouse_lemur) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFVPK musMus Mus musculus (mouse) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK ratNor Rattus norvegicus (rat) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK cavPor Cavia porcellus (guinea_pig) VSCGKCGHGLGHEFLNDGPKPGQSRFuIFSSSLKFIPK oryCun Oryctolagus cuniculus (rabbit) VFCGKCGHRFGHEFLNDGLKPGQSRFuIFSNTLKFVPK ochPri Ochotona princeps (pika) VSCGKCGNGLGHEFLNDGPKPGKSRFuIFSSSLKFIPK canFam Canis familiaris (dog) VSCGRCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFIPK felCat Felis catus (cat) VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK bosTau Bos taurus (cow) VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK susScr Sus scrofa (pig) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFVPK equCab Equus caballus (horse) VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPR eriEur Erinaceus europaeus (hedgehog) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK loxAfr Loxodonta africana (elephant) VSCGKYGHGLGHEFLNDGPNWGQSRFuIFSSSLKFIPK echTel Echinops telfairi (tenrec) VSCGKCGNGLGHEFLNDGPKKGQSRFuIFSNTLKFVPK triVul Trichosurus vulpecula VSCGKCGNGLGHEFLNDGPRRGQSRFuIFSSSLKFIPK ornAna Ornithorhynchus anatinus (platypus) VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK galGal Gallus gallus (chicken) VLCGKCGNGLGHEFINDGPKKGQSRFuIFSSSLKFVPK anoCar Anolis carolinensis (lizard) VSCGKCGNGLGHEFINDGPKKGQSRFuIFSSSLKFIPK xenTro Xenopus tropicalis (frog) VRCGKCGNGLGHEFVNDGPKHGLSRFuIFSSSLKFIPK danRer Danio rerio (zebrafish) VRCGKCGNGLGHEFLNDGPSRGLSRFuIFSSSLKFIPK tetNig Tetraodon nigroviridis (pufferfish) VRCGKCGNGLGHEFVNDGPSRGLSRFuIFSSSLRFIPK takRub Fugu rubripes (fugu) VRCGKCGNGLGHEFVNDGPAKGVSRFuIFSSSLKFIPK gasAcu Gasterosteus aculeatus (stickleback) VRCGKCGNGLGHEFVNDGPSKGLSRFuIFSSSLKFIPK oryLap Oryzias latipes (medaka) IRCGKCNNGLGHEFLNDGPKHGLSRFuIFSSSLKFV.. ictFur Ictalurus furcatus (fish) VRCGKCGNGLGHEFVGDGPKKGLSRFuIFSSSLKFV.. oncMyk Oncorhynchus mykiss (trout) VRCGKCGNGLGHEFVGDGPKKGLSRFuIFSSSLKFV.. salSal Salmo salar (salmon) .SCGKCGNGLGHEFLNDGLKAGQSRYuIFSNSLKFVPK calMil Callorhinchus milii (elephantfish)
SEPHS1 and SEPHS2: selenophosphate synthetases
Selenophosphate synthetases are biosynthetic enzymes that capture selenium in a chemical form suitable for selenoprotein biosynthesis. One of the two mammalian selenophosphate synthetase paralogs, SEPHS2, itself contains a selenocysteine in a conserved UGCK motif several dozen residues in from the amino terminus. This motif contains threonine as TGCK in SEPHS1 which apparently functions in a selenium salvage system recycling catabolic selenocysteine (tRNA is charged with precursor serine), whereas the SEPHS2 enzyme functions in selenite assimilation (selenophosphate from selenide and ATP). Unsurprisingly only SEPHS2 has a selenocysteine insertion sequence (SECIS) element in 3' UTR.
This gene family has a curious evolutionary history: every additional level of phylogenetic resolution adds another twist -- even 29 genomes from cnidarian to placental do not provide adequate genomic saturation. The original single-copy gene (itself an ancient internal tandem duplicatation fold with active site on dimeric cleft) procedes uneventfully from ur-bacteria to hagfish/lamprey divergence.
However, prior to teleost fish divergence, block duplication of the 8-exon gene took place. No flanking synteny persists in extant species. The copy that would remain closest to the ancestral gene then lost its selenocysteine to nucleophile threonine, its 3' UTR SECIS insertion element decayed, and the old function strayed to salvaging selenide from free selenocysteine (arising from selenoprotein catabolism or diet). The copy we call SEPHS2 diverged rapidly ( rather unconstrained amino terminus in both length and sequence) but with conservation about the selenocysteine active site motif and beyond.
Between monothere and marsupial divergence, a fully processed retrogene (no coding region introns) carrying the 3' UTR SECIS insertion element became functionally established, remarkably displacing the previous 8-exon gene (now an undetectable decayed pseudogene or deletion), to the extent that every species of placental mammal retains only the processed retrogene and threonine copies. Opossum, but not platypus nor elephant or tenrec, retains all 3 gene copies.
Here the processed retrogene mechanism (begin at 3' end of UTR but not extend necesarily to initial methionine or 5' regulatory regions) brings along the SECIS selenocysteine insertion loop making a new selenoprotein feasible and while explaining non-homologous start peptides. Humans have an assortment of fragmentary and processed pseudogenes for which SEPHS1 is usually the parent, most notably a mis-spliced processed pseudogene omitting exon 7 (build 35, chr7:63756925-63757879).
SELO: rapid change in last exon
The terminal exon containing the selenocysteine is evolving very erratically in length, suggesting that is unimportant.
VRRVLKLLETPYHCEAGAATDAEATEADGADGRQR SYSSKPPLWAAELCVTuSS* SELO_homSap Homo sapiens (human) VRRVLKLLETPYHCESGAATDAEATEANGADGRQR SYSSKPPLWAAELCVTuSS* SELO_panTro Pan troglodytes (chimp) VRRVLKLLETPYHCEAGAATDAEATEADGADGRQR SYSSKPPLWAAELCVTuSS* SELO_ponPyg Pongo pygmaeus (orang_sumatran) VRRVLKLLETPYHCEAGAATDAEATEADGADGRQR SYSSKPPLWAAELCVTuSS* SELO_macMul Macaca mulatta (rhesus) VQRVLKLLETPYDNGGGAAAEPKDGSRAASRRP SYSSKPPLWAAELCVTuSS* SELO_otoGar Otolemur garnettii (bushbaby) VRRVLKLLESPYHSEEEATGPEAVARSTEEQS SYSNRPPLWAAELCVTuSS* SELO_musMus Mus musculus (mouse) VRRVLKLLESPYHSEEEATGPEAVARTTDEQS SYSSRPPLWAAELCVTuSS* SELO_ratNor Rattus norvegicus (rat) VRRVLKLLESPYQHEGEHAEALEVAGPEGAATGASRRP SYSSKPPLWAAELCVTuSS* SELO_cavPor Cavia porcellus (guinea_pig) VRRVLELLETPYHRAEEAARVPEATEPEGASGADSGGH SYSSKPPLWAAELCVTuSS* SELO_canFam Canis familiaris (dog) VRRVLKLLETPYGGEAEAEAAEPAEASEAAEETGGAAGRRR SYSSKPPLWAAELCVTuSS* SELO_bosTau Bos taurus (cow) VRRVLKLLETPYHREGEAAEPAEPEAAEGRL SYSSKPPLWAAELCVTuSS* SELO_susScr Sus scrofa (pig) VRRVLKLLEAPYHRAEEAAEVSEAAEPEGAGGASGRRR SYSSKPPLWAAELCVTuSS* SELO_equCab Equus caballus (horse) VRRVLKLLEAPYHREGEAAEVLEAAEPEGAGSTAERRR SYSSKPPLWAAELCVTuSS* SELO_myoLuc Myotis lucifugus (microbat)) VRRVLKLLETPYPQEREAPEALEAAEPQGAGRQD SYSSRPPLWAAELCVTuSS* SELO_eriEur Erinaceus europaeus (hedgehog) VRRVLKLLEAPYSREEPTEALELGEAAGAVGLQR SYSSRPPPWAAELCVTuSS* SELO_loxAfr Loxodonta africana (elephant) VQRVLRLLEKPYGEPWEDDADGLLAAAAAADSGEAESRR SYGRKPPLWAAELCVTuSS* SELO_monDom Monodelphis domestica (opossum) VRKVAKLLEHPYREEEGPDDGEGTADRSVRGL AYGGKPPHWAAQLCVTuSS* SELO_ornAna Ornithorhynchus anatinus (platypus) VRNVLKLLENPFQETEDSTEMETKEEEATATAAACAQATRSRL SYCSKPPLWASELCVTuSS* SELO_galGal Gallus gallus (chicken) VKRVLQMLENPYQEGESCQSIADKSPEEDVAVAASSVSTNPSRL PYNSKPPLWATELCVTuSS* SELO_xenTro Xenopus tropicalis (frog) VQRVLKVLEKPFSVQEGLEQPGWMGRGGAAIPGERDETEEEGSNSSGAGARGLVPYDSKPPVWANEICVTuSS* SELO_danRer Danio rerio (zebrafish) VQEERLRVMEGTNPEFPAWVGGSGEAANQGERDEGEEQQQAMASSSTPRNP VSYDSKPPAWAGEICVTuSS* SELO_takRub Takifugu rubripes (fugu) VHLLQKTLRHPFHKQREAEEA GYSSRPPLWARELRVSCSS* SELO_calMil Callorhinchus milii (elephantfish)
Selenoprotein pseudogenes: SELW
Processed pseudogenes of selenoproteins are of special interest. First, they represent fossil transcripts within germline tissue (somatic cell insertions would not be heritable) offering clues to ancestral functionality, continuity of germline expression and potentially validation of contemporary alternative splicing. Second, retrogenes occasionally retain expression and functionality in descendent clades, thus extending the selenoproteome repertoire. Finding a complete set of selenoproteins in each species has long been a critical research goal.
Since the mechanism of processed retrogene insertion begins with the 3' end, the critical SECIS element and polyadenylation signal would be brought along. Although the process does not always extend to full 5' length, many selenoproteins are quite short, so the prospects of retropositioning a complete intronless copy with a suppressible TGA codon are rather favorable. However regions necessary for initiation of transcription are left behind, making expression and its control subject to properties of pre-existing sequence upstream of the insertion site.
The status of a given retroprocessed locus is not always clear, especially for recent insertions for which insufficient time has elapsed for accrual of definitive pseudogene signatures (such as in-frame non-TGA stop codons, pronounced Kd/Ks ratio, or structurally implausible substitutions at conserved residues). Conventional gene duplicates commonly diverge rapidly away from the parent gene as they neo- or sub-functionalize.
Because few vertebrate genome projects are accompanied by an extensive transcript program (eg mouse, human), absence of supporting expression is not a reliable guide to pseudogene character. Even in human, some well-established coding genes still lack GenBank transcripts as of December 2008, presumably because these are rare or restricted to cell types not represented in studied libraries. Conversely, untranslated transcripts covering non-genes can be found, indeed most of the genome is transcribed at some level despite only 1-2% coding for protein.
Two processes further complicate the characterization of pseudogenes: they become obliterated past the point of reliable recognizability by point mutations and small indels over a few tens of millons of years and they are subject to repeat element insertion (eg Alu elements) that may follow initial rounds of speciation, complicating the establishment of orthologous position especially in intergenic regions not providing stable flanking markers.
These Alu insertions result in pseudo exons which in general lack GT-AG splice sites, conserved reading frame, or parental gene position but nonetheless cause the pseudogene to appear segmentally duplicated or incompletely retro-processed. These factors limit ability to date an initial event and track it forward (reliably interpreted) in all descendent clades.
Each of the 25 human selenoproteins can be used as initial Blat query. Typically the secondary matches (if any) are retrogenes, though a recent pseudogene may score higher because introns of authentic selenogenes interrupt alignment and so lower score. This process tallies the number of pseudogenes back to the phylogenetic depth commensurate with Blat sensitivity -- use of Blast or profile querying may detect still older debris.
Blat suffices here because the primary purpose is finding functional retrogenes paralogous to well-characterized genes. The target genome can be varied to place the event between consecutive phylogenetic tree divergences. Both query protein and target genome can be changed to search for retrogenes in other clades (eg artiodactyl or marsupial).
By this method, some 81 retroprocessed loci can be found for 11 human selenoproteins, suggesting the other 14 selenoproteins are not as frequently expressed in germline tissue. Most of these loci have so many deleterious accrued changes that they can be immediately classified as pseudogenes. Others are not so clear and require detailed case-by-case curation:
SELW gives a single secondary match on a different chromosome from the parental gene, at coordinates chr1:31,339,873-31,341,525 of NCBI assembly 36. By examining syntenic regions from other species established by whole genomic multiz alignment, the event can be dated to post-marmoset, pre-macaque. The retrogene in all old world apes has two apparent exons whereas the parent gene has five.
The RepeatMasker track shows an AluSg retroposon interrupting the reading frame, suggesting (invoking parsimony) a single insertion event prior to macaque divergence. Close examination of the putative exons shows they wholly lack potential splice donor and acceptors (GT-AG or less common alternatives). Consequently the AluSg cannot be spliced out. However if it were included in a transcript, that would introduce non-TGA stop codons as well as grossly disrupt protein structure with extraneous residues (as Alus derive from 7S RNA).
Consequently this feature is a pseudogene in all apes, despite constituting nearly a full length copy of SELW with just 7 conservative substitutions, only frame-preserving indels, Ka/Ks ratios mildly indicative of positive selection, and a SECIS element capable of a stem-loop (though missing two key bases GA at position 78-79).
In support of this, none of numerous ape transcripts align to this feature. However GenBank does contain an unusual macaque cDNA from placenta, CN804216, with two internal stop codons and only 67% identity to SELW from that species. While that might reflect a previously functional selenoretrogene with continuing relic transcription, it fails to match any of the 31 million genomic traces from macaque. Further, the private contractor providing the 2004 sequence reports only 97 Q20 bases. Consequently the sequence is an artefact, though an instructive one.
tBlastn alignment of SELW of M. mulatta with cDNA CN804216
SELW MALAVRVVYCGACGYKSKYLQLKKKLEDEFPGRLDICGEGTPQATGFFEVMVAGKLIHSKKKGDGYVDTESKFLKLVAAIKAALAQG*
MAL V VYCGA Y SKYLQL KKL++EFPGRLDIC E T A+GF E++ G LI S KKGDGYV T S FLKLVA IKAAL G
CN804216 MALVVSCVYCGA*CYYSKYLQLPKKLKNEFPGRLDICCESTRWASGFVEILGTGTLIRSTKKGDGYVHT*STFLKLVADIKAALG*G*
Macaque contains a second contiguous retroprocessed SELW locus at chr11:99257376-99257637 that can be detected by Blast but not Blat. As expected, it is considerably more decayed (3 stop codons, a frameshift, numerous substitutions) than the retroprocessed feature discussed above. This feature is found from marmoset to human but is missing in tarsier, lemurs, and earlier diverging mammals Using the UCSC 44-species genomic alignment). Consequently it is an older feature predating new world monkey divergence.
SELW MALAVRVVYCGACGYKSKYLQLKKKLEDEFPGRLDICGEGTPQATGFFE----VMVAGKLIHSKKKGDGYVDTESKFLKLVAAIKAALAQG
M L +++YCGA G KSKY QLKK LED+ P LDIC TP TGFFE MVAG L KK DGY++ ESKFLKLVA IKA LAQG
Retro2 MPLMFQIIYCGA*G*KSKYFQLKK-LEDKSPRYLDICSNRTP*VTGFFEGMVVFMVAGSLQEDKKISDGYMNGESKFLKLVATIKAGLAQG
Clade-specific SELW pseudogenes are most conveniently survey by tBlastn of NCBI database wgs, which contrary to its name contains assembled contigs from all genome projects, including short contigs omited from final genome assemblies. Restricting the query to mammals, many species exhibit retroprocessed products of SELW including Dasypus (4), Choloepus (2), Loxodonta, Echinops, Tupaia, Bos (2), Felis (2), Myotis, Rattus, Microcebus (2), and Tarsius in addition to the ape retroprocessed loci discussed above. While not all of these situations can be definitively declared pseudogenes, it can be concluded that SELW is transcribed in germline tissue in a wide variety of extant placentals and thus in their last common ancestor.
The locus in armadillo contig AAGV020179194 is noteworthy. It is either a very recent pseudogene or functional (being 85% identity to sloth SELW). A retrogene under selection should have a subsitution site mismatch with conserved residue profile because the latter sites would still mostly be important despite putative re-functionalization. The structure of mouse SELW has recently been determined to be a thioredoxin-like fold (PDB: 2NPB), similar to SelV, SelT, SelH and Cys-containing Rdx12 (aka C17orf37) as shown in Fig.4 of that paper (below).
SELW MALAVRVVYCGAUGYKSKYLQLKKKLEDEFPGRLDICGEGTPQATGFFEVMVAGKLIHSKKKGDGYVDTESKFLKLVAAIKAALAQG 87
MALAVR+VYCGA GYK KY+QLKK+LEDEFPG LDI GEGTPQ TGFFEVMVAGKL+HSKK+GDG+VD ESKFL+LV AIKA LA G
dasNov MALAVRMVYCGA*GYKPKYIQLKKRLEDEFPGCLDISGEGTPQTTGFFEVMVAGKLVHSKKRGDGFVDMESKFLRLVPAIKATLAHG 619
Microarrays for human SELW show it predominantly expressed in heart. Available macaque transcripts (tBlastn of est_others restricted to Macaca) have been recovered from lens, thymus, kidney, liver, brain, ovary, lymphocytes, ileum, jejunum and testis (DR771274).
Alternative splices of selenoproteins: introduction
In the search for significant alternate splice forms of selenoproteins, it is very important not to take transcript data at face value. Even if multiple transcripts support a given alternative splice, that does not imply that such transcripts exist in vivo in healthy cells (millions of GenBank entries come from cancer cell lines), nor that any such protein is ever made (nonsense-mediated decay), nor if translated that the protein is stably folded (rather than immediately degraded at the protosome), nor that stable protein fragments (such as selenoproteins missing their redox site) are functional. Due to post-translational trimming of amorphous termini, many alternative splices in distal regions would be irrelevent to mature protein.
On the experimental quality side, human and mouse transcript entries at GenBank contain millions upon millions of defective entries, notably from the early years of EST factories. Transcripts can be riddled with direct sequencing error and not align satisfactorily to any assembled genome. Others are accurate but represent cross-species laboratory contamination. A vast number do not correspond to plausible coding genes, remain unvalidated by later work, and seem merely to have been primed off naturally occuring non-genic genomic polyA. It appears that many partially processed transcripts (5' caps not required, intron-retaining) entered the sequencing queue. Consequently, each alternative splice transcript must be individually evaluated for various parameters affecting its quality.
Despite vast expenditures on proteomics mass spectroscopy, not 1 in 10,000 proposed alternatively spliced products has been validated to date in vertebrates. Indeed for fully half of all human proteins, no product -- much less an alternative splice form -- has ever been reported (according to SwissProt, November 2008). Some protein structure scientists question whether the vast majority alternative splice forms can stably fold as it makes no sense from a folding energetics perspective to omit a beta strand from a sheet or insert a helix into an existing sandwich.
In this view, vertebrate transcription is inherently an error-prone and wasteful process with imperfect splicing accuracy mitigated downstream by a series of quality controls. This is perhaps inevitable give intrinsically weak splice signals (the core GT and AG occur by random every 16 bp) dwarfed by the vast expansion of many internal introns resulting from a retroposon explosion that masks splice donors and acceptors with the effect of intron retention and exon skipping, respectively. For example the 79 coding exons of dystrophin (the largest human gene) occupies 11 kbp of a 2,400,000 bp transcript meaning introns average some 30,000 bp.
Alternate splicing is often invoked to explain why humans are "special" yet this process is vastly more extensive in fruit flies and early-diverging life forms. Human proteins are among the slowest evolving and least innovative of any mammalian species, evolving at a third the rate of mouse. Take any cartilaginous fish protein, tblastn against all vertebrate genomes (thus avoiding annotation effort bias), and find human is very often the top hit. Consequently anthropomorphic justification of alternate splicing is wholly unwarranted.
Thus screening the nominal selenotranscriptome for functionally significant alternative splices requires multiple types of strong supporting data to overcome the null hypothesis (artefact). It is extremely time-consuming to pursue candidates further experimentally, so prioritization by quality is imperative.
Because documented human innovation past chimpanzee divergence is so minimal, the vast majority of valid alternate splices should exhibit strong comparative genomics support at significant phylogenetic depth. Unlike transcript support, whose availability and quality differ radically by species, comparative genomics support is uniformly available in a large set of phylogenetically spaced genomes and readily intepretable.
Alternative splicing in 5', coding and 3' regions are separate topics. Of these, coding has the best prospects for bioinformatic validation especially when a determined 3D structure permits modelling of the alternatively spliced product and can supplement phylogenetic aligment. However comparative genomics is still applicable in some other cases at least to the extent of dating the phylogenetic origin of and persistence of the splice donor and acceptor.
Evaluating an alternative 3' splice of SELS
SELS has a well-known 3' non-coding alternative splice (RefSeq NM_203472) which skips the SECIS element to terminate downstream in a spearate non-coding exon. Since the selenocysteine is near-terminal position, a nearly full-length non-functional protein could be produced, yet the transcript might be subject to nonsense-mediated decay, seemingly a very wasteful and atypical way (compared to regulation at the upstream promoter) of down-regulating functional SELS production. Another option is that SELS forms an essential homodimer -- having one monomer sparing a need for selenium could provide a selective advantage (even as a quarter of the dimers would be duds).
It might be inferred from extensive and available microarray data how SELS is expressed, yet that data is irrelevent if NMD is operative (making transcript level an invalid proxy for protein level). Since the function of SELS is not known, experimental conditions in which a mouse model might down-regulate SELS are unclear; the alternate splice donor could be knocked out in mouse yet no phenotypic effect might be observable.
Conservation depth of the splice donor and acceptor are readily established at the UCSC 44-species whole genome alignment. Here we find that the GT donor in homologous position applies to most euarchontoglires and laurasiatheres (5 exceptions) but earlier diverging species lacks it. The extent of 3' sequence homology (chimp, macaque, bar, mouse, cow) extends to the region down to the SECIS but not strikingly to the alternate acceptor region. The AG acceptor site is similarly only feasible in euarchontoglires (at precise homological position).
Consequently, comparative genomic support is fairly shallow, not greatly beyond random conservation expectations. There is more to splice junctions than just GT-AG and other species need further testing with predictive tools (eg GeneScan) for junctional validity. It is worth noting that a Papio anubis EST GE878368 dated 2008-10-19 supports this splice (as do numerous human transcripts):
3' UTR donor and acceptor splice match of baboon transcript GE878368 to human SELS chr15:99630478-99629374 GGCGGATGAgGCTAAgAATCTTgttagtg...atctcctgcagGTAGAATATTCC Papio GGCGGATGAGGCTAAGAATCTTgttagtg...atctcctgcagGTAGAATATTCC Homo
Overall, this raises more questions than answers. The alternate splice cannot be dismissed as an artefact yet cross-species support for it is underwhelming. Supposing the selected function is to down-regulate this gene product, would not other mammals have a similar need? If they utilized another mechanism present for many million years of last common ancestor, euarchontoglires then lost that well-established mechanism and gained a novel one over a fairly brief evolutionary span.
Evaluating an exon-skipping, frame-shifting alternative splice of SEP15
The human SEP15 locus has two recognized splice forms at RefSeq resulting in proteins that differ markedly in distal sequence. The main splice form yields five exons. These exons, the seleoncysteine position, and the conventional stop codon are extraordinarily conserved back to lamprey, indeed throughout bilatera. This is clearly the ancestral form of SEP15.
>SEP15A_homSap length=165 NM_004261 0 MAAGPSGCLVPAFGLRLLLATVLQA 0 0 VSAFGAEFSSEACRELGFSSNLLCSSCDLLGQFNLLQLDPDCRGCCQEEAQFETKK 0 0 LYAGAILEVCGuKLGRFPQVQ 1 2 AFVRSDKPKLFRGLQIK 0 0 YVRGSDPVLKLLDDNGNIAEELSILKWNTDSVEEFLSEKLERI* 0 >SEP15B_homSap length=124 NM_203341 0 MVAMAAGPSGCLVPAFGLRLLLATVLQA 0 0 VSAFGAEFSSEACRELGFSSNLLCSSCDLLGQFNLLQLDPDCRGCCQEEAQFETKK 0 0 LYAGAILEVCGuKLGRFPQVQ 1 2 VCPWFRPCIKAFGRQWEHC* 0
The second splice form skips the fourth exon, jumping from the splice donor of the third exon to the splice acceptor of the fifth exon. The donor phases are such that the reading frame is shifted, causing the final exon to have altogether different amino acid sequence from the main splice form. The new reading frame reaches a stop codon after 20 amino acids but much earlier in 6 other species of mammals including old world monkeys and lemurs:
Premature stop codon in macaque final exon of exon-skipping splice form
Homo VCPWFRPCIKAFGRQWEHC*
VCPWFRP IKAFG QWEHC
Macaca VCPWFRPYIKAFG*QWEHC
A goodly proportion of human ESTs skip exon 4 and a 2002 Riken mouse transcript BY235176 provides some comparative genomics support. However exon-skipping is not observed in a very large number of other mammalian transcripts:
391 of 392 mammalian transcripts in 9 species do not skip exon 4 Mus musculus (mouse) 198 (only BY235176 skips) Papio anubis (Doguera baboon) 85 Macaca mulatta (rhesus macaque) 30 Macaca fascicularis (cynomolgus monkey) 22 Cavia porcellus (guinea pig) 2 Spermophilus tridecemlineatus (squirrel) 2 Bos taurus (cow) 11 Ovis aries (domestic sheep) 34 Equus caballus (equine) 8
The structure of the non-skipping form of SEP15 was recently determined. Omission of exon 4 would obliterate almost all secondary structure, causing complete loss of helix 1, helix 2 and beta strand 2; the reading frameshift would take out almost all the rest (though the structure if any of the short form in this region is not known). The short form would be left only with its signal peptide, a single (ie dysfunctional) beta strand, and the cysteine-rich region.
Clearly the long form of SEP15 is the only relevent protein species at this locus. It is not feasible to have selection acting simultaneously on different translated reading frames; it is implausible that the short form could fold stably but wholly differently given a very brief evolutionary history. Conceivably the cysteine-rich region could still function though the protein would likely be subject to very rapid decay. The role of the selenocysteine without its supporting tertiary structure would be unclear.
Thus the alternative splice forms here, though too abundant to be experimental artifact, are misleading. They do not in all likelihood encode an alternative functioning protein. Instead, they may merely reflect a human polymorphism in the intron preceding exon 4 that has the effect of weakening it as a splice acceptor (alternatively a polymorphism enhancing the splice acceptor preceding exon 5). This amounts to the final splice acceptor out-competing the fourth exon acceptor in a substantial fraction of transcripts.
The net effect of is a certain inefficiency but nothing particularly detrimental to the cell: functional SEP15 selenoprotein is still produced and the fragmentary mis-spliced polypeptide is rapidly recycled. While this down-regulates SEP15 production, as an evolved adaptive mechanism, it lacks any depth of comparative genomics support.
An AluJb-translating splice form of primate SEPN1
Two splice forms of SEPN1 have gained prominence, the 12 exon NM_206926 and the 13-exon NM_020451. Both have GenBank status of "Reviewed RefSeq: This record has been curated by NCBI staff" but that process has never been described and is never accompanied by explanatory comments. The difference between the two RefSeqs is an alternately included exon 3 which supposedly contains an additional TGA selenocysteine codon.
However, far more likely, this is gross annotation error. As noted but not genetically analyzed in a publication, the extra exon is nothing but an AluJb retroposon that inserted in minus strand orientation, with the polyA stretch serving as the pyrimidine stretch (polyT) augmenting a random GT to create a splice acceptor. This introduced a downstream inframe stop codon that happened to be TGA and so was "written off" as a disabling mutation because the later valid selenocysteine codon would be accompanied by a SECIS insertion element. As a further coincidence, a GT donor past the stop codon (but still well within the 296 bp AluJb) restored reading phase.
The insertion event is readily dated using the 44-species genomic alignment at UCSC to the post-lemur, pre new world monkey stem. This implies that homologous amino acids will not exist in the SEPN1 genes of any other primate (or vertebrate). That is readily validated by blast searches of all genomes and transcripts at GenBank.
Although this new exon could represent a dramatic primate innovation, other lines of reasoning strongly suggest this extra exon is not functional. Retroposons are indeed occasionally co-opted for functional purposes; however 99.999% have no remotely plausible gene role and simply represent junk dna (as proven by various engineered megabase deletions in mice and massively reduced but still satisfactorily advanced fugu genome).
Alu-inactivating mutations are far more commonly a source of genetic disease; however this particular AluJb does not inactivate the gene but simply results in a minor level of transcriptional noise from the occasional mis-splice. It cannot be argued that natural selection would have rapidly removed it from the genome in view of the other 1.1 million SINE elements in the human genome -- and these continue to accrue faster than they are being deleted. Statistical arguments concerning Ks/Ka cannot be applied to a mere 96 bp because nothing is known about evolutionary processes or rates in this very localized region of chromosome 1.
AluJb SINE elements arise from 7S RNA, not proteins as in LINE elements. This distinction is exceedingly important. It means the extra exon was never vetted by selection for its compositional or folding properties. Indeed the reading frame here arose randomly from the three available. Random peptides of 32 amino acids have near-zero propensity for useful structural properties.
Normally transcripts containing an inframe stop codon resulting from occasionally spicing in an Alu would be dismissed as junk rapidly degrading via nonsense-mediated decay. However an inframe TGA stop codon here must be given special consideration because SEPN1 already has a long-established selenocysteine site in exon 9 accompanied by a SECIS element in 3' UTR. There is precedent for a single SECIS element to substitute selenocystein at multiple TGA sites, notably the selenocysteine storage protein.
However there is much more to a selenocysteine-substitutable site than just TGA. It appears that a cis-acting selenocysteine redefinition element (SRE) is also required for any efficiency. This small stem-loop immediately downstream has been specifically studied in SEPN1 exon 9 where it was determined to be a phylogenetically conserved stem–loop structure that starts 6 bp downstream from the UGA codon of exon 9 of SEPN1. It provides measurable but low levels of read-through via both selenocysteine and cysteine tRNAs. No such SRE can be detected in the AluJB-derived exon region.
Although SRE elements are successfully predicted by genomewide bioinformatic EvoFold searches for conserved RNA folds, no such element is anticipated at the putative novel AluJb site. Selenocysteine insertion is inefficient to begin with. A second site lacking an SRE would result in multiplicative inefficiency, resulting in hardly any production of full length protein even if transcripts escaped the nonsense-mediated decay machinery. Since only a fraction of the transcripts encode the AluJb exons, it is implausible that significant in vivo levels of alternatively translated protein would occur.
offset 0 1 2 3 hg18.chr1 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC monDom4 GGGCGGACTCTCCGGGAAACAGTCCTGGAAAGTTCGCCC echTel1 -GGCGGACTCTCCGGGAGACAGTCCTGGAGAGCCCG-CC loxAfr1 GGGCGGACTCTCCGGGAGACTGTCCTGGAGAGTTCGCCC dasNov1 GGGCGGACTCTCCGGGAGACCGTCCTGGAAAGTTCGCCC canFam2 GGGCGGACTCTCCGGGAGACCGTCCTGGAAAGTTCGCCC bosTau2 GGGCGGACTCTCCGGGAGACCGTCCTGGAAAGTTCGCCC oryCun1 GGGCGAACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC mm8 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGCCCGCCC rn4 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGCCCGCCC rheMac2 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC panTro1 GGGCGGACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC galGal2 GGGCGAACTCTCCGGGAGACTGTCCTGGAAAGTTCGCCC xenTro1 GGGCGAACTCTCCGGGAGACCGTCCTAGAGAGTTTGCCC danRer3 GGGCGGACTCTTCGGGAGACAGTTCTCGAGAGTTCGCCC tetNig1 GGGCGGACTCTCCGGGAGACGGTGCGGGAGAGTTCGCCC fr1 GGGCGAACTCTCCGGGAGACAGTCCTGGAAAGTTCGCCC SS anno (((((((((.(((((((.....))))))).))))))))) pair symbol abcdefghi jklmnop ponmlkj ihgfedcba score 999999999799999989999989999997999999999
The UGA codon of exon 9 of SEPN1 is remarkable for occuring at the very end of the exon (human ...DDQSCu 12 GSGRTLRETVLESSP... denotes a complete TGA codon followed by 1 bp of the next codon which is completed to glycine by the first 2 bp of the next exon). The SRE, here a 39 bp stemloop occurs just after the splice acceptor and initial 2 residues of the next exon. The stemloop nucleotides thus selectively compete with whatever role the encoded 13 amino acids have, though there is opportunity to evolve independently at 11 third codon positions.
The fold of SEPN1 is unclear. It has no known Pfam domain other than an EF hand at position 67-102, no paralogs within vertebrate genomes and no resemblance to any previously determined 3D structure by Blastp against PDB. A thioredin domain cannot be detected in the expected SCUG region by SuperFamily, SMART, or Pfam search.
The primary sequence is moderately well conserved and full-length orthologs (all selenocysteine encoding in exon 9) are readily curated back to lamprey. The intron pattern is completely invariant and never contains supplemental exons in any lineage. The selenocysteine is evidently critical because no species has substituted cysteine, though homologs outside of deuterostomes are quite difficult to locate and do not encompass fungi or bacteria. The earliest diverging species, sponge, does not have muscle cells. None of the species are good model organisms; the gene appears to have been lost in drosophila and nematode.
Nematostella vectensis (cnidaria) TGA-confirmed selenocysteine and phase 1 of exon 9
homSap VSYLPFTEAFDRAKAENKLVHSILLWGALDDQSCCGSGSGRTLRETVLESSPILTLLNESFISTWSLVKELE
V YLPF EAF+RAKAE KLVH I+LWGALDDQSC G+GSGRTLRE LES P+L LL +++++ WSLV+EL+
nemVec VDYLPFKEAFNRAKAEKKLVHHIVLWGALDDQSC*GNGSGRTLREGPLESRPVLQLLQDNYVNCWSLVEELK
Aplysia californica (mollusc) TGA-confirmed selenocysteine
homSap QFVFEEIKWQQELSWEEAARRLEVAMYPFKKVSYLPFTEAFDRAKAENKLVHSILLWGALDDQSCCGSGRTLRETVLESSPILTLLNESFISTWSLVKELE
+F +++I W+QE+S +EA LE+ ++PFK+V Y +EAF A+AE+KLVH I+LWGALDDQSC GS RTLRE+ L+ ++ LL++ F+S+W+L+ +L+
aplCal EFSWDDIAWEQEISEQEARDALELKLFPFKQVNYHNLSEAFTVAQAESKLVHCIVLWGALDDQSC*GSARTLRESALQGPKVMALLHQRFVSSWTLLVDLK
Ixodes scapularis (tick) TGA-confirmed selenocysteine
aplCal QVNYHNLSEAFTVAQAESKLVHCIVLWGALDDQSCXGSARTLRESALQGPKVMALLHQRFVSSWTLLVDLK
QV YHNL+ A +A+ E K+VH +VLWG LDDQSC GS RTLR L PKV LL + F+S W++ LK
ixoSca QVPYHNLTTAAALARQERKMVHSVVLWGNLDDQSC*GSGRTLRRGPLASPKVQELLLKHFISMWSITAQLK
Oopsacas minuta (sponge) TGA-confirmed selenocysteine
homSap FEEIKWQQELSWEEAARRLEVAMYPFKKVSYLPFTEAFDRAKAENKLVHSILLWGALDDQSCCGSGRTLRETVLESSPILTLLNESFISTWSLVKELEELQNN
FE W E + EEA L+ +YPFK++ Y+ +A D A+ ENKLVHSI++WG+LDDQSC GSGRTLR+ L S + L E+FIS+WSL+ LE+L N
oopMin FEIPAWDHEKTKEEATIELDRVLYPFKRIPYIDIRDAVDTARVENKLVHSIIMWGSLDDQSC*GSGRTLRDGPLASDVVQLTLKENFISSWSLIVHLEDLMAN
Because the region containing the alternatively spliced exon is strongly conserved over many billions of years of vertebrate branch length, it is exceedingly likely to have a precisely defined stable functional fold. Because intron position almost never corresponds to natural domain boundaries (even in proteins with multiply interated domains such as actinin), introducing 32 gratuitous residues via a new exon is very likely to be structurally disruptive. Here the final two residues of the calcium responsive EF hand are disrupted.
High conservation of exons 2-3 flanking the putative new exon in 22 species of vertebrate: SEPN1_homSap ELALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT GSTPAASCEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG SEPN1_panTro ELALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT GSTPVASYEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG SEPN1_ponAbe ELALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT GSTPAASYEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG SEPN1_calJac ESALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT GSTPEASYEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG SEPN1_micMur ESALKALGTEGLFLFSSLDTDQDMYISPEEFKPIAEKLT GSTPAASYEDEGLPADPSEETLTIEARFQPLLLETMTKSKDGFLG SEPN1_tupBel EAARKVLGTDGLFLFSSLDTDRDMYISPEEFRPIAEKLT GSTPAASYEE--PAPDPSEETLTIEARFQPLLPETMTKSKDGFLG SEPN1_musMus ESALKVLGTDGLFLFSSLDTDQDMYISPEEFKPIAEKLT GSVPVANYEEEELPHDPSEETLTIEARFQPLLMETMTKSKDGFLG SEPN1_ratNor ESALKVLGTDGLFLFSSLDTDQDSYISPEEFKPIAEKLT GSVPVASYEEEELPHDPSEETLTIEARFQPLLTETMTKSKDGFLG SEPN1_cavPor ESALKALGTEGLFLFSSLDTDQDMRLSPEEFKPIAEKLT GSTLTASYEEEELPLNPSEETLTIEARFQPLLPETMTKSKDGFLG SEPN1_ochPri ETVRKTLGTKSFFLFSSLDTDQDLYISPEEFKPIAEKLT GSAAAPSYEEEELTADPSEETLTIEARFQPLRPETMTKSKDGFLG SEPN1_turTru DSALKALGTEGLFLFSSLDTDGDMYLSPEEFKPIAEKLT GSTPSASYEEEDLPPDPSEETLTIEARFQPLLPESMTKSKDGFLG SEPN1_bosTau DSALKTLGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT GSTPTANYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG SEPN1_equCab ESALKALGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT GSTPAASYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG SEPN1_canFam ESALKALGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT GSTPAASYEEEELPLDPSEETLTIEARFQPLIPESMTKSKDGFLG SEPN1_myoLuc ESAMKALGTEGVFLFSSLDTDRDLYLSPEEFGPLAEKLT GSTPTTSYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG SEPN1_eriEur KSARKVLGTEGLFLFSSLDTDQDMSINPEEFRPIVEKLI GSTPKPSYEEEELSSDPREEKLTIETRFQPLLLNSMTKSNSRFPG SEPN1_loxAfr --ALKALGTEGLFLFSSLDTDQDMYISPEEFKPIAEKLT GSTPATSYEDEDLPPDPSEETLTIEARFQPLLPETMTKSKDGFLG SEPN1_proCap ESALKALGTEGLFLFSSLDTDQDMYISPEEFKPIAAKLT GSTPATSYEDEELPPDPSEETLTIEARFQPLIPETMTKSKDGFLG SEPN1_galGal ELALKSLGSEGLFLFSSLDTNNDLYLSPEEFKPIAEKLT GVTPVSDFEEDA--PDPNGETLSIVAKFQPLVMETMTKSKDGFLG SEPN1_xenTro EAALRTLGAEGLFLFSSLDTDNDMHISPEEFKPISEKLT GISTTSDYEEEE-LLDPNGETLSVASRFQPLLMETMTKSKDGFLG SEPN1_danRer EAGLKALGADGLFFFSSLDTDHDLYLSPEEFKPIAEKLT GVAPPPEYEEEI-PHDPNGETLTLHAKMQPLLLESMTKSKDGFLG SEPN1_petMar EMALRTLGNDGLFLFTSLDTNMDMQISPEEFRPIVDKII GPPPSEYE--GTQEADPQGEGLTMLARFEPLLMETMSKSRDGFLG
While the function of SEPN1 (an endoplasmic reticulum glycoprotein with early developmental expression) is still obscure, it evidently involves a redox role for the selenocysteine (which occurs thioredoxin-like immediately adjacent to an invariant cysteine as SCUG rather than as UxC or UxxC). Since the redox function is already met, what purpose is served by introducing a gratuitous additional selenocysteine at a site far removed from the active site or evolved interaction sites with other enzymes? Although the new site is of the UxxC form common in other functional selenoproteins, this is likely an accidental outcome of ancestral AluJb translation (rather than subsequently evolved) because it is present in all descendent clades.
Finally it's worth noting that SEPN1 is a much-studied disease gene,rigid spine with muscular dystrophy type 1. None of the known disease alleles (notably M1V, G273E, G315S, R466Q, H293R, N340I, U462G, U462x, W453S, R466Q) map to the region of the putative new exon. While two separate disease alleles modify the selenocysteine codon in exon 9 none effect the putative new selenocysteine in exon 3.
No physical evidence has ever been put forward for the existence of the extra exon in stable protein -- it is all based on genomic inference and transcripts. There is however solid experimental evidence for the short form of the protein. The long form cannot even be detected under transfection conditions in which it should be greatly over-expressed.
In summary, the evidence clearly refutes folklore of AluJb co-optively recruited as a coding exon of an alternatively spliced protein. Instead of representing a primate innovation, the AluJb instead creates a subclinical disease state, partially knocking out an essential gene by causing a fraction of its expression to be mis-spliced to a deviant untranslatable transcript. The GenBank entry NM_020451 -- which cites but fails to read the key 2003 paper -- is incompetently annotated: no 590 residue protein with two selenocysteines exists. This error has propagated irrevocably into hundreds of bioinformatic databases as mis-numbering of disease alleles and related problems.
Selenoprotein sequences available from genome project species
...... DIO1 DIO2 DIO3 GPX1 GPX2 GPX3 GPX4 GPX5 GPX6 GPX7 MSRB1 MSRB2 MSRB3 SELH SELI SELK SELM1 SELM2 SELO SELS SELT SELU1 SELU2 SELU3 SELV SEPHS1 SEPHS2 SEPN SEPP1 SEPP2 SEPW TXNRD1 TXNRD2 TXNRD3 homSap DIO1 DIO2 DIO3 GPX1 GPX2 GPX3 GPX4 GPX5 GPX6 GPX7 MSRB1 MSRB2 MSRB3 SELH SELI SELK SELM1 SELM2 SELO SELS SELT SELU1 SELU2 SELU3 SELV SEPHS1 SEPHS2 .... SEPP1 ..... SEPW TXNRD1 TXNRD2 TXNRD3 panTro DIO1 .... .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... SELS .... ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... ponPyg DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... macMul DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... SELK ..... ..... .... SELS SELT ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... calJac DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... tarSyr .... .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... otoGar DIO1 DIO2 .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... SELS SELT ..... ..... ..... SELV ...... ...... .... ..... ..... .... ...... ...... ...... micMur DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... tupBel DIO1 DIO2 .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... musMus DIO1 .... DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 MSRB2 MSRB3 SELH .... SELK ..... SELM2 SELO SELS SELT ..... ..... ..... SELV SEPHS1 SEPHS2 .... SEPP1 ..... SEPW ...... ...... ...... ratNor DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... SELK ..... SELM2 SELO SELS SELT ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... cavPor DIO1 .... DIO3 .... GPX2 .... .... .... .... .... MSRB1 ..... ..... .... .... SELK ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... speTri DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... dipOrd DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... oryCun DIO1 .... .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... ochPri DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... canFam DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 MSRB2 MSRB3 SELH .... SELK ..... SELM2 .... SELS SELT ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... felCat DIO1 .... DIO3 GPX1 GPX2 .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... equCab DIO1 DIO2 .... GPX1 GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... SEPP1 ..... .... ...... ...... ...... myoLuc DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... SELS .... ..... ..... ..... SELV ...... ...... .... ..... ..... .... ...... ...... ...... pteVam DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... .... ...... ...... ...... bosTau DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 MSRB2 ..... SELH .... SELK ..... SELM2 .... SELS SELT ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... oviAri .... .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... susScr DIO1 DIO2 DIO3 .... .... .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... SELM2 .... SELS .... ..... ..... ..... SELV ...... ...... .... ..... ..... SEPW ...... ...... ...... turTru DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... vicVic DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... eriEur DIO1 .... .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... SELS .... ..... ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... sorAra .... DIO2 .... GPX1 GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... dasNov .... .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... choHof DIO1 .... .... .... GPX2 .... .... .... .... .... ..... ..... ..... SELH .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... SEPP1 ..... SEPW ...... ...... ...... loxAfr DIO1 .... .... .... GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... SELV SEPHS1 SEPHS2 .... ..... ..... .... ...... ...... ...... proCap DIO1 .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... SELV ...... ...... .... ..... ..... .... ...... ...... ...... echTel DIO1 DIO2 DIO3 .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... monDom DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 ..... ..... SELH .... .... ..... ..... .... SELS SELT SELU1 ..... ..... .... SEPHS1 SEPHS2 .... ..... SEPP2 SEPW ...... ...... ...... macEug .... .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... SELU1 ..... ..... .... ...... SEPHS2 .... ..... SEPP2 .... ...... ...... ...... ornAna DIO1 DIO2 DIO3 GPX1 GPX2 .... GPX4 .... .... GPX7 MSRB1 MSRB2 MSRB3 SELH SELI SELK SELM1 SELM2 SELO SELS SELT SELU1 ..... ..... .... SEPHS1 SEPHS2 .... SEPP1 SEPP2 SEPW ...... ...... ...... tacAcu .... .... .... GPX1 .... .... GPX4 GPX5 .... GPX7 ..... ..... ..... SELH .... SELK SELM1 ..... SELO SELS .... SELU1 ..... ..... .... SEPHS1 SEPHS2 .... SEPP1 ..... .... ...... ...... ...... galGal .... DIO2 DIO3 GPX1 GPX2 GPX3 .... .... .... .... MSRB1 ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... ..... .... ...... ...... ...... taeGut .... .... .... .... .... .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... .... .... SELU1 ..... ..... .... ...... ...... .... ..... SEPP2 .... ...... ...... ...... anoCar DIO1 DIO2 .... GPX1 .... .... .... .... .... .... MSRB1 ..... ..... .... .... SELK ..... ..... .... .... .... SELU1 ..... ..... .... ...... ...... .... ..... SEPP2 SEPW ...... ...... ...... xenTro DIO1 DIO2 DIO3 GPX1 GPX2 .... .... .... .... .... MSRB1 MSRB2 ..... .... .... SELK ..... SELM2 .... SELS .... ..... ..... ..... .... SEPHS1 SEPHS2 .... SEPP1 SEPP2 SEPW ...... ...... ...... danRer DIO1 DIO2 DIO3 GPX1 .... .... .... .... .... .... MSRB1 ..... ..... SELH .... .... ..... SELM2 .... SELS .... ..... ..... ..... .... SEPHS1 ...... .... SEPP1 SEPP2 SEPW ...... ...... ...... tetNig .... DIO2 DIO3 GPX1 .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... SEPP2 .... ...... ...... ...... takRub DIO1 DIO2 DIO3 GPX1 .... .... .... .... .... .... ..... ..... ..... SELH .... .... ..... ..... .... .... .... SELU1 ..... ..... .... ...... ...... .... ..... ..... SEPW ...... ...... ...... gasAcu .... DIO2 DIO3 GPX1 .... .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... SEPP2 .... ...... ...... ...... oryLap .... DIO2 DIO3 GPX1 .... .... .... .... .... .... ..... ..... ..... SELH .... SELK ..... ..... .... .... .... ..... ..... ..... .... ...... ...... .... ..... SEPP2 .... ...... ...... ...... calMil DIO1 DIO2 DIO3 .... GPX2 .... .... .... .... .... ..... ..... ..... .... .... .... ..... ..... SELO SELS .... SELU1 ..... ..... .... SEPHS1 ...... .... ..... SEPP2 SEPW ...... ...... ...... petMar .... DIO2 .... .... GPX2 .... .... .... .... .... ..... ..... MSRB3 .... .... .... SELM1 ..... SELO SELS SELT ..... ..... SELU3 .... SEPHS1 ...... SEPN ..... ..... SEPW ...... ...... ...... braFlo .... .... DIO3 .... .... .... .... .... .... .... ..... ..... ..... .... .... SELK ..... ..... .... .... .... SELU1 ..... ..... .... SEPHS1 ...... .... ..... ..... .... ...... ...... ......
Reference set of 656 vertebrate selenoproteins
The sequences below reflect the full range of deuterostome species for which reliable full-length orthologs could be collected. The protein sequences display position and phase of each exon. The selenocysteine is displayed as lower case u for visibility. That residue is often displaced in tetrapods on with cysteine, typically as phyloSNP though sometimes with erratic persistence within mammals.
SELH: 25 vertebrate sequences
>SELH_homSap Homo sapiens (human) NP_734467 Selenoprotein H exons chr11 80% identity musMus 0 MAPRGRKRKAEAAVVAVAEKREKLANGGEGMEEATVVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPTKPRRGSFEVTLLRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_ponPyg Pongo pygmaeus (orang_sumatran) 0 MAPRGRKCKAEATVVAVAEKrEKLTNGGEGMEEATIVIEHC 2 1 TSuRVYGRNAAALSQVLCLEAPELPVKVNPTKPRRGSFEVTLLRPDGS 1 2 SVELWTGIKKGPPCKLKFPEPQEVVEKLKKYLS* 0 >SELH_macMul Macaca mulatta (rhesus) 0 MAPRGRKRKAEAAMVAAAEKQEKLANSGEGMEETTVVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPSKPRRGSFEVTLLRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_micMur Microcebus murinus (mouse_lemur) 0 MAPRGRKRKAEASVVATAEKREKLENGGEAVEEATVVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPAKPRRGSFEVTLQRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQVVVKELKKYL.* 0 >SELH_tupBel Tupaia belangeri (tree_shrew) 0 MAPRGRKRKAEAAVVATAEKQEKLQNGGEGVKEASIVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNSAKPRRGSFEVTLLRPDGS 1 2 SVELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_musMus Mus musculus (mouse) 0 MAPHGRKRKAGAAPMETVDKREKLAEGATVVIEHC 2 1 TSuRVYGRHAAALSQALQLEAPELPVQVNPSKPRRGSFEVTLLRSDNS 1 2 RVELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_ratNor Rattus norvegicus (rat) TGA verified 77% 0 MAPLGRKRKAGAAPIESADKREKLAEGAAVVIEHC 2 1 TSuRVYRRHAAALSQALQLEAPEISVQVNRSKPRRGSFEVTLLRPDNS 1 2 RVELWTGIKKGPPRKLKFSEPQEMVEELKKYLS* 0 >SELH_speTri Spermophilus tridecemlineatus (squirrel) 0 MAPRVRKRKAEAAAVSTSEKREKLENGKEQVEEAVVIEHc 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPSKPRRGSFEVTLLRRDGTS 1 >SELH_oryCun Oryctolagus cuniculus (rabbit) 0 MAPGKRKRKAEAAPVASAEKREKLANGGQGVEEIVIEHc 2 1 tSuRVYGRNAAALSQALRLQAPELPVTVNPSKPRRGSFEVTLLRPDGS 1 2 gAELWTGIKKGPPRKLKFPEPQQVVEELKKYLS* 0 >SELH_ochPri Ochotona princeps (pika) 0 MAPNRRKRKAEAVADAAAEKREKQAKQANGVGGGEEIVIEHc 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPAKPRRGSFEVTLQRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQQVVEELKKYLS* 0 >SELH_canFam Canis familiaris (dog) 0 MASRGRKRKAEAAGVAAAEKRDKPASGRKAVEEATVVIEHC 2 1 TSuRVYGRNAAALSQALRLETPELPVEVNPAKPRRGSFEVTLLRPDGS 1 2 SVELWTGIKKGPPRKLKFPEPQEVVKALKQHLS* 0 >SELH_bosTau Bos taurus (cow) TGA verified 0 MASRGRKRKAEAALAAAAEKREKPAGGQEGGVEGPSVVIEHC 2 1 TSuRVYGRNAAALSQALRLQAPELTVKVNPARPRRGSFEVTLLRADGS 1 2 SAELWTGLKKGPPRKLKFPEPHVVLEELKKYLS* 0 >SELH_oviAri Ovis aries (sheep) 0 MASRGRKRKAEAALAAAAEKREKPAGSREGEVAGPSVVIEHC 2 1 TSuRVYGRNAAALSQALRLQAPELAVKVNPSRPRRGSFEVTLLRADGS 1 2 AELWTGLKKGPPRKLKFPEPHVVLEELKKYLS* 0 >SELH_susScr Sus scrofa (pig) 0 MASRGRKRKAETALGAAAEKQETPASGRKGMEEPSVVIEHC 2 1 TSuRVYGRNAAALSQALRVEAPELPVRVNPTKPRRGSFEVTLMRPDGS 1 2 SAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_equCab Equus caballus (horse) 0 MASRGRKRKAEAVVAVAEKREKLTSGGKGVEEVTVVIEHc 2 1 TSuRVYGRNAAALSQALRLEAPELPVKVNPAKPRRGSFEVTLLRPDGS 1 2 sAELWTGIKKGPPRKLKFPEPQEVVEELKKYLS* 0 >SELH_choHof Choloepus hoffmanni (sloth) 0 MASRGRKRKAEAVVLAAAEKQEKVASGKEGEKEAVVVIEHC 2 1 TSuRVYGRNAAALSQALRLEAPEIPVKVNFAKPRRGSFEVTLLRPDGS 1 2 TELWTGIKKGPPRKLKFLQPLKVVEELKKYLS* 0 >SELH_loxAfr Loxodonta africana (elephant) 0 MASRGRKRKAEAAVAAAAAEKREKPVGGQAAAEEVVVIIEHC 2 1 TSuRVYGRNAAALSQALRLEAPELSVKVNPSKPRRGSFEVTLQRPDGS 1 2 GAELWTGIKKGPPRKLKFPEPQEVVEELKKYL* 0 >SELH_monDom Monodelphis domestica (opossum) 0 MAPRGRKRKADVAAAALTEKPEKLAQGGEEGAGEARVVIEHC 2 1 QSuRVYARHAEAVGQALRLARPGLPVLLNPAKPRRSSFEVTLLRPDGS 1 2 RVELWSGIKKGPPRKLKFPEPAQVVEELKARLV* 0 >SELH_ornAna Ornithorhynchus anatinus (platypus) E5DI3CH10F50JR 0 DAGGAEVGEGLHVVIEHC 2 1 RSuGVYGRRAEALSRALSLAAPDLPVLLNPTKPRRNSFEVTLLRPDGT 1 2 RTELWSGIKKGRP >SELH_galGal Gallus gallus (chicken) tga confirmed 0 MAPRGRKRAARRPAEPEARADPPEKRPRDEAEGSPGDAGGPRVVIEHC 2 1 RSuRVYGRNAAALSEALRGAVASLAVEINPRQPRRNSFEVSLVKEDGS 1 2 TVQLWSGIGKGPPRKLKFPEPAAVVEALRSSLA* 0 >SELH_danRer Danio rerio (zebrafish) AI877878 BM037625 CKSU tga verified exons fused 0 MATRGKSARKRKADSDEKEKLDDAKKEKLEDKDEETGLRVVIEHC 2 1 KSuRVYGRNADVVREALADSHPELKVMINPHKPRRNSFEITLMDGERADVLWSGIKKGPPRKLKFPEPAEVVTALKQALEKE* 0 >SELH_takRub Takifugu rubripes (fugu) tga confirmed, no Ests 3 exons 0 MTPRVLMTGRRGTKRKAEEDEKPKEEKKEKQREDDQGGPRVAIEHC 2 1 KSuRVYGRNAEAVKSALLAAHPGLTVVLNPEKPRRNSFEITLLDEGK 1 2 ETSLWTGIKKGPPRKMKFPQPDVVVTALQEALKTE* 0 >SELH_ictPun Ictalurus punctatus (catfish) CB940790 tga verified 0 MATRAKAGRGAKRKADVIAAAEPVAKQDKGNKGEREDDEGQRVIIEHC 2 1 KSuRVYGRNADAVREALLSAHPELHVVLNPEKPRRNSFEVTLIEGK 1 2 KELVLWTGLKKGPPRKLKFPEPAEVVTALEEALKSK* 0 >SELH_salSal Salmo salar (salmon) CA043802 tga verified 0 MASRNKAGRVLKRKASVKEESVEEKRGKGEDDQPEIVTEGRRVVIEHC 2 1 KSuRVYGRNAEGVRVALLAACPDLTVVLNPQKPRSKSFEVILVEGE 1 2 KEVCLWSGIKKGPPRKLKFPEPEVVVSALEKALKTE* 0 >SELH_oryLat Oryzias latipes (medaka) BJ026077 tga verified 0 MASKAGRRGTKRKVEAKKEEDKTSTEEKKARGENAHEEAGLKVLIEHC 2 1 KSuRVYGRNAEEVKSALLAARPELTVVCNPEKPRRNSFEITLLDGAK 1 2 ETSLWTGIKKGPPRKLKFPQPDDVVAAFKDALKTE* 0 >SELH_oncMyk Oncorhynchus mykiss (trout) BX312781 0 MASLTKAGRVLKRKVETEESSVEGKRGKGEDDHPEIVTEGQRVVIEHC 2 1 KSuRVYGRNAEGVRVALLAACPDLTVVLNPQKPRSKSFEVILFEGE 1 2 KEVCLWSGIKKGPPRKLKFPEPEVVVSALEKALKTE* 0
SELL: 10 pre-tetrapod sequences
>SELL_danRer Danio rerio (zebrafish) 9 exons note illegal gene name, serious genome misassembly 0 MAEVDESTLVGALKELTIFGRTSLEYVEKDTAN 1 2 GCTLQEVNGKVLPLLGLMSAGAKFCNS 2 1 LGVKNQKEAESLWERFFQ 2 1 NTEVRESVEGLLQLE 0 0 VEWDRFLKRLDVQMQMSDTVLSQHPAAQTLSSDMRFLHVQTKE 2 1 NVSLGQYLGKGENLLLVLLRHFGuLPuRDHLTELKNSK 0 0 MLLEAQSLRVLVVSYGSLEGATFWLEQTGFEFDMLLDTERT 0 0 VYKMFGLGSSMSKVMKFKLMFHYSEIMAMNRTLPEMPPQFIEDLFQ 0 0 MGGDFVLEQDGKVIFSYRCKSPVDRPSATQILATVSAHYAH* 0 NVSLGQYLGKGENLLLVLLRHFGuLPuRDHLTELKNSK Danio rerio NVSLGQYLGKGENLLLVLLRHFGcLPcRDHLTELKNSK Tetraodon nigroviridis SVTLGEFLGQNQKLLLVLIRHFGuLPuRDHVAELEASE Oryzias latipes SVTLGRYLGQGQKPLLVLIRHFGuLPuRDHVAELEANQ Gasterosteus aculeatus SVSLGRYFGKGENLLLVLIRHLGuLPuRDHLAQMETNQ Osmerus mordax NVTLGLYFGKGENLLLVLIRHFGuLPuREHVAELATQQ Salmo salar NVTLGMYFGKGENLLLVLIRHFGuLPuREHVAELATQQ Oncorhynchus kisutch EVLLEEFLQRQEKLLLVLLRHFAuLPuRDHLAQLQKNQ Squalus acanthias EIQLEEFLHRREKLLLVLLRHFAuLPuRDHLVQLQAQQ Leucoraja erinacea EVLLNNYLGKEYLVLLVMLRHLAuVPuREHVAQLQAHQ Eptatretus burgeri FDGNLLFSSNHRNCLVILLRHFAuLPuRKHVTEIQEKQ Ciona intestinalis AVEL-RSLWREHACVVAGLRRFGCVVCRWIAQDLSSLA Homo sapiens C1orf93 RVPF-GALFRERRAVVVFVRHFLCYICKEYVEDLAKIP Homo sapiens C9orf21 TFKA-KELWEKNGAVIMAVRRPGCFLCREEAADLSSLK Homo sapiens C10orf5
SELI: 10 deuterostome sequences
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_homSap Homo sapiens (human)
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_ponPyg Pongo pygmaeus (orang_sumatran)
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_macMul Macaca mulatta (rhesus)
VKQLSSHFQIYPFSLKKPNSDuLGMEEKNIGL* SELI_otoGar Otolemur garnettii (bushbaby)
VKQLSSHFQIYPFSLRKPNSDuLGVEEKSIGL* SELI_tupBel Tupaia belangeri (tree_shrew)
VKQLSRHFQIYPFSLRKPNSDuLGMEEQNIGL* SELI_musMus Mus musculus (mouse)
VKQLSRHFQIYPFSLRKPNSDuLGLEEKSIGL* SELI_ratNor Rattus norvegicus (rat)
VKQLSSHFQIYPFSLRKPNSDuLGTEEKNIGL* SELI_cavPor Cavia porcellus (guinea_pig)
VKQLSSHFQIYPFSLKKPNSDuLGMEEKNIGL* SELI_speTri Spermophilus tridecemlineatus (squirrel)
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIAL* SELI_oryCun Oryctolagus cuniculus (rabbit)
VKQLSSHFQIYPFSLRKPNSDuLGMEAKNIVV* SELI_ochPri Ochotona princeps (pika)
VKQLSNHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_canFam Canis familiaris (dog)
VKQLSNHFQIYPFSLRKPNSDuLGTEEKSIGL* SELI_felCat Felis catus (cat)
VKQLSNHFHIYPFSLRKPNSDuLGVEEKNIGL* SELI_bosTau Bos taurus (cow)
VKQLSSHFQIYPFSLRKPNSDuLGM.......* SELI_susScr Sus scrofa (pig)
VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* SELI_equCab Equus caballus (horse)
VKQLSSHFHIYPFSLRKPNSDuLGAEE-ITGL* SELI_eriEur Erinaceus europaeus (hedgehog)
VNQLSNHFQIYPFSLKKPNSDuLEMEEKNIGL* SELI_sorAra Sorex araneus (shrew)
VKQLSSHFQIYPFSLKKPNSDuLGVEEKKIGL* SELI_dasNov Dasypus novemcinctus (armadillo)
VKQLSSHFQIYPFSLRKPNSDuLGMEVYKIRQ* SELI_loxAfr Loxodonta africana (elephant)
VYQLSNHFKILPFSLKKPSSDuLEVEEEKIGLQSTEVL* SELI_monDom Monodelphis domestica (opossum)
VSQLSRHFNIRPFSLKKPTPDuLGMEEEKISLRSAEVL* SELI_galGal Gallus gallus (chicken)
VSQLSQHFHIRTFSLRKPSSDuLGVEEAMAAGAEEKVALRSAEEL* SELI_anoCar Anolis carolinensis (lizard)
VNQLSKHFNILPFSLRKPSTDuL--EEEKIGLQSAQVL* SELI_xenTro Xenopus laevis (frog)
VKQLSDHFNIFAFSLKKPNSDuQ--DEEKIGLKAAEV* SELI_danRer Danio rerio (zebrafish)
VQQLSDHFNIKAFSLKKPSADuQ--EEERIGLTEAEV* SELI_tetNig Tetraodon nigroviridis (pufferfish)
VQQLSNHFSILAFSLKKPNSDuQ--EEDQIGLKAAEV* SELI_gasAcu Gasterosteus aculeatus (stickleback)
VQQLSKHFNIFAFSLKKPSSDuQ--EEERIGLTGAEV* SELI_oryLap Oryzias latipes (medaka)
VNQLSKHFNICAFSLKKPSTEuLEEEQKICLQASEVL* SELI_leuEri Leucoraja erinacea (skate)
VIQMADHFNINIFTLKKQ--DuPKVQTTKYLLYVCGVMSWQC... SELI_braFlo Branchiostoma floridae (amphioxus)
VNKLAAHFGIKVFSIKENS-DuHAAPSVAANAAVHAGV* SELI_cioInt Ciona intestinalis (tunicate)
>SELI_homSap Homo sapiens (human) chr2
0 MAGYEYVSPEQLAGFDKYK 0
0 YSAVDTNPLSLYVMHPFWNTIVK 0
0 VFPTWLAPNLITFSGFLLVVFNFLLMAYFDPDFYAS 1
2 APGHKHVPDWVWIVVGILNFVAYTL 1
2 DGVDGKQARRTNSSTPLGELFDHGLDSWSCVYFVVTVYSIFGRGSTGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0
0 TISFVYIVTAVVGVEAWYEPFLFNFLYRDLFTAMII 1
2 GCALCVTLPMSLLNFFR 2
1 SYKNNTLKLNSVYEAMVPLFSPCLLFILSTAWILWSPSDILELHPRVFYFMVGTAFANST 0
0 CQLIVCQMSSTRCPTLNWLLVPLFLVVLVVNLGVASYVESILLYTLTTAFTLAHIHYGVRV 0
0 VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* 0
>SELI_macMul Macaca mulatta (rhesus)
0 MAGYEYVSPEQLAGFDKYK 0
0 YSAVDTNPLSLYVMHPFWNTIVK 0
0 VFPTWLAPNLITFSGFLLVVFNFLLMAYFDPDFYAS 1
2 APGHKHVPDWVWIVVGILNFVAYTL 1
2 DGVDGKQARRTNSSTPLGELFDHGLDSWSCVYFVVTVYSIFGRGSTGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0
0 TISFVYIVTAVVGVEAWYEPFLFNFLYRDLFTSMII 1
2 GCALCVTLPMSLLNFFR 2
1 SYKNNTLKHNSVYEAVVPLFSPCLLFILSTAWILWSPSDILELHPRVFYFMVGTAFANST 0
0 CQLIVCQMSSTRCPTLNWLLVPLFLVVLVVNLGVASYVESILLYTLTTAFTLAHIHYGVRV 0
0 VKQLSSHFQIYPFSLRKPNSDuLGMEEKNIGL* 0
>SELI_bosTau Bos taurus (cow) NM_001075257
0 MASYEYVSPEQLAGFDKYK 0
0 YSAVDTNPLSLYVMHPFWNTVVK 0
0 VFPTWLAPNLITFSGFLLVVFNFLLLAYFDPDFYAS 1
2 APGHKHVPDWVWIVVGILNFMAYTL 1
2 DGVDGKQARRTNSSTPLGELFDHGLDSWSCVYFVVTVYSIFGRGSSGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0
0 TISFVYIVTAVVGVEAWYEPFLFNFLYRDLFTAMIF 1
2 GCALCVTLPMSLLNFFR 2
1 SYKNNTLKHNSVYEAVVPFFSPCLLFILSTAWILLSPSDILELHPRVFYLMVGTAFANST 0
0 CQLIVCQMSSTRCPTLNWLLLPLFVIVMVVNVGVTPYVESILLFTLTAAFTLAHIHYGVRV 0
0 VKQLSNHFHIYPFSLRKPNSDuLGVEEKNIGL* 0
>SELI_monDom Monodelphis domestica (opossum)
0 MAGAIENRSMGIRDDL 0
0 YRAVDTNPLSLYIMHPFWNAIVK 0
0 IFPTWLAPNLITLSGFLLLIFNFLLMAYFDPDFFAS 1
2 APGGKHVPDWVWILVGILNFVAYTL 1
2 DGVDGKQARRTNSSTPLGELFDHGLDSWSCVYFVVTVYSIFGRGSTGVSVVVLYILLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0
0 TISLVYIVTAIVGVEAWYEPFLFNFLYRDLFTAMII 1
2 GCASCVTLPMSLYNFFK 2
1 AYKNNSLKHSSVYETMLPFISPCLLFILSTTWVLWSPSDILETHPRLFYYMVGTAFANIT 0
0 CRLIVCQMSNTRCQTLNLLLIPLALVVLLVRLELPLHVEPILLYLLSIIITLAHIHYGIQV 0
0 VYQLSNHFKILPFSLKKPSSDuLEVEEEKIGLQSTEVL* 0
>SELI_ornAna Ornithorhynchus anatinus (platypus) 53% LOC100086265
0 MAGYEYVSAEQLAGFDKYK 0
0 YSALDTNPLSLYVMHPFWNTIVKV 0
0 IFPTWLAPNLITFSGFLLLVFNFLLMAYFDPDFYAS 1
2 APGQKHVPDWVWIVVGILNFTAYTL 1
2 DGVDGKQARRTNSSTPLGELFDHGLDSWACVYFVVTVYSIFGRGRTGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDMSQV 0
0 TISIVYIVTAVVGVEAWYEPFLFNFLYRDLFTTMII 1
2 GCAVTVTLPMSLYNFYR 2
1 NKTLKYSSVYETMLPFVSPCLLFTLSTTWIFLSPSNILETHPRLFYFMVGTLFANIT 0
0 CQLIVCQMSNTRCQPLNWLLMPLALVILVVHSGLAPHSETFLLYSLTALVTVAHIHYGIRV 0
0 VNQLSKHFKILPFSLRKPSSDuLGLEEEKIGL* 0
>SELI_galGal Gallus gallus (chicken)
0 MEYVTAEQLAGFSKYK 0
0 YSAVDSNPLSLYVMHPFWNTIVK 0
0 IFPTWLAPNLITFSGFLLLVFNFFLMAYFDPDFYAS 1
2 APDHQHVPNGVWVVVGLLNFIAYTL 1
2 DGVDGKQARRTNSSTPLGELFDHGLDSWACVYFVVTVYSTFGRGSTGVSVFVLYLLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0
0 TISIVYIVTAIVGVEAWYAPFLFNFLYRDLFTTMII 1
2 ACALTVTLPMSLYNFYK 2
1 AYKNNTLKHHSVYEIMLPLVSPVLLFALCTTWIFVSPMDILEVHPRLFYFMVGTAFANIS 0
0 CQLIVCQMSSTRCQPLNWMLLPIALVLFMVMSGFAPSSETLLLYLLTAFLTLAHIHYGVVV 0
0 VSQLSRHFNIRPFSLKKPTPDuLGMEEEKISLRSAEVL* 0
>SELI_xenTro Xenopus tropicalis (frog)
0 MAWEYVSPEQLAGFDKYK 0
0 YSAVDTNPLSLYVMHPFWNSIVR 0
0 FFPTWLAPNLITFSGFLLLVFTFLLMAFFDPDFYAS 1
2 APGQEHVPNWVWIVAGLLNFTAYTL 1
2 DGVDGKQARRTNSSTPLGELFDHGLDSWACIFFVVTVYSIFGRGETGVSAVVLYFLLWVVLFSFILSHWEKYNTGILFLPWGYDLSQV 0
0 TISCVYLVTAVVGAETWYQPVVFNILYRDLFTTMIV 1
2 GCAVCVTLPMSLYNVFK 2
1 GYRNNTLKHTSLYEAMLPIISPVLLFILSTLWILASPSKILEHHPRLFYFMVGTTFANIT 0
0 CQLIVCQMSNTRCRPLSWLLIPLTLAVVVSLSTVTPRDETLALVLLTVLVTLAHIHYGVNV 0
0 VNQLSKHFNILPFSLRKPSTDuLEEEKIGLQSAQVL* 0
>SELI_danRer Danio rerio (zebrafish)
0 MALYHYVTQEQLSGFDKYK 0
0 YSAVDSNPLSIYVMHPFWNSVVK 0
0 VLPTWLAPNLITFTGFMFLVLTFTMLSFFDFDFYAS 1
2 GEGHTHVPSWVWIAAGLFNFLAYTL 1
2 DGVDGKQARRTNSSTPLGELFDHGLDSWACVFFVATVYSVFGRGETGVSVVTLYYLLWVVLFSFILSHWEKYNTGILFLPWGYDISQV 0
0 TISIVYIITAVVGVETWYKPVFLNIHYRDLFTVMII 1
2 GCLFVVTLPMSLYNVFK 2
1 AYRNNTLKHSSLYEALLPFFSPVLLFVLSTLWISFSPTNIVQKQPRIFYLMVGTAFSNVT 0
0 CKLIVCQMSNTRCKPLSLLLLPMSIVVLLVISGTVQGGESLVLFIWTAVVLLTHIHYGVSV 0
0 VKQLSDHFNIFAFSLKKPNSDuQDEEKIGLKAAEV* 0
>SELI_petMar Petromyzon marinus (lamprey) seleno exon not available
0 MAAEHAYITAEQLAGFDKYK 0
0 YSAVDTNPLSVYILHPFWNFVVK 0
0 LFPKWVAPNLLTFSGFLLLVFNFFILAYYDWDFYAS 1
2 APGQVHVPGWVWILAGILSFLAYTL 1
2 GIDGKQARRTGSSTPLGELFDHGLDSWSCLFFGSTVYSIFGRGQTGVSLATMHMLLWVVLFSFVLSHWEKYNTGVLFLPWGYDISQV 0
0 TISIVYIVTAVVGVEAWHQDLPFGLLYRDLFLFMLL 1
2 GGAFLVTLPQSLFNLHK 2
1 AHKAGTLKKDSLWEAMLPMVSPTLLFVLTSAWVWLSPERVVDSQPRVFYWLVGTLFSNIT 0
0 CKLIVCQMSSTRCQPLNWLLAPLACAVAFVATFQPKVEVETAVLFGMTALVTAAHIHYGVCVV 0
0 * 0
>SELI_braFlo Branchiostoma floridae (amphioxus) frag
0 RLPRWLAPNLMTFVGFLLLIFNFALLTWYDPQFRAS 12 SSQHPDDPLVPNWVWLVCGFAHFLSHTL 1
2 DGVDGKQARRTNSSTPLGELFDHGLDSWATMFFPTAIYSMFGRGTEYGITVTQMYMVVWVVMIGFIMSHWEKYNTGTLFLPWGYDMSQV 0
0 CMLVMYIAAFFCGQDLWRTHLPMGLRFHYLFLFSFY 1
2 GGFLFISLPHTIGNLYR 2
1 STSEGTGRNLSLYEGMLPLLSPVLLFLLCVAWLKMSPTDILEQQPRLFYFMSGTVFANIA 0
0 CRLIIAQMSNTRCEGVNALLWPLMLIVAGVCAVPLGQWELVLLIGYTVMATLLHIHFGVCV 0
0 VIQMADHFNINIFTLKKQDuPKVQTTKYLLYVCGVMSWQC
>SELI_cioInt Ciona intestinalis (tunicate) 1 exon fusion, 1 split
0 MYITEQELSGFDTYK 0
0 YSCKDTSPLSNYVMHPFWNEAVK 0
0 LFPRWLAPNVMTFGGFVLLILQYILLWYFDPTYHAS 12 TTDIAYPSIPTWVWWFSLFAQFFSHTL 1
2 DGCDGKQARRTGTSSPLGELFDHGIDSWCVSLFTLNVLSVFGRDLAP 1
2 VSLMYSVQCMTLMAFLLSHWEKYNTGILFLPWAYDLSQV 0
0 LMAIVYLITAVSGVGVWRGTLLGYKVTTWFKGVLYV 1
2 ATFGFAVPMCLYNIYI 2
1 ANKNGTGKNLTFYEGSLPLLSPLLSFTILTTWIYISPSNVLSTNPRIFMLIISVLFSNIL 0
0 CRLIVSQMSNTRCQVFNKFVFFIGAGFACTLYFNNPVIENYILLALAIGLTVAHIHYGITV 0
0 VNKLAAHFGIKVFSIKQDSDuHAAPSVAANDAVHAGV* 0
SELK: 32 vertebrate sequences
>SELK_homSap Homo sapiens (human) chr3 last coding bp and stop codon split off into 5th exon 0 MVYISN 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 1 FKTLLQQDVKKRRSYGNSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_panTro Pan troglodytes (chimp) 0 MVYISN 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 1 FKTLLQQDVKKRRSYGNSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_ponPyg Pongo pygmaeus (orang_sumatran) 0 MVYISN 1 2 AQVLDSWSQSPWRLSLITDFFWGIAEFVVLF 2 1 FKTLLQQDLKKRRSYGNSSDFRYDDDGRG 2 1 PPGNPPGRMGPINHLGGPSPPPMAGGuGR 2 1* 0 >SELK_macMul Macaca mulatta (rhesus) 0 MVYISN 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 1 FKTLLQQDVKKRRSYGNSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_otoGar Otolemur garnettii (bushbaby) 0 MVYISN 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 1 FKTLLQQDVKKRRGYGNSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLQGPSPPPMAGGuGR 2 1* 0 >SELK_micMur Microcebus murinus (mouse_lemur) 0 MSYISN 1 2 GQLLDSRSQSPWRLSLITDFFWRKPEFVVLF 2 1 FKTLLQQDVKKRRGCGNSSDSRYDDGRG 2 1 PPGNPFQRMGQINHLHGPSPPMAGGuGR 2 1* 0 >SELK_tupBel Tupaia belangeri (tree_shrew) 0 MVYISN 1 2 GRVSDSWSQSPWRLSLITDFFWGMAKFVVLS 2 1 FKTLLQQDVRKRKGYGNSSDSRYDDGRG 2 1 PPGNPPRRMGRISHLRGPSPPPVAGGuGR 2 1* 0 >SELK_musMus Mus musculus (mouse) 0 MVYISN 1 2 GQVLDSRNQSPWRVSFLTDFFWGIAEFVVFF 2 1 FKTLLQQDVKKRRGYGSSSDSRYDDGRG 2 1 PPGNPPRRMGRISHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_ratNor Rattus norvegicus (rat) 0 MVYISN 1 2 GQVLDSRNQSPWRLSFITDFFWGIAEFVVFF 2 1 FKTLLQQDVKKRRGYGGSSDSRYDDGRG 2 1 PPGNPPRRMGRISHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_speTri Spermophilus tridecemlineatus (squirrel) 0 MVYISK 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVILF 2 1 FKTLLQQDVKKRRGYGNSSDSRYDDGRG 2 1 PPGNPPRRMGGINHLQGPSPPPMGGGuGR 2 1* 0 >SELK_cavPor Cavia porcellus (guinea_pig) 0 MVYISN 1 2 GQVLDNRSQSPWRLSLITDFFWGIAEFVVLF 2 1 FKTLLQQDVKKGRGYRNSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLQGPSPPPMAGGuGR 2 1 * 0 >SELK_oryCun Oryctolagus cuniculus (rabbit) 0 MVYISN 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLL 2 1 FKTLLQQDVKRGRGYRNSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_ochPri Ochotona princeps (pika) 0 MVYISN 1 2 GQVLDSRSQSPERLSLITDFFWGIAEFVVLF 2 1 FKTLLQQDVKRRRDYRSSSDSRYDDGRG 2 1 LPGNPPRRMGRIYHLRCPSPPPPPGGGuGR 2 1* 0 >SELK_canFam Canis familiaris (dog) 0 MVYISN 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 1 FKTLLQQDVKKRRGYGNSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_neoVis Neovison vison (mink) 0 MVYISN 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLF 2 1 FKTLLQQDVKKRRGYGSSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 1* 0 >SELK_felCat Felis catus (cat) 0 MVYISN 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVVLL 2 1 FQTLLQQDVKKRRGYGNSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_equCab Equus caballus (horse) 0 MVYISN 1 2 GQVLDSRSQSPWRLSFITDFFWGIAEFVVLF 2 1 FKTLLQQDVKKRRGYGSSSDSRYDDGRG 2 1 PPGNPRRRMGRINHLHGPSPPPMAGGuGR 2 1 * 0 >SELK_myoLuc Myotis lucifugus (microbat) 0 MVYIEN 1 2 RQVLDSRSQSPwRLSLITDFFWGIAEFVVLF 2 1 FKTLLQKDMKKGRGYGNSSDSRYDDGRG 2 1 PPGNPPRRMGRIDHLRGPSPPPMAGGuGR 2 1* 0 >SELK_bosTau Bos taurus (cow) 0 MVYISN 1 2 GQVLDSRSQSPWRLSFITDFFWGIAEFVVLF 2 1 FRTLLQQDVKKRRGYGSSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLQGPNPPPMAGGuGR 2 1 * 0 >SELK_susScr Sus scrofa (pig) 0 MVYISN 1 2 GQVLDSRSQSPWRLSFITDFFWGIAEFVVLF 2 1 FRTLLQQDVKKRRGYGGSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLRGPNPPPMAGGuGR 2 1 * 0 >SELK_dasNov Dasypus novemcinctus (armadillo) 0 MVYISN 1 2 GQVLDSRSQSPWRLSLITDFFWGIAEFVILL 2 1 FRTLLQQDVKKKRGYGSSSDSRYDDGRG 2 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_loxAfr Loxodonta africana (elephant) 0 MVYISN 1 2 GQVLDSRSQSPWRLSFITDFFWGIAEFVVLF 2 1 FKTLLQQDVKKRGGYGSSSDFRYDDGRG 2 1 PPGNPPRRMGRINHLRGPSPPPMAGGuGR 2 1 * 0 >SELK_ornAna Ornithorhynchus anatinus (platypus) EUEMSW407EBWK3 454 0 MVYISN 1 2 GHVLNGQNRSLWSLSFIKDFFWGILDFIIMF 2 1 FKSMIHPNVKRGCRNSSSDSKYDDGRG 2 1 PPGYPRRGMGRINHSNGPSPPPMAGGGuGR 2 1 * 0 >SELK_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01DCGU7 454 0 MVYISN 1 2 GHVLNGQNRSPWSLSYIKDFFWGILDFIIMF 2 1 FKSMIHPNVKRGCRNSSSDSKYDDGRG 2 2 PPGYPRRGMGRINHSNGPNPPPMAGGuGR 2 1 * 0 >SELK_anoCar Anolis carolinensis (lizard) 0 MVYISN 1 2 GQVLDNQSRAPWSFSSITDFFWGIADFVVMF 2 1 FQSIIHPDLTRGSTSSSSSRYDDGRG 2 1 PPGLPRRRMGRISHFGGPSPPPMAGGuGR 2 1 * 0 >SELK_galGal Gallus gallus (chicken) 0 MVYISN 1 2 GQVLDNRSRAPWSLSAITDFFWSIADFVVMF 2 1 FQSIIQPDLRRRGYTSSSYLGQSDGRG 2 1 PPGNPRRRMGRINHWGGGPSPPPMAGGGuGR 2 1 * 0 >SELK_taeGut Taeniopygia guttata (finch) 0 MVYISN 1 2 GQVLDSQSRAPWRLSSITDFFWSIADFVVLF 2 1 FQSIIQPDLRRRGYTSSSYSGYHDRRG 2 1 PPANPRRRMGRINHWGGGPSPPPMAGGGuGR 2 1* 0 >SELK_xenTro Xenopus tropicalis (frog) 0 MVYISN 1 2 GQVLDGQSRSPWRLSFLTDMFWGITDFVVMF 2 1 FQSIIHPNVTRRGCQNSSSSTRYDDGRG 2 1 PPGHPRRMGRINHGSGPSAPPMAGGGGuGR 2 1 * 0 >SELK_oncMyk Oncorhynchus mykiss (trout) 0 MVYVAN 1 2 GQVLDNRSQSPWRMSFLTDLFWGAVEFIGLF 2 1 FQSLVQPDLTKRGNSGSSSTGYSDGRG 2 1 PPGPPGGRRRMGRINHGGGPSAPPMGGGGuGR 2 1 * 0 >SELK_salSal Salmo salar (salmon) 0 MVYVAN 1 2 GQVLDNRSQSPWRMSFLTDLFWGAVEFIGLF 2 1 FQSLVQPDLTKRGNSGSSSAGYSDGRG 2 1 PPGPPGGRRRMGRINHGGGPSAPPMGGGGuGR 2 1 * 0 >SELK_oryLap Oryzias latipes (medaka) BJ003636 tag confirmed 0 MVYVSN 1 2 GQVLDSRAQSPWRLSLLVDLFWDALEFFRLF 2 1 FKTMFHPDLTKDGNSASSRFSDGRG 2 1 PPGPPGGRRRIGRINHGAGPNAPPMGGGGuGR 2 1 * 0 >SELK_braFlo Branchiostoma floridae (amphioxus) tga confirmed 0 MVYVTG 1 2 GQMLENRSPWRLSIIPEIFWGFINFIVLF 2 1 FQTMYNPSLSKHGSDTPSSYSRGDGRG 2 1 PPPPPPRRRFGGVRRGGAPGPPPMAGGGuGR 2 1 * 0
SELO: 6 vertebrate sequences
>SELO_homSap Homo sapiens (human) tga-sel taa-stop 0 MAVYRAALGASLAAARLLPLGRCSPSPAPRSTLSGAAMEPAPRWLAGLRFDNRALRALPVEAPPPGPEGAPSAPRPVPGACFTRVQPTPLR QPRLVALSEPALALLGLGAPPAREAEAEAALFFSGNALLPGAEPAAHCYCGHQFGQFAGQLGDGAAMYLGEVCTATGERWELQLKGAGPTPFSR 2 1 QADGRKVLRSSIREFLCSEAMFHLGVPTTRAGACVTSESTVVRDVFYDGNPKYEQCTVVLRVASTFIR 2 1 FGSFEIFKSADEHTGRAGPSVGRNDIRVQLLDYVISSFYPEIQAAHASDSVQRNAAFFRE 0 0 VTRRTARMVAEWQCVGFCHGVLNTDNMSILGLTIDYGPFGFLDR 2 1 YDPDHVCNASDNTGRYAYSKQPEVCRWNLRKLAEALQPELPLELGEAILAEEFDAEFQRHYLQKMRRKLGLVQVELEEDGALVSKLLETMHLT 1 2 GADFTNTFYLLSSFPVELESPGLAEFLARLMEQCASLEELRLAFRPQMDPR 2 1 QLSMMLMLAQSNPQLFALMGTRAGIARELERVEQQSRLEQLSAAELQSRNQGHWADWLQAYR 2 1 ARLDKDLEGAGDAAAWQAEHVRVMHANNPKYVLRNYIAQNAIEAAERGDFSE 0 0 VRRVLKLLETPYHCEAGAATDAEATEADGADGRQRSYSSKPPLWAAELCVTuSS* 0 >SELO_musMus Mus musculus (mouse) 0 MAVYRAALGASLAAARLLPLGRCSPSPAPRSTLSGAAMEPAPRWLAGLRFDNRALRALPVEAPPPGPEGAPSAPRPVPGACFTRVQPTPLR QPRLVALSEPALALLGLGAPPAREAEAEAALFFSGNALLPGAEPAAHCYCGHQFGQFAGQLGDGAAMYLGEVCTATGERWELQLKGAGPTPFSR 2 1 QADGRKVLRSSIREFLCSEAMFHLGVPTTRAGACVTSESTVVRDVFYDGNPKYEQCTVVLRVASTFIR 2 1 FGSFEIFKSADEHTGRAGPSVGRNDIRVQLLDYVISSFYPEIQAAHASDSVQRNAAFFRE 0 0 VTRRTARMVAEWQCVGFCHGVLNTDNMSILGLTIDYGPFGFLDR 2 1 YDPDHVCNASDNTGRYAYSKQPEVCRWNLRKLAEALQPELPLELGEAILAEEFDAEFQRHYLQKMRRKLGLVQVELEEDGALVSKLLETMHLT 1 2 GADFTNTFYLLSSFPVELESPGLAEFLARLMEQCASLEELRLAFRPQMDPR 2 1 QLSMMLMLAQSNPQLFALMGTRAGIARELERVEQQSRLEQLSAAELQSRNQGHWADWLQAYR 2 1 ARLDKDLEGAGDAAAWQAEHVRVMHANNPKYVLRNYIAQNAIEAAERGDFSE 0 0 VRRVLKLLETPYHCEAGAATDAEATEADGADGRQRSYSSKPPLWAAELCVTuSS* 0 >SELO_ratNor Rattus norvegicus (rat) 0 MASFRAAFGASLAVARTRPQCVGLELQSSAPWSAWAAAMEPTPRWLARLRFDNRALRALPVETPPPGPEDSLSTPRPVPGACFSRARPAPLR QPRLVALSEPALALLGLEVSEEAEVEAEAALFFSGNALLPGTEPAAHCYCGHQFGQFAGQLGDGAAMYLGEVCTAAGERWELQLKGAGPTAFSR 2 1 QADGRKVLRSSIREFLCSEAMFHLGIPTTRAGACVTSESTVMRDVFYDGNPKYEKCTVVLRIAPTFIR 2 1 FGSFEIFKPPDELTGRAGPSVGRNDIRVQMLDYVISSFYPEIQAAHTCDTDNIQRNAAFFRE 0 0 VTRRTARMVAEWQCVGFCHGVLNTDNMSIVGLTIDYGPFGFLDR 2 1 YDPDHVCNASDNAGRYTYSKQPQVCRWNLQKLAEALEPELPLVLAEAILKEEFDTEFQRHYLQKMRKKLGLVRVEKEDETLVAKLLETMHQT 1 2 GADFTNTFCVLSSFPAEPSDTAEFLTQLTSQCASLEELKLAFRPQMDPR 2 1 QLSMMLMLAQSNPQLFALIGTQANVTKELERVEHQSRLEQLSPSELQSKNRDHWETWLQEYR 2 1 ERLDKEKEGVGDIAAWQAERVRIMHANNPKYVLRNYIAQKAIEAAENGDFSE 0 0 VRRVLKLLESPYHSEEEATGPEAVARTTDEQSSYSSRPPLWAAELCVTuSS* 0 >SELO_ornAna Ornithorhynchus anatinus (platypus) uc003bjx.1 0 2 1 HADGRKVLRSSIREFLCSEAMFHLGIPTTRAGTCVTSESAVIRDVHYDGNPKYEKCAVVLRIASTFLR 2 1 FGSFEIFKPRDEHTGRQGPSVGRNDIRIRMLDYVIGTFYPEIEEANADDTVRRNAAFFRE 0 >SELO_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01DJRM3 454 0 YGGHQFGTWAGQLGDGRAHLIGIYTNRHGEQWELQLKGSGRTPYSR 2 1 NGDGRAVFRSSVREFLGSEAMHYLRIPTSR >SELO_galGal Gallus gallus (chicken) 0 MQRSGGVLRRGRADTERGETGGGWLSALRFDNLAMRSLPVDPFEDCAPRAVPGACFARVRPTPLRNPRLVAMSAPALALLGLEAGGPEAERE AEAALYFSGNRLLPGSEPAAHCYCGHQFGSFAGQLGDGAAIYLGEVRGPRGARWELQLKGAGITPFSR 2 1 QADGRKVLRSSIREFLCSEAMFHLGIPTTRAGTCVTSDSEVVRDIFYDGNPKKERCTVVLRIASTFIR 2 1 FGSFEIFKPPDEYTGRKGPSVNRNDIRIQMLDYVIGTFYPEIQEAHADNSIQRNAAFFKE 0 0 ITKRTARLVAEWQCVGFCHGVLNTDNMSIVGLTIDYGPFGFMDR 2 1 YDPEHICNGSDNTGRYAYNRQPEICKWNLGKLAEALVPELPLEISELILEEEYDAEFEKHYLQKMRKKLGLIQLELEEDSKLVSELLETMHLT 1 2 GGDFTNIFYLLSSFSVDTDPSRLEDFLEKLISQCASVEELRVAFKPQMDPR 2 1 QLSMMLMLAQSNPQLFALIGTKANINKELERIEQFSKLQQLTAADLLSRNKRHWTEWLEKYR 2 1 VRLHKEVESISDVDAWNTERVKVMNSNNPRYILRNYIAQNAIEAAENGDFSE 0 0 VRNVLKLLENPFQETEDSTEMETKEEEATATAAACAQATRSRLSYCSKPPLWASELCVTuSS* 0 >SELO_calMil Callorhinchus milii (elephantfish) frag tgt-cys tga-sel tga-stop? P taa-stop 0 LNFDNLALRSLPVDSSGERSCRRVPGACFSLAGATPVDNPRLVASSR 0 0 QSDGRKVLRSSIREFLCSEAMFHLGIPTTRAGTCVTSDTKVLRDVFYDGNSKHENCTIILRIAPTFLRF 2 1 FGSFEILKPEDELTGRQGPSSNRNDIRIQMLDYVIGTFYPEAQQAHPENQVQRNAAFFRE 0 0 VTQRTARLVAEWQCVGFCHGVLNTDNMSIMGLTIDYGPFGFMDR 2 1 FDPSYICNASDNRGRYAYNQQPEICKWNLGKLAEVLVPELPLKDSQSIIDEEYDTEFQRHYLQKMRKKLGLLQCEQEDDDKLVSELLDIMYRT 1 2 GADFTNTFYLLSSFPVELESPGLAEFLARLMEQCASLEELRLAFRPQMDPR 2 1 QLSILLMLSQSNPQLFEVIGSKEGIAKELDLIERSSKLQQATAEDIHSNNAKVWTEWLQKYR 2 1 SRLATEAEGVDDVDEQNAERVKVMNLNNPKFILRNYIAQNAIEAAEKGDFSE 0 0 VHLLQKTLRHPFHKQREAEEAGYSSRPPLWARELRVSCSSuR* 0 >SELO_leuEri Leucoraja erinacea (skate) DT725503 frag sel-tga taa-stop 1 NPQLFELIGDKRGVAKEMDRIERFSELQQMTTEELLSKLKNAWTDWLQKYR 2 1 SRLENEAEGVDDVAELKAERVKVMNANNPKFILRNYIAQNAISAAEDGDFSE 0 0 VRRVLKLLEHPYSEDACVEEELMQEATVEAPASCEPLSNKRIPYHSKPPDWSGKLLVTuSS* 0 >SELO_petMar Petromyzon marinus (lamprey) frag 0 GASMCSLYVLPLGRN 1 2 APRQVPGAVFSRVRPSPVERPRVVAISVPALRLLGLRDPEAEAARPEAAEFLSGNRVPPGAQPAAHCYCGHQFGSFAGQLGDGAAMYLGEVQPGPGQRWEVQLKGAGPTPYS 2 1 HSDGRKVLRSSLREFLCSEAMHHLGVPTTRAGSCVTSHSTVLRDVHYDGNARPEQCSVVLRIAPSFLR 0 1 FGSFEIFKSTDKDTGRTGPSAGREDIKVTMLDYVIDTFYPELLEGHGDGASHKYTAFFRE 0 0 VVRRTAHLVAEWQCVGFCHGVLNTDNMSILGLTIDYGPFGFMDR 2 1 GADFTNSFRLLSRLTLEPGSVEELATLLCQQCATVEEMKRAYKPRIDPR 2
SEPN1: 14 metazoan sequences
>SEPN1_homSap Homo sapiens (human) 0 MGRARPGQRGPPSPGPAAQPPAPPRRRARSLALLGALLAAAAAAAVRVCARHAEAQAAARQ 0 0 ELALKTLGTDGLFLFSSLDTDGDMYISPEEFKPIAEKLT 1 2 GSTPAASCEEEELPPDPSEETLTIEARFQPLLPETMTKSKDGFLG 0 0 VSRLALSGLRNWTAAASPSAVFATRHFQPFLPPPGQELGEPWWIIPSELSMFTGYLSNNRFYPPPPKGKE 0 0 VIIHRLLSMFHPRPFVKTRFAPQGAVACLTAISDFYYTVMFR 2 1 IHAEFQLSEPPDFPFWFSPAQFTGHIILSKDATHVRDFRLFVPNHR 2 1 SLNVDMEWLYGASESSNMEVDIGYIPQ 0 0 MELEATGPSVPSVILDEDGSMIDSHLPSGEPLQFVFEEIKWQQELSWEEAARRLEVAMYPFKK 0 0 VSYLPFTEAFDRAKAENKLVHSILLWGALDDQSCu 1 2 GSGRTLRETVLESSPILTLLNESFISTWSLVKELEELQ 0 0 NNQENSSHQKLAGLHLEKYSFPVEMMICLPNGTV 0 0 VHHINANYFLDITSVKPEEIESNLFSFSSTFEDPSTATYMQFLKEGLRRGLPLLQP* 0 >SEPN1_musMus Mus musculus NM_029100 0 MGQARPAARRPHSPDPGAQPAPPRRRARALALLGALLAAAAAVAAARACALLADAQAAARQ 0 0 ESALKVLGTDGLFLFSSLDTDQDMYISPEEFKPIAEKLT 1 2 GSVPVANYEEEELPHDPSEETLTIEARFQPLLMETMTKSKDGFLG 0 0 VSRLALSGLRNWTTAASPSAAFAARHFRPFLPPPGQELGQPWWIIPGELSVFTGYLSNNRFYPPPPKGKE 0 0 VIIHRLLSMFHPRPFVKTRFAPQGTVACLTAISDSYYTVMFR 2 1 IHAEFQLSEPPDFPFWFSPGQFTGHIILSKDATHIRDFRLFVPNHR 2 1 SLNVDMEWLYGASETSNMEVDIGYVPQ 0 0 MELEAVGPSVPSVILDEDGNMIDSRLPSGEPLQFVFEEIKWHQELSWEEAARRLEVAMYPFKK 0 0 VNYLPFTEAFDRARAEKKLVHSILLWGALDDQSCu 1 2 GSGRTLRETVLESPPILTLLNESFISTWSLVKELEDLQ 0 0 TQQENPLHRQLAGLHLEKYSFPVEMMICLPNGTV 0 0 VHHINANYFLDITSMKPEDMENNNVFSFSSSFEDPSTATYMQFLREGLRRGLPLLQP* 0 >SEPN1_bosTau Bos taurus NM_001114976 0 MGRTRPGERGPPGSGPAAPPAPPPRRARALALLGALLAAAAAAAAARAFARYSEAQAAARQ 0 0 DSALKTLGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT 1 2 GSTPTANYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG 0 0 VSRLALSGLRNWTTAASPSAVFAARHFRPFLPPQGHVELGEPWWIIPSELSVFTGYLSNNRFYPPPPKGKE 0 0 VIIHRLLSMFHPRPFVKTRFAPQGAVACLTAISDFYYTVMFR 2 1 IHAEFQLSEPPDFPFWFSPGQFTGHVVLSKDATHVRDFRLFVPNHR 2 1 SLNVDMEWLYGASESSNMEVDIGYIPQ 0 0 MELEALGPSVPSVILDEDGNLIDSRLPSGEPLQFVFEEIQWQQELSWEEAARRLEVAMYPFKK 0 0 VTYLPFTEAFDQAKAKNKLVHSILLWGALDDQSCu1 2 GSGRTLRETVLESSPILALLNESFISTWSLVKELEDLQ 0 0 NNQENPSHQKLAGLHLEKYSFPVEMMICLPNGTV 0 0 VHHINANYFLDITSMKPEDIENHVFSFSSTFEDPSTATYMQFLKEGLRRGLPLLQP* 0 >SEPN1_equCab Equus caballus NM_001917067 0 MEKAGVCCAQ 0 0 ESALKALGTEGLFLFSSLDTDRDMYISPEEFKPIAEKLT 1 2 GSTPAASYEEEELPPDPSEETLTIEARFQPLLPESMTKSKDGFLG 0 0 VSRLALSGLRNWTTAASPSAVFAARHFQPFLPPRGHVELGEPWWIIPSELSVFTGYLSNNRFYPPPPKGKE 0 0 VIIHRLLSMFHPRPFVKTRFAPQGAVACLTAISDFYYTVMFR 2 1 IHAEFQLSEPPDFPFWFSPGQFTGHIILSKDATHVRDFRLFVPNHR 2 1 VSDVDMEWLYGASESSNMEVDIGYIPQ 0 0 MELEALGPSVPSVILDEDGNLIDSRLPSGEPLQFVFEEIKWQQELSWEEAARRLEVAMYPFKK 0 0 VTYLPFTEAFDQAKAKNKLVHSILLWGALDDQSCu 1 2 GSGRTLRETVLESSPILALLNESFISTWSLVKELEDLQ 0 0 NNQDNPSHKKLADLHLEKYSFPVEMMICLPNGTV 0 0 VHHINANYFLDITSMKPEDIESNVFSFSSTFEDPSTATYMQFLKEGLRRGLPLLQP* 0 >SEPN1_galGal Gallus gallus NM_001114972 0 MAVPGAAPSRLALALAALAALAAVKYYRDAEAARQQ 0 0 ELALKSLGSEGLFLFSSLDTNNDLYLSPEEFKPIAEKLT 1 2 GVTPVSDFEEDAPDPNGETLSIVAKFQPLVMETMTKSKDGFLG 0 0 ISHVALSGLRNWTAPVSPKSVMLARQFKAFLPPKNKLDLGDPWWIIPSELNIFTGYLSNNRFYPPPPKGKE 0 0 IIIHRLLSMFHPRPFVKTRFAPQGSVACIQAISTYYYTIAFR 2 1 IHAEFQLNEPPDFPFWFSPGQFTGYIVLSKDSSHVREFKLFVPNKR 2 1 SLNVDMEWLYGASEGSNMEVDIGYLPQ 0 0 MELESTGPSVPSVIYDENGNVIDSRDPSGEPIQFVFEEITWQQEIPWEEAAQKLEVAMYPFKK 0 0 VSYLPFTEAFERAKAEKKLVHSILLWGALDDQSCu 1 2 GSGRTLRETVLESSPILALLNESFISSWSLVKELEELQ 0 0 TNRENEFYSKLADLHLEKYNFPVEMIICLPNGTV 0 0 IHHINANYFLDITSMKPEDVESSIFSFSANFDDPSTATYLQFLKEGLQRAKAYLQN* 0 >SEPN1_xenTro Xenopus tropicalis NM_001077433 0 MSADLRKRKKDTTADNDAPQEAQAEEENKKEKPSSCRYRFLWKLLLGLLLIALLFGIKIHRDNELVRQQ 0 0 EAALRTLGAEGLFLFSSLDTDNDMHISPEEFKPISEKLT 1 2 GISTTSDYEEEELLDPNGETLSVASRFQPLLMETMTKSKDGFLG 0 0 ITHSALSGLRNWTTPVVPNNVFYAGQFKAFLPPKNKLEVGNPWWIIPSELSIFTGYLPNNRIYPPPPKGKE 0 0 VIIHKLLSMFHPRPFIKTRFAPQGSVACIRAISDFYYDIVFR 2 1 IHAEFQLNEPPNFPFWFSPGQFTGNIVISKDAAHVRHFKLFVPNNR 2 1 TLNVDMEWLYGASESSNMEVDIGYLPQ 0 0 MEIESLGPSIPSTIYDENGNIMESRNAEGEAIEFVFEDINWQSEMTFEEAARKLEVTMYPFKR 0 0 VSYYPFPEAFDRALAEKKLVHSVLLWGALDDQSCu 1 2 GSGRTLRETVLESLPVLALLNESFISTWSLVKELEELQ 0 0 SNKDSYATFASLHLEKYNFPVEMMICLPNGTV 0 0 VHHINANYFLDITSLKPDEVETNLFSFSDGVEDLSTVTYIKFLKEGLEKAKPFLHS* 0 >SEPN1_danRer Danio rerio NM_001004294 0 MAADVDKTPAGEQKDDHEDRGTPSSRRARSRFTHISSLFIIAAIPVIGFCIKYCLDIQFVKRH 0 0 EAGLKALGADGLFFFSSLDTDHDLYLSPEEFKPIAEKLT 1 2 GVAPPPEYEEEIPHDPNGETLTLHAKMQPLLLESMTKSKDGFLG 0 0 VSHSSLSGLRSWKRPAISSSTFYASQFKVFLPPSGKSAVGDTWWIIPSELNIFTGYLPNNRFHPPTPRGKE 0 0 VLIHSLLSMFHPRPFVKSRFAPQGAVACIRATSDFYYDIVFR 2 1 IHAEFQLNDVPDFPFWFTPGQFAGHIILSKDASHVRDFHIYVPNDK 2 1 TLNVDMEWLYGASETSNMEVDIGYLPQ 0 0 MELSAEGPSTPSVIYDEQGNMIDSRGEGGEPIQFVFEEIVWSEELKREEASRRLEVTMYPFKK 0 0 VPYLPFSEAFSRASAEKKLVHSILLWGALDDQSCu 1 2 GSGRTLRETVLESSPVLALLNQSFISSWSLVKELEDLQ 0 0 GDVKNVELSEKARLHLEKYTFPVQMMVVLPNGTV 0 0 VHHINANNFLDQTSMKPEDEGPGLSFSAGFEDPSTSTYIRFLQEGLEKAKPYLES* 0 >SEPN1_petMar Petromyzon marinus (lamprey) tga-sel taa-stop frag 0 0 0 EMALRTLGNDGLFLFTSLDTNMDMQISPEEFRPIVDKII 1 2 GPPPSEYEGTQEADPQGEGLTMLARFEPLLMETMSKSRDGFLG 0 0 VGQSCLAGLRGWKKAEAPSQHFGANQFKVFLPPKSDLELGEAWWLVPNDLNLFTGYLPNSRYYPPPPVAKE 0 0 IIIFKLLSMFHPRPFVKSRFAPQGSVACIRAQSDMYYDIVFR 2 1 VHAEFQLNEPPAFPFWFTPAQFTGHVTIARDSSHVRAFHMFVPNNR 2 1 SLNVDMEWLFGSMDQGNMEVDIGFMPK 0 0 MELVAEGPSVPALIYDENGNAINTSDPDVEPIQFVFENIEWRSEISFQEAYRQMEVAMYPFKK 0 0 IQYHPFTEAFEKAKAEDKLVHSILLWGALDDQSCu 1 2 KVLTQSLKFDFNITSPVLALLSENFISSWSLVKDLEDL 0 0 KQEEQAEHAKWATLHLAKYTFPVEMMIALPNGTV 0 0 VHCINANDFLDATAVKAEDLTPDLPAEFLDPTSTTYLKFLKEGLQKAQTYLQA* 0 >SEPN1_braFlo Branchiostoma floridae genomic fragment ABEP01021665 TGA verified 0 MASEGKRDPPDKRGGGGDAAGTDHRPSVTTKRSTLRSCCFWVCAPAAVLLAALGVGMYLDYRRWVRK 0 0 EVGMMTLGPDATSLFLAYDTNEDGYLSMEEFKPLLDRLQ 1 2 GNETSYPVDEEIDQAGEIIVLKTHFEPFVMQSMSKNKNPFGFLG 0 0 NSNLQGLREWREPVRELASFGANHFAPFLPPPAAYSVLGAAYEIIPSTLTPFSGTLSSNRYFPPKVSNKE 0 0 VILHRLLGLFHPRPFVHMRFAPQGAVATVRAENERWVDVVFR 2 1 IHAEFQLNEPPLYPFWFTPAQFTGNIVLSRDASRVRHFHLYVPTNRSLNV 2 1 DMEWLHGPGHENMEVDIGYIPR 0 0 MELTSVAPSTPVLIYQEDGSYIDQREEELSEVCSVYEPTIFLEEDSGDVRWNSEISMEEAMKSLEVKMYPFKT 0 0 VPYYNFTEAFSRAEAENKLVHSILLWGALDDQSCu 1 2 GSGRTLREGPLESSPIVKMLMDHFISTWVLVAELEGIK 0 0 FPVEMMVSLPNGTVVHHLNANDLLDMSSGESFDVASNM 0 0 * 0 >SEPN1_cioInt Ciona intestinalis (tunicate) XR_053040 TGA confirmed for selenocysteine MLRNRKSRIKENVKREEKVTANDEKQQADKPTEVPHSISENKKKEP NDKCLTFVVVFAAAVIAIALGLQVYVYSIIQENAKNAAVRQLGQHGLKLFKEYDINKDMQ IDPKEFRQLYDFILSEDILKASHEVEEEITKEIFDPSTDEIVQLKADMIPLVLSSMSQTN KNPAFGTPLFHYSKDFSGLVAWKSVKTEQKDLYAKEFKAFLPNNSSQLVGEPYWLIQRPN NPEELTSNRYRYPIPKTKIEKLLHNLLVMFHPRPFVTMRFSPQGAAAVIRAQNQVYLEIV FRFHSEFQLNEPPYLPYWFTPAQFTGRLIIARDGTHVRGFQLYVPNNKKLNVDMEWETND PNAPENNMEVDIGFLPLMTINQTQPSSPMILNNDDGTSIDRRELIKQGKDIVVEFDEIQW KDDAITTQYAHGQLEKLFFPFKKVDYLPFNEAFKRASVENKLIHQVVLWGALDDQSC-GS GRTLRETALESSPVIQLLNQSFISTWSLLKDLEVISNDKQSPLSNVANLSIEAYKFPVQM MVILPNGTVISSLNANDWLAQANGEVPVDEKWLSLGSKDYNTFLSNALEASKNL* >SEPN1_strPur Strongylocentrotus purpuratus (urchin) TGA confirmed for selenocysteine MMPSKDRKKPPDRGEGRSAASAAGANCVATATQTTEHYPPNFGRSNRGRCGWKSLTFLSLFVAVVAVMAGNLA NRIMMETFVMEEMKLTIGEEGVSIFNAYDRDGDGHLNVIEFEPLLERLNIEDTAERGDFT YGDEDLDPAEEVLTIEANFIPLQQDTMSKAKDQSFLPLLTHAEFQLNEPPSHPFWFTPAQ FIGHLIIRKDASHVKYFNMHVPSNRSLNVDMEWMNGPNEVENMEVDIGFMPKMELNAPNP SSQVTIYSESGEVLYEPSNQRETSSGIIHWDEEISMEEALVLLEKEMYPFKKIEYMPLNA AFKRAHAEKKLVHHVLLWGALDDQSCuGSGRTLRETSLESPPVLQLLGSHYVSSWSLVAE LEVIKQNKSDPEYAALASMSLEKYNFPVEMLVSLPNGTVIHHINANDLLDASNTENTSIL DTFTDPLVTNYAKFLKEGISKAEAVGF* >SEPN1_nemVec Nematostella vectensis (cnidaria) some exon differences TGA confirmed and phase 1 0 MTESDEDDGDTATSSKEKVNGSPFEEESVLSSRTKVLTVSLLVALLSISCYVFFIEHPILEGTGDKIEEEIMGLFVSYDTNADGRLDPYEFVPVAHRILGQK 0 0 ESRSEKQAVVHQNWDDSGEHLTMKALFKPLELSTMTKTSDSYF 0 0 LSDSALTGLKSWKVVSIPSMTLPVNAFAAFLPPSHASTGMLGTPWFIVEPQIAKF 1 2 SPQLSSNRYNPPDVSGNNAVIHQLLAMFHPRPFIFTRFGPQGTAACIEAQNNEYLAIQFR 2 1 IHAEFQLNEPPLHPFWFSPAQFAGRLVISRDGKHIASFHMGVPTNKSLNV 1 2 DMEWLLGKPQVTTKQ 1 2 ENDASETTSSNMGVDI 1 2 GFLPQMEIVDQGESCSVTAGGCDSGTSGKHSDNIEWNDQIPNWAAEKKLEQHFYPFKK 0 0 VDYLPFKEAFNRAKAEKKLVHHIVLWGALDDQSCu 1 2 GSGRTLREGPLESRPVLQLLQDNYVNCWSLVEELK 0 0 KISKDGSDQEMANIAQISLDNYRFPVESMVMHANGSL 0 0 VARMNANEMMEVPEHDSLPKKIVETTFHQGFEDDISYTYFNFLKNSLRTE* 0 >SEPN1_oopMin Oopsacas minuta (sponge) AM761280 transcript TGA confirmed ARGEIYNSNRYNAPQPSGREIVFFKLLQQFHHNVFLQNRFGPRGGVAMIRAKAGPIIEVV FRIHAEYQLNEHPLHPFWFSPAHFTGNLVFNTDTDEVLQFELYLPSIRKLNVDMEWITGS DELEGEKMEVDIGYTSAMKLSTNQTFEIPAWDHEKTKEEATIELDRVLYPFKRIPYIDIR DAVDTARVENKLVHSIIMWGSLDDQSCuGSGRTLRDGPLASDVVQLTLKENFISSWSLIV HLEDLMANSSVKQEVKDL >SEPN1_oscCar Oscarella carmela (sponge) EC367053 not quite to TGA ENSNLEGLRTWKKPHKRFNTYNAPNFQSFLPYRGASLGEVYVIVAKPIQFGNQINSVKYF PPYPEGREVLMHMLLNQFHRRPFVRMRFEPQGTIGLIRAENEEYVEIMFRCHAEFQLNEP PMFPFWFTPGQFTGHVIMKKNGSHVRHFKMFVPTNKSLNVDMEWMTGPATSLDPNDPDGS FQGNNQVDIGYMAEMRLESTRSSYFPMLPDENEEDYFKRASKEDEEIKWNKEISRQEALE KLERYFFPFKKSHTCLSKSVSWLS
SELS: 18 vertebrate sequences
>SELS_homSap Homo sapiens (human) 0 MERQEESLSARPALETEGLRFLHTT 1 2 VGSLLATYGWYIVFSCILLYVVFQKLSARLRALRQRQLDRAAAAV 1 2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLKQ 0 0 LEEEKRRQKIEMWDSMQEGKSYKGNAKKPQ 0 0 EEDSPGPSTSSVLKRKSDRKPLRGG 1 2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 >SELS_panTro Pan troglodytes (chimp) 0 MERQEESLSARPALETEGLRFLHTT 1 2 VGSLLATYGWYVVFSCILLYVVFQKLSARLRALRQRQLDRAAAAV 1 2 EPDVVVKRQEAVAAARLKMQEELNAQVEKHKEKLKQ 1 0 LEEEKRRQKIEMWDSMQEGKSYKGNAKKPQ 0 0 EEDSPGPSTSSVLKRKSDRKPLRGG 1 2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 >SELS_macMul Macaca mulatta (rhesus) 0 MEREEDSLSARPALETEGLRFLHVT 1 2 VGSLLASYGWYIVFSCILLYVVFQKLSARLRALRQRQLERAAASV 1 2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLKQ 0 0 LEEEKRRQKIEMWDSMQEGKSYKGNAKKPQ 0 0 EEDSPGPSTSSVVKRKSDRKPLRGG 1 2 GYNPLSGEGGGSCSWRPGRRGPSSGGuS* 0 >SELS_otoGar Otolemur garnettii (bushbaby) 0 MEEEAEPLPARPALEAEGLRFLHVT 1 2 VGSLLAAYGWYIVFSCVLLYVVFQKFSTRLRALRQRQLDRAAAAV 1 2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLKQ 0 0 LEEEKRRQKIEMWDSMQEGKSYKGNTKKPQ 0 0 EEDNSGPSVIPKRKSDRKPLRGG 1 2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 >SELS_musMus Mus musculus (mouse) 0 MDRDEEPLSARPALETESLRFLHVT 1 2 VGSLLASYGWYILFSCILLYIVIQRLSLRLRALRQRQLDQAETVL 1 2 EPDVVVKRQEALAAARLRMQEDLNAQVEKHKEKLRQ 0 0 LEEEKRRQKIEMWDSMQEGRSYKRNSGRPQ 0 0 EEDGPGPSTSSVIKGKSDKKPLRGG 1 2 GYNPLTGEGGGTCSWRPGRRGPSSGGuG* 0 >SELS_ratNor Rattus norvegicus (rat) 0 MDRGEEPLSARPALETESLRFLHVT 1 2 VGSLLASYGWYILFSCVLLYIVIQKLSLRLRALRQRQLDQAEAVL 1 2 EPDVVVKRQEALAAARLRMQEDLNAQVEKHKEKLRQ 0 0 LEEEKRRQKIEMWDSMQEGRSYKRNSGRPQ 0 0 EEDGPGPSTSSVIPKGKSDKKPLRGG 1 2 GYNPLTGEGGGTCSWRPGRRGPSSGGuS* 0 >SELS_canFam Canis familiaris (dog) tga confirmed 0 MERDGQQLSARPALETEGLRYLHVT 1 2 VGSLLATYGWYIVFSCILLYVVFQKLSTRLRALRQRQLDRAEAAV 1 2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 0 LEEQKRRQKIEMWDSMQEGKSYKGNARKHP 0 0 EEDSPGPSTSSVLPKRKPDRKPLRGG 1 2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 >SELS_equCab Equus caballus (horse) 0 MEPDEERLSARPAVEAEGLRFLHVT 0 2 GSLLAAYGWYLVFSCILLYVLFQKLSSRLRALRQRRLDRAAAAV 1 2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 0 LEEEKRRRKIEMWDSMQEGKSYKRNARGPQ 0 0 EEDSPGPSTSSVIKRKSDRKPLRGG 1 2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 >SELS_bosTau Bos taurus (cow) tga confirmed 0 MERDGDQLSARPTLETEGLRFLHVT 1 2 VGSLLATYGWYIVFSCILLYVVFQKLSTRLRALRQRHLDQAAAAL 1 2 EPDIVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 0 LEEEKRRQKIEMWDSMQEGKSYKGNTRKPQ 0 0 EEDSPGPSTSSVIPKRKSDRKPLRG 1 2 GGYNPLSGEGGGTCSWRPGRRGPSSGGuG* 0 >SELS_susScr Sus scrofa (pig) 0 MEQDGDQLSARPALETESLRFLHVT 1 2 VGSLLATYGWYIVFCCILLYVVFQKLSTRLRALRQRHLDGAAAAL 1 2 EPDVVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 0 LEEEKRRQKIERWDSVQEGRSYRGDARKRQ 0 0 EEDSPGPSTSSVIPKRKSDKKPLRGG 1 2 GYNPLSGEGGGACSWRPGRRGPSSGGuG* 0 >SELS_eriEur Erinaceus europaeus (hedgehog) 0 MEADTEPLPLRPALETEGLRLLHAT 1 2 VGSLLASYGWYIVLCGVVLYVAFQKLSPRLRALRRRQLDRAEAAV 1 2 EPDVVVKRQEALAAARLKMQEELNAQAEKHKEKLKQ 0 0 LEEERRRQKIEMWDSMQEGKSYRGNSRRAQ 0 0 EDDSPGPSTSSAVKRKPDRKPLRGG 1 2 GYNPLSGEGGGSCSWRPGRRGPSSGGuG* 0 >SELS_dasNov Dasypus novemcinctus (armadillo) 0 1 2 LGSLLATYGWYIVFSCVLLYVVFQKLSTRLRALRQRQLDRAAAAV 1 2 EPDIVVKRQEAVAAARLKMQEELNAQAEKHKEKLKQ 0 0 LEEEKRRQKIEMWDSIQEGKYGNAKKP 0 0 KGDCPGSSTSLVMKCQSAKLRGG 1 2 GYNPLSGEGGGTCSWRPGRRGPSAGGuG* 0 >SELS_monDom Monodelphis domestica (opossum) 0 MDLDEEEQQQLLAKPALELEGIRFLQET 1 2 VGWLLSNYGWYLLFSFIILYVIFQKLFFRVMRQRALESVASSI 1 0 EPDTVVKQQEALAASRLRMQEELNTQAEKYKEKLKQ 0 2 LEEEKRKQKIEMWESMQEGKSYKGKAKKPQ 0 0 QEPEPGPSTSSNIPKPKPDRKSLRGS 1 2 GYNPLTGEGSGTCSWRPGRRGPSSGGuG* 0 >SELS_ornAna Ornithorhynchus anatinus (platypus) 0 LFI 1 2 VGSVLSAYGWYILFGCAVLYLIFQKLSGSLRVMRRRYSDTTGAAI 1 2 DPEVVVKRQEALAASRLRMQEELNAQAEKYREKQKQ 0 0 LEEAKRRQKIEIWESMQEGKSYKGNSRLQPQ 0 0 QETDPGPSTSSVIPKPKPARKPLRG 1 2 GYNPLSGEGGGTCSWRPGRRGPSSGGuG* 0 >SELS_tacAcu Tachyglossus aculeatus (echidna) EUEMSW404CHB0V 454 0 1 2 LRVMRQRYSDTTGAAI 1 2 DPEVVVKRQEALAASRLRMQEELNAQAEKYREKQKQ 0 0 LEEAKRRQKIEIWESMQEGKSYKGNSRLRPK 0 0 QETDPGPSTSSVIPKPKPARKPLRGG 1 2 SYNPLSGEGGGTCSWRPGRRGPSSGGuG* 0 >SELS_galGal Gallus gallus (chicken) 0 MELGDRGGAGPGPGKPALEREGLRFLHVT 1 2 VGSLLATYGWYIVFSCILLYVVFQKLSTRLRALRQRHLDQAAAAL 1 2 EPDIVVKRQEALAAARLKMQEELNAQVEKHKEKLRQ 0 0 LEEEKQRRQKIEMWDSMQEGKSYKGNTRKPQ 0 0 EEDSPGPSTSSVIPKRKSDRKPLRGG 1 2 GYNPLSGEGGGTCSWRPGRRGPSSGGuG* 0 >SELS_xenTro Xenopus tropicalis (frog) tga confirmed 0 MELGNQPGPGNRPEIELEWYQYLQNT 1 2 VGVVLSSYGWYILLGCILIYLLIQKLPQNFTRAGTSNHSTVT 1 2 DPDEIVRRQEAVTAARLRMQEELNAQAELYKQKQVQ 0 0 LQEEKRQRNIETWDRMKEGKSSKVACRLGQEPS 0 0 PSTSTSAATKPKQEKQERKTLRGS 1 2 GYNPLTGDGGGTCAWRPGRRGPSSGGuG* 0 >SELS_danRer Danio rerio (zebrafish) 0 MEAEDGARVRNEDVPPQNQDLSFLQPS 1 2 VTAFMSEYGWYLLFGCVGVYLLIQHLRKSRSSTQTRSSSGSAEAH 1 2 DVGSVVRRQEALEASRRRMQEEQDARAAEFREKQRM 0 0 LEEEKRRQKIEMWDSMQEGKSYKGSAKVAQ 0 0 QNTEEAASSSSLRPKTEKKPLRSS 1 2 GYSPLSGDAGGSCSWRPGRRGPSAGGuG* 0 >SELS_calMil Callorhinchus milii (elephantfish) 0 1 2 1 2 EPLLIVKQQEAMEAARRKMQAELDTQAVKYKEKQNL 0 0 LEEAKRRQKIEAWESIQEGRSYRANTRFNQ 0 0 QEPGQPSSSNLTKPKSGKKSLAT 1 2 GFNALTGDGGGSCTWRPNRRGPSSGGuG* 0 >SELS_squAca Squalus acanthias (spiny_dogfish) ES606337 511 756 tga-sel taa-stop 0 1 2 ILSEYGWYILFSGVAGYLVLQRISGMLQARSRKWTQASEAQL 1 2 EPSLIVRRQEAMDAARHRMQQELDAQAAKYREKQKE 0 0 IEEEKRREKIEAWDNMQEGKSSRKKANLNPE 0 0 PESSTSSATKPKSDKKSLRG 1 2 GFNPLTGDGGGSCLWRPGRRGPSAGAuG* 0 >SELS_petMar Petromyzon marinus (lamprey) cDNA EB082976 39% identity homSap tga-sel taa-stop full 0 MDPRNEEPAVIRGVSDAVSELLARFGWPL 1 2 LLFCALLYFTVRRAAPWLRWGRSSSSSTSISNP 1 2 DLLMNMHSAMERSRIRMQQELDARAAEHQARVKQ 0 0 LEEERQKQKMESWERGRSLRPRRDPQSQQ 0 0 EDNSTPSTSLPRTERQRLRDN 1 2 NYSPLSGGGGPTCLWRPGRRGPASGGGuG* 0
SELT: 11 vertebrate sequences
>SELT_homSap Homo sapiens (human) 5 exons recent pseudogenes on chr 5, chr9 0 MQYATGPLLKFQIC 2 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 >SELT_macMul Macaca mulatta (rhesus) 0 MRLLLLLLVAASAMVRSEASANLGGVPSKRLKMQYATGPLLKFQIC 2 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 >SELT_otoGar Otolemur garnettii (bushbaby) 0 MRLLLLLLVAASAVVRSEASANLGGVPSKRLKMQYATGPLLKFQIC 2 1 VSuGYRRVFEEYMRVISQRYPDIRIIGKIYSLHHSYR 2 1 HIASFLSIFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 >SELT_musMus Mus musculus (mouse) 0 MRLLLLLLVAASAVVRSEASANLGGVPSKRLKMQYATGPLLKFQIC 2 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 >SELT_ratNor Rattus norvegicus (rat) 0 MRLLLLLLVAASAVVRSEASANLGGVPSKRLKMQYATGPLLKFQIC 2 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 >SELT_bosTau Bos taurus (cow) 0 MRLLLLLLVAASAVVRSDASANLGGVPGKRLKMQYATGPLLKFQIC 2 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 >SELT_canFam Canis familiaris (dog) 0 MRYLLVLLVAASAVIRSDASANLGGVPSKRLKMQYATGPLLKFQIC 2 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 >SELT_monDom Monodelphis domestica (opossum) 0 MRFLLLLLMAAAAL-QGEASADTGGVPGKRLKMQYATGPLLKFQIC 2 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 0 IYACMMVFFLSNMIENQCMSTGAFEITLN 1 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHH*SQ * 0 >SELT_ornAna Ornithorhynchus anatinus (platypus) 0 MRLLLLVVVAAAAGGRSEASADLGGLPSKRLKMQYATGPLLKFQIC 2 1 VSuGYRRVFEEYMRVISQRYPDIRIEGENYLPQPIYR 2 1 HIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENK 0 0 VYACMMVFFLSNMIENQCMSTGAFEITLN 1 2 DVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS* 0 >SELT_petMar Petromyzon marinus (lamprey) cDNA EE737658 tga-sel tag-stop full 0 MRTHLFAGPVLKVEYC 2 1 VSuGYRRVFEEYSRVIAERFPDIRVEGDNYLPQPLYR 2 1 YIASFFSVFKLVLIGLVLSGKNLFPMLGVDTPGVWTWSQENK 0 0 LYACLMIFFVSNMVETQCMSTGAFEVSLN 1 2 DVPVWSKLQSGRVPSPQEILQILDNHVKLSGGSAGRMQPS* 0 >SELT_eptBur Eptatretus burgeri (hagfish) cdna BJ650136 tga-sel tag-stop full 0 MKSKMYSGPELLFQYC 2 1 ISuGYRRVFEEYSQALRERYPDIRIEGSNYPPPPLYS 2 1 TCASVLSVLKVMLIVLVVSGRNPFPLLGLDTPNAWNWSQNNK 0 0 IYACLMIFFLTNMIENQCLSTGAFEVVFN 1 2 DVPIWSKLQSGRVPSLPELAQILDNHLAMGGQASPSNTHGPQ* 0
SEPHS1: 13 chordate sequences
>SEPHS1_homSap Homo sapiens (human) PHYH- SEPHS1- DUF1172- PRFF18+ FRMD4A- 0 MSTRESFNPESYELDKSFRLTRFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL 1 2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKMTDR 0 0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPGATS* 0 >SEPHS1_musMus Mus musculus (mouse) 0 MSTRESFNPETYELDKSFRLTRFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL 1 2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKMTDR 0 0 ERDKVIPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPGATS* 0 >SEPHS1_loxAfr Loxodonta africana (elephant) frag 0 MSVRESFNPESYELDKSFRLTKFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRLGM 1 2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 0 GRIACANVLSDLYAMGVTECDNMLMLLGISNKMTDR 0 0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 2 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTP 1 >SEPHS1_monDom Monodelphis domestica (opossum) 0 MSVRESFNPESYELDKSFRLTRFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL 1 2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 0 GRIACANVLSDLYAMGVTECDNMLMLLGISNKMTDR 0 0 ERDKVVPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPEMAATALYTLDNNVLTLEQRQFYEDNGFLVIGATS* 0 >SEPHS1_ornAna Ornithorhynchus anatinus (platypus) virtual selenocys 0 MSVRESFNPESYELDKSFRLTRFTELKGTGCKVPQDVLQKLLEALQENHFQEDEQFLGAVMPRL 1 2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 0 GRIACANVLSDLYAMGVTECDNMLMLLGISNKMTDR 0 0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPGATS* 0 >SEPHS1_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01BJUY7 454 0 AEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQ 0 2 AKQQRSEVSFVIHNLPIIAKMAAITKACGNRFGLLQGTSSETS 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPRIIEVTPRGAAAPPPDNNSSA >SEPHS1_galGal Gallus gallus (chicken) 0 MSvREtFNPESYELDKSFRLTRFTELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL 1 2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKMTDR 0 0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWIVLGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNPTPGATS* 0 >SEPHS1_xenTro Xenopus tropicalis (frog) frag 0 MSVRESFNPESYELDKSFRLTRFAELKGTGCKVPQDVLQKLLESLQENHFQEDEQFLGAVMPRL 2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKLTDR 0 0 ERDKVMPLIIQGFKDAAEEAGTSVTGGQTVLNPWVVLGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRT 1 2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGSCPETS 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVASQNVNPTPGATS* 0 >SEPHS1_danRer Danio rerio (zebrafish) first occurence of threonine form 0 MSVRESFNPESYELDKNFRLTRFTELKGTGCKVPQDVLQKLLESLQENHYQEDEQFLGAVMPRL 2 GIGMDTCVIPLRHGGLSLVQTTDYIYPIVDDPYMM 0 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKLTEK 0 0 ERGKVMPLVIQGFKDASEEAGTSVTGGQTVINPWIVLGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMLNMARLNRT 1 2 AAGLMHTFNAHAATDITGFGILGHAQNLARQQRTEVSFVIHNLPVLAKMAAVSKACGNMFGLMHGTCPETS 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNRTARIIDKPRIIEVAPQVATQNVNTTPGATS* 0 >SEPHS1_calMil Callorhinchus milii (elephantfish) frag 0 MSVRETFNPENYELDKNFRLTRFAELKGTGCKVPQDVLHKLLEALQENHYQEDEQFLGAVMPRL 1 2 GIGMDSCVIPLRHGGLSLVQTTDFFYPLVDDPYMM 0 0 GRIACANVLSDLYAMGVTECDNMLMLLGVSNKMADK 0 0 ERDKVMPLIIQGFKDAADEAGTAVTGGQTVLNPWIILGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMFNMARLNRT 1 2 1 2 GGLLICLPREQAARFCAEIKSPKYGEGYQAWIIGIVEKGNRTARIIDKPRIIEVAPTVATQNVNPTPGATS* >SEPHS1_petMar Petromyzon marinus (lamprey) cDNA LyEST8076 FD729689 tga-sel tgc-cys tag-stop not so virtual selenocys full 0 MSVHRYFDPEDHDLDKSFRLTKFSELKGuGCKVPQETLLKLLEGLEQDGPYQDEHQQFMGAVMPRL 1 2 GIGMDACVIPLRHGGLSLVQTTDFFYPLVDDPYMM 1 2 GKIACANVLSDLYAMGVTECDNMLMLLAISQKLSEK 0 0 ERDKVIPLMIRGFKDAAEEAGTTVTGGQTVVNPWIVIGGVATTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVNAHQWLDM 0 0 PEKWNKIKLVVTQEDVELAYQEAMFNMARLNRTAAGLMHTFNAH 2 1 AATDITGFGILGHAQNLAKQQRNEVSFVIHNLPVIAKMAAISKACGNLFGLLQGTSAETSG 1 2 GLLICLPREQAARFCAEIaKYGEGHQAWIIGIVEKGGRTARIIDKPRIIEVAPRGASPTGVASPDTPTGPPLT* 0 >SEPHS1_eptBur Eptatretus burgeri (hagfish) cdna BJ649814 BJ654390 tga-sel taa-stop full 0 MSVHRYFDPEDHELDKSFRLTKFSDLKGuGCKVPQESLLKLLEGLEQDSPFQDEHQQFMGAVMPRL 1 2 GIGMDSCVIPLRHGGLSLVQTTDFFYPLVDDPYMM 0 0 GKIACANVLSDLYAMGVTDCDNMLMLLGISQKLSEK 0 0 ERDKVVPLMVRGFKDAAEEAGTTVTGGQTVMNPWIIIGGVASTVCQPNEFIM 2 1 PDNAVPGDVLVLTKPLGTQVAVAVHQWLDI 0 0 PEKWNKIKLVVTQEDVELAYQEAMMNMARLNRTAAGLMHTFNAH 1 2 AATDITGFGILGHAQNLARQQRNEVAFVIHNLPRNEVAFVIHNLPVVAKMAAISKACGNLFGLLQGTSAETSG 1 2 GLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGARTARIIDKPRIIEVAPRGAPPPTVTTPTTPTAPLS* 0 >SEPHS1_braFlo Branchiostoma floridae (amphioxus) tga confirmed 73% hsa BW738890 BW748096 frag 0 MAAPEQVQQAVRPGRVFDPVAHGLDKSFRLTRFADLKuCCKVPQEVLLNLLEGLQNQTIPEEQGQFLPPHFAPI 1 2 GIGMDSCVVPLRQGGLSLVQTTDFFYPLVDDPYMQ 0 0 GKIACANVLSDLYAMGVKTCDNMLMLLGVSNKMSDK 0 0 ERDTVVPLIIRGFKDLAEEAGTMVQGGQTVLNPWVTIGGVATTVCPPNEFIE 2 1 PGNAVVGDVLVLTKPLGTQVAVNAHQWLDE 0 0 PERWNRIRLVVSEEDVERAYQESMFLMSKLNRT 1 2 AAKLMHKYNAHGATDITGFGLLGHAKNLVQHQKNEVSFTIHNLPVIAKMAAVAKACGNMFQLLQGYSAETS 1 2 * 0 >SEPHS1_strPur Strongylocentrotus purpuratus (sea_urchin) tga confirmed one fused exon, one new 0 MAGLPQDVVQQQHNQAILVQQQPQFEVTEPVVVQQQQQVVRRFDPEAHELHKDFRLTRYTELKGuGCKVPREALLNLLKGLEPNEGEQGQGDNAEYMQVQSGVPKF 1 2 GIGMDCSVTPLRHGGLCLVQTTDFFFPLVDDPYMQ 0 0 GQIACANVLSDLCAMGVTECDNMLMLLGVPKAFTDK 0 0 ERDAIMPLIMRGFK DKAAEAETSVTGGQTVINPWCLVGGVATCVCLPTDIVS 2 1 PNNAVVGDVLVLTKPLGTQIAVNAHQWLDQ 12 PEKWKKIQSVVSPEEVERAYQEAMYNMSLLNRT 1 2 AASLMRKFNAHGSTDVTGFGILGHAKNLVENQKNEVRFVIHNLPVIANMQNVAKTCGNQFQLLQGYSAETS 1 2 GGLLIAFPREQAAAFCREMEKSTGHTAWIIGIVDRGSRTAQIIEKPRVIAVTTKMN* 0
SEPHS2: 8 vertebrate sequences
>SEPHS2_homSap Homo sapians (human) 1 exon but intronated like SEPHS1 for comparison MAEASATGACGEAMAAAEGSSGPAGLTLGRSFSNYRPFEPQALGLSPSWRLTGFSGMKGuGCKVPQEALLKLLAGLTRPDVRPPLGRGLVGGQEEASQEAGLPAGAGPSPTFPAL GIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM GRIACANVLSDLYAMGITECDNMLMLLSVSQSMSEEE EKVTPLMVKGFRDAAEEGGTAVTGGQTVVNPWIIIGGVATVVCQPNEFIM PDSAVVGDVLVLTKPLGTQVAVNAHQWLDN PERWNKVKMVVSREEVELAYQEAMFNMATLNRT AAGLMHTFNAHAATDITGFGILGHSQNLAKQQRNEVSFVIHNLPIIAKMAAVSKASGRFGLLQGTSAETS GGLLICLPREQAARFCSEIKSSKYGEGHQAWIVGIVEKGNRTARIIDKPRVIEVLPRGATAAVLAPDSSNASSEPSS* >SEPHS2_musMus Mus musculus (mouse) 1 exon MAEAAAAGASGETMAALVAAEGSLGPAGWSAGRSFSNYRPFEPQTLGFSPSWRLTSFSGMKGuGCKVPQETLLKLLEGLTRPALQPPLTSGLVGGQEETVQEGGLSTRPGPGSAFPSL SIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM GRIACANVLSDLYAMGITECDNMLMLLSVSQSMSEKER EKVTPLMIKGFRDAAEEGGTAVTGGQTVVNPWIIIGGVATVVCQQNEFIM PDSAVVGDVLVLTKPLGTQVAANAHQWLDN PEKWNKIKMVVSREEVELAYQEAMFNMATLNRT AAGLMHTFNAHAATDITGFGILGHSQNLAKQQKNEVSFVIHNLPIIAKMAAISKASGRFGLLQGTSAETS GGLLICLPREQAARFCSEIKSSKYGEGHQAWIVGIVEKGNRTARIIDKPRVIEVLPRGASAAAAAAPDNSNAASEPSS* >SEPHS2_loxAfr Loxodonta africanus (elephant) 1 exon MAEAAATGASGEAMAVAEGSSGPAGFSLGRGFSSYRPFEPQALGLNPSWRLTGFSGMKGuGCKVPQETLLKLLAGLTRPDMRPPLGRTLVEGHEEAIQEAGLPAGSGPSPTLPSL GIGLDSCVIPLRHGGLSLIQTTDFFYPLVEDPYMM GRIACANVLSDLYAMGITECDNMLMLLSVSQNMSEE EREKVTPLMIKGFRDAAEEGGTAVTGGQTVINPWIIIGGVATVVCQPNEFIM PDSAVVGDVLVLTKPLGTQVAVNAHQWLDN PERWNKIKMVVSREEVELAYQEAMFNMATLNRT AAGLMHTFNAHAATDITGFGILGHSQNLARQQRNEVSFVIHNLPIIAKMAAISKASGRFGLLQGTSAETS GGLLICLPREQAARFCSEIKSSKYGEGHQAWIVGIVEKGNRTARIIDKPRVIEVLPRGTTATTLVPDNFNASSEPTL* >SEPHS2a_monDom Monodelphis domestica (opossum) processed 1 exon processed MAAAAAATVGNGACTPATGGSLAGSYRPFEPQALGLSPNWRLTSFSDMKG uGCK VPQETLLTLLAGLTRPEERPPRAPGLNLGFGDVVAEEAAGPAVLEPGPDAGPFLPSAAGPSLPSPTL GIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM GRIACANVLSDLYAMGITECDNMLMLLSVSQKMNEE EREKVMPLMIKGFRDAAEEGGTSVTGGQTVVNPWIIIGGVATVVCQPNEFIM PDSAVPGDVLVLTKPLGTQVAVNAHQWLDN PEKWNKIKLVVTREDVELAYQEAMFSMAMLNRT AAGLMHTFNAHAATDVTGFGILGHAQNLAKQQRSQVSFVIHNLPIIAKMAAISKASGRFGLLQGTSAETS GGLLICLPREHAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPGIIEVTPRGATVPLHPNNSHASLEPGS* >SEPHS2a_macEug Macropus eugenii (wallaby) 1 exon processed MRRRQRTVRTEAVTPAVGGSFAGSYSPFEPQSLGLSPNRRLTSFSDMKGuGCKVPQEALLTLLAGLTRPEERPPRAPGLGLGFGDSAAEEAAGPAAPEPGPDAGPFLAPAAGASPSPPTL GIGMDSCVIPLRHGGLSLVQTTDFFYPLMEDPYMM GRIACANVLSDLYAMGITECDNMLMLLSVSQKMNEE EKEKIMPLMIKGFRDAAEEGGTSVTGGQTVINPWIIIGGVATVVCQPNEFIM PDSAVPGDVLVLTKPLGTQVAVNAHQWLDNPERWNKIKLVVTREDVELAYQEAMFNMAMLNRT AAGLMHTFNAHAATDVTGFGILGHAQNLAKQQRSQVSFVIHNLPIIAKMAAISKASGRFGLLQGTSAETS GGLLICLPREHAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPGIIEVTPRGATVPLHPNNSHASLEPGS* 0 >SEPHS2b_monDom Monodelphis domestica (opossum) unprocessed 8 exons no syntenty to 1 frag 0 MASVPPRAPAAEGAGAASPGAYRPFEPKALGLSPSWRLTGFSELKGuGCKVPPEALLTLLASLTAAPAPAADRGN 1 2 GIGMDSCVVPLRHGGLSLVQTTDFFYPLVEDPYMM 0 0 GRIACANVLSDLYAMGITECDNMLMLLSVSQKMSEE 0 0 EREKIMPLMVRGFRDAAEQGGTSVTGGHTVINPWLIIGGVATSVCQPNEFIM 2 1 0 0 PEKWNKIKLVVSKEDVELAYQEAMFSMAMLNRT 1 2 AARLMHTFNAHAATDITGFGILGHAQNLAKQQRSEVSFVIHNLPIIAKMAAVSKASGCFGLLQGTSAETS 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPRIIEVMPRGATLHQDNSNTSPEPAL* 0 >SEPHS2b_macEug Macropus eugenii (wallaby) unprocessed 0 MAAAPPPPPPPLAAEGASAGAGARSQGAYRPFEPQALGLSPSWRLTGFSELKGuGCKVPQEALLTLLAGLARPPVAPAPDGGD 1 2 GIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM 0 0 GRIACANVLSDLYAMGITDCDNMLMLLSISQKMNEE 0 0 EREKIMPLMVKGFRDAAEEGGTSVTGGHTVINPWIIIGGVATVVCQSNEFIM 2 1 0 0 PERWNKIKLVVSKEDVELAYQEAMFSMAMLNRT 1 2 AARLMHTFNAHAATDITGFGILGHAQNLAKQQRSEVSFVIHNLPIIAKMAAVSKASGCFGLLQGTSAETS 1 2 GGLLICLPREQ AARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPRIIEVMPRGAALHHDSSNTSPEPAS* 0 >SEPHS2_ornAna Ornithorhynchus anatinus (platypus) tga confirmed 0 FDPELLGLSPSWRLTGFSELKGuGCKVPQETLLKLLAGLTHPDPRPGAAPPDPQDPPPDPQDRTPHHAGPAPPAL 1 2 GIGMDSCVIPLRHGGLSLVQTTDFFYPLVEDPYMM 0 0 GRIACANVLSDLYALGITECDNMLMLLSVSQKMNEE 0 0 EREKIMPLMIKGFRDAAEEGGTSVTGGQTVVNPWIIIGGVATVVCQPNEFIM 2 0 PDNAVPGDVLVLTKPLGTQVAVNAHQWLDN 0 0 PEKWNKIKLVVSREDVELAYQEAMFSMAMLNRT 1 2 AAGLMHTFNAHAATDITGFGILGHAQNLAKQQRSEVSFVIHNLPIIAKMAAITKACGNRFGLLQGTSSETS 1 2 GGLLICLPREQAARFCAEIKSPKYGEGHQAWIIGIVEKGNQTARIIDKPRIIEVTPRGAAAPPPDNNSSASPETGS* 0 >SEPHS2_tacAcu Tachyglossus aculeatus (echidna) EUPZL4S02I1W5B 454 0 KLVVSREDVELAYQEAMFSMAMLNRT 1 2 AAGLMHTFNAHAATDI >SEPHS2_galGal Gallus gallus (chicken) frag 0 MAAPPGPSVPSPTPGPSPGSGCALGPSTSYRPFDPTSLGLDPNWRLTSYSELRGuGCKVPEETLLRLLEGLGGPRSGAAAAAAAAGGGDGEGAEPGGAPTL 1 2 GIGTDCCVIPLRHGGLSLVQTTDFFYPLVDDPYMM 0 0 GRIACANVLSDLYAMGITECDNMLMLLSVSQKMTDE 0 0 ERDKVMPLIVRGFRDAAEAAGTSVTGGQTVLNPWVIVGGAASVVCQRQEFIV 2 1 PDSAVPGDVLVLTKPLGTQVAVTAHQWLDN 0 0 PERWNKIKLVVTREEVEAAYREAMFSMATLNRT 1 2 AAGLMHTFNAHAATDITGFGILGMHKNLAKQQRNEVSLLFTSLPVLAKNGCCQMAAVSKGLWQYCLGLYSLELVPETS 1 2 GGLLICLPREQAARFCAE >SEPHS2_xenTro Xenopus tropicalis (frog) 8 exons in genome 0 maAPGASHLSAACPFFPGYRPFDPVSVGLEASFRLTSFSDLKGuGCKVPRETLLRLLTGLTEEEAAVSGGAIRDPDVPCAGQDGGQNRL 1 2 GIGLDSCVIPLRHRGLSLVQTTDFFYPSVEDPYMQ 0 0 GRIACANVLSDLYAMGITECDNMLMLLSVSQKMTEE 0 0 EREKVTPLMIKGFRDAAEEGGTSVTGGQTVVNPWIIIGGVATVVCQANEFIM 2 1 PQNAVAGDVLVLTKPLGTQVAVNAHQWLDN 0 0 PERWNKIRLVVSREDVELAYQEAHVNMATLNRT 1 2 AAALMHSFNAHAATDVTGFGILGHAQNLAQEQQNEVSFVIHNLPIIAKAAITKACGNRFgLLQGTSPETS 1 2 GGLLICLPREQAARFCAEIKSPKHGEGHQAWIIGIVEKGNRSARIIEKPRiiEVTPRGAaTSDNTA* 0
TXNRD1 (TR1): 27 chordate terminal exons
>TXNRD1_homSap Homo sapiens (human) 13 exons tga-sel taa-stop 0 MNGPEDLPKSYDYDLIIIGGGSGGLAAAK 0 0 EAAQYGKKVMVLDFVTPTPLGTRW 1 2 GLGGTCVNVGCIPKKLMHQAALLGQALQDSRNYGWKVEET 1 2 VKHDWDRMIEAVQNHIGSLNWGYRVALREKKVVYENAYGQFIGPHRIK 0 0 ATNNKGKEKIYSAERFLIATGERPRYLGIPGDKEYCISS 0 0 DDLFSLPYCPGKTLVVGASYVALECAGFLAGIGLDVTVMVRSILLRGFDQDMANKIGEHMEEHGIKFIRQFVPIK 0 0 VEQIEAGTPGRLRVVAQSTNSEEIIEGEYNT 0 0 VMLAIGRDACTRKIGLETVGVKINEK 2 1 TGKIPVTDEEQTNVPYIYAIGDILEDKVELTPVAIQAGRLLAQRLYAGSTVK 0 0 CDYENVPTTVFTPLEYGACGLSEEKAVEKFGEENIE 0 0 VYHSYFWPLEWTIPSRDNNKCYAKIICNTKDN 0 0 ERVVGFHVLGPNAGEVTQGFAAALKCGLTKKQLDSTIGIHPVCAE 0 0 VFTTLSVTKRSGASILQAGCuG* 0
VFTTLSVTKRSGASILQAGCuG TXNRD1_homSap Homo sapiens (human) VFTTLSVTKRSGASILQAGCuG TXNRD1_panTro Pan troglodytes (chimp) VFTTLSVTKRSGASILQAGCuG TXNRD1_ponPyg Pongo pygmaeus (orang_sumatran) VFTTLSVTKRSGASILQAGCuG TXNRD1_macMul Macaca mulatta (rhesus) VFTTLSVTKRSGGSVLQAGCuG TXNRD1_otoGar Otolemur garnettii (bushbaby) VFTTLSVTKRSGGSILQAGCuG TXNRD1_tupBel Tupaia belangeri (tree_shrew) IFTTLSVTKRSGGDILQSGCuG TXNRD1_musMus Mus musculus (mouse) IFTTLSVTKRSGGDILQSGCuG TXNRD1_ratNor Rattus norvegicus (rat) VFTTLSVTKRSGASVLQSGCuG TXNRD1_cavPor Cavia porcellus (guinea_pig) VFTTLSVTKRSGGDILQAGCuG TXNRD1_oryCun Oryctolagus cuniculus (rabbit) VFTTLSVTKRSGASILQAGCuG TXNRD1_canFam Canis familiaris (dog) IFTTLSVTKRSGGNILQTGCuG TXNRD1_equCab Equus caballus (horse) VFTTLSVTKRSGGNILQTGCuG TXNRD1_bosTau Bos taurus (cow) VFTTLSVTKRSGASILQAGCuG TXNRD1_susScr Sus scrofa (pig) VFTTLSVTKRSGGSILQAGCuG TXNRD1_eriEur Erinaceus europaeus (hedgehog) VFTTLSVTKRSGGSILQTGCuG TXNRD1_echTel Echinops telfairi (tenrec) VFTTLSVTKRSGGSILQAGCuG TXNRD1_monDom Monodelphis domestica (opossum) IFTTLTVTKRSGGNVIQAGCuG TXNRD1_ornAna Ornithorhynchus anatinus (platypus) VFTTLSITKRSGENTLQSGCuG TXNRD1_galGal Gallus gallus (chicken) IFTTLSVTKRSGENILQSGCuG TXNRD1_anoCar Anolis carolinensis (lizard) IFTTLTVTKRSGGNILQSGCuG TXNRD1_xenTro Xenopus tropicalis (frog) IFTTMEVTKSSGGDITQSGCuG TXNRD1_danRer Danio rerio (zebrafish) IFTTLEVTKSSGKSIIQTGCuG TXNRD1_tetNig Tetraodon nigroviridis (pufferfish) IFTTMEVTKSSGGNINQSGCuG TXNRD1_takRub Takifugu rubripes (fugu) VFTTLDVTKRSGGNITQSGCuG TXNRD1_gasAcu Gasterosteus aculeatus (stickleback) IFTTLQVTKSSGGDIKQAGCuG TXNRD1_oryLap Oryzias latipes (medaka) IFTMLAVTKRSGADIKQTGCuG TXNRD1_petMar Petromyzon marinus (lamprey) IFTTMDITKGSGEDPTKTGCuG TXNRD1_braFlo Branchiostoma floridae (amphioxus)
TXNRD3 (TGR): 29 chordate terminal exons
>TXNRD3_homSap Homo sapiens (human) 12 exons tga-sel tga-stop 0 MVLDFVVPSPQGTSW 1 2 GLGGTCVNVGCIPKKLMHQAALLGQALCDSRKFGWEYNQQ 1 2 VRHNWETMTKAIQNHISSLNWGYRLSLREKAVAYVNSYGEFVEHIK 0 0 ATNKKGQETYYTAAQFVIATGERPRYLGIQGDKEYCITS 2 1 DDLFSLPYCPGKTLVVGASYVALECAGFLAGFGLDVTVMVRSILLRGFDQEMAEKVGSYMEQHGVKFLRKFIPVM 0 0 VQQLEKGSPGKLKVLAKSTEGTETIEGVYNT 0 0 VLLAIGRDSCTRKIGLEKIGVKINEK 2 1 SGKIPVNDVEQTNVPYVYAVGDILEDKPELTPVAIQSGKLLAQRLFGASLEK 0 0 CDYINVPTTVFTPLEYGCCGLSEEKAIEVYKKENLE 0 0 IYHTLFWPLEWTVAGRENNTCYAKIICNKFDH 0 0 DRVIGFHILGPNAGEVTQGFAAAMKCGLTKQLLDDTIGIHPTCGE 0 0 VFTTLEITKSSGLDITQKGCuG* 0
VFTTLEITKSSGLDITQKGCuG TXNRD3_homSap Homo sapiens (human) VFTTLEITKSSGLDITQKGCuG TXNRD3_panTro Pan troglodytes (chimp) VFTTLEITKSSGLDITQKGCuG TXNRD3_macMul Macaca mulatta (rhesus) VFTTLEITKSSGLDITQKGCuG TXNRD3_musMus Mus musculus (mouse) VFTTMEITKSSGLDITQKGCuG TXNRD3_ratNor Rattus norvegicus (rat) VFTTLDITKSSGLDITQRGCuG TXNRD3_cavPor Cavia porcellus (guinea_pig) VFTTLEITKSSGLAVTQQGCuG TXNRD3_oryCun Oryctolagus cuniculus (rabbit) VFTTLEITKSSGLDITQKGCuG TXNRD3_canFam Canis familiaris (dog) VFTTLEITKSSGLDITQKGCuG TXNRD3_felCat Felis catus (cat) VFTTLEITKASGLDITQKGCuG TXNRD3_equCab Equus caballus (horse) VFTTLEITKSSGLDITQKGCuG TXNRD3_myoLuc Myotis lucifugus (microbat) VFTTLEITKASGLDITQKGCuG TXNRD3_bosTau Bos taurus (cow) IFTTLEITKASGLDILQRGCuG TXNRD3_sorAra Sorex araneus (shrew) VFTTLEITKSSGLDITQKGCuG TXNRD3_dasNov Dasypus novemcinctus (armadillo) VFTTLEITKSSGLDITQKGCuA TXNRD3_loxAfr Loxodonta africana (elephant) A=single trace VFTTLEITKSSGLDITQKGCuG TXNRD3_proCap Procavia capensis (hyrax) VFTSLEITKSSGLDISQKGCuG TXNRD3_echTel Echinops telfairi (tenrec) VFTTLEITKSSRTVISPRGCuG TXNRD3_monDom Monodelphis domestica (opossum) IFTTLTVTKRSGGNVIQAGCuG TXNRD3_ornAna Ornithorhynchus anatinus (platypus) VFTTMDITKSSGQDITQRGCuG TXNRD3_galGal Gallus gallus (chicken) VFTTMDITRASGKDIAQSGCuG TXNRD3_anoCar Anolis carolinensis (lizard) VFTTMDVSKSSGGDITQKGCuG TXNRD3_xenTro Xenopus tropicalis (frog) IFTTMEVTKSSGGDITQSGCuG TXNRD3_danRer Danio rerio (zebrafish) IFTTLEVTKSSGKSIIQTGCuG TXNRD3_tetNig Tetraodon nigroviridis (pufferfish) IFTTMEVTKSSGGNINQSGCuG TXNRD3_takRub Takifugu rubripes (fugu) VFTTLDVTKRSGGNITQSGCuG TXNRD3_gasAcu Gasterosteus aculeatus (stickleback) IFTTLQVTKSSGGDIKQAGCuG TXNRD3_oryLap Oryzias latipes (medaka) VFTTLSVSKRSGGNIQQSGCuG TXNRD3_calMil Callorhinchus milii (elephantfish) IFTMLAVTKRSGADIKQTGCuG TXNRD3_petMar Petromyzon marinus (lamprey) IFTTMDITKGSGEDPTKTGCuG TXNRD3_braFlo Branchiostoma floridae (amphioxus)
TXNRD2 (TR3): 33 vertebrate terminal GCUG* exons
>TXNRD2_homSap Homo sapiens (human) 17 exons tga-sel taa-stop 0 MAAMAVALRGLGGRFRWRTQAVAGGVRGAARGAA 1 2 AGQRDYDLLVVGGGSGGLACAKE 1 2 AAQLGRKVAVVDYVEPSPQ 1 2 GTRWGLGGTCVNVGCIPKKLMHQAALLGGLIQDAPNYGWEVAQPVPHDW 2 1 RKMAEAVQNHVKSLNWGHRVQLQDR 2 1 KVKYFNIKASFVDEHTVCGVAKGGKE 0 0 ILLSADHIIIATGGRPRYPTH 0 0 IEGALEYGITSDDIFWLKESPGKT 2 1 LVVGAS 1 2 YVALECAGFLTGIGLDTTIMMRSIPLRGFDQ 0 0 QMSSMVIEHMASHGTRFLRGCAPSRVRRLPDGQLQVTWEDSTTGKEDTGTFDTVLWAI 1 2 GRVPDTRSLNLEKAGVDTSPDTQKILVDSREATSVPHIYAIGDVVE 0 0 GRPELTPIAIMAGRLLVQRLFGGSSDLMDYDN 0 0 VPTTVFTPLEYGCVGLSEEEAVARHGQEHVE 0 0 VYHAHYKPLEFTVAGRDASQCYVK 0 0 MVCLREPPQLVLGLHFLGPNAGEVTQGFALGIK 2 1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0
1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_homSap Homo sapiens (human) 1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_ponPyg Pongo pygmaeus (orang_sumatran) 1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_macMul Macaca mulatta (rhesus) 1 CGASYVQVMQTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_calJac Callithrix jacchus (marmoset) 1 CGASYAQVMRTVGIHPTCSEEVVRLRISKRSGLDPTVTGCuG* 0 TXNRD2_otoGar Otolemur garnettii (bushbaby) 1 CGASYAQVTRTVGIHPTCAEEVVRLRISKRSGLDPTVTGCuG* 0 TXNRD2_micMur Microcebus murinus (mouse_lemur) 1 CGASYAQVMQTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_tupBel Tupaia belangeri (tree_shrew) 1 CGASYAQVMQTVGIHPTCSEEVVKLHISKRSGLEPTVTGCuG* 0 TXNRD2_musMus Mus musculus (mouse) 1 CGASYAQVMQTVGIHPTCSEEVVKLHISKRSGLDPTVTGCuG* 0 TXNRD2_ratNor Rattus norvegicus (rat) 1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_cavPor Cavia porcellus (guinea_pig) 1 CGASYAQVMRTVGIHPTCSEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_ochPri Ochotona princeps (pika) 1 CGASYAQVMRTVGIHPTCAEEVAKLRITKRSGLDPTVTGCuG* 0 TXNRD2_canFam Canis familiaris (dog) 1 CGASYAQVMRTLGIHPTCAEEVAKLRITKRSGLDPTVTGCuG* 0 TXNRD2_equCab Equus caballus (horse) 1 CGASYAQVMRTVGIHPTCAEEVTKLRISKRSGLDPTVTGCuG* 0 TXNRD2_myoLuc Myotis lucifugus (microbat) 1 CGASYQQLMRTVGIHPTCAEEVAKLRISKRSGLDPTVTGCuG* 0 TXNRD2_bosTau Bos taurus (cow) 1 CGASFQQVIRTVGIHPTCAEEVAKLRISKRSGLDPTVTGCuG* 0 TXNRD2_oviAri Ovis aries (sheep) 1 CGASYEQVMRTVGIHPTCAEEVAKLRISKRSGLDPTVTGCuG* 0 TXNRD2_susScr Sus scrofa (pig) 1 CGASYTQVMQTVGIHPTCAEEVAKLRISKRSGLDPTVTGCuG* 0 TXNRD2_eriEur Erinaceus europaeus (hedgehog) 1 CGASYAQVMQTVGIHPTCAEEVAKLHISKRSGLDPTVTGCuG* 0 TXNRD2_sorAra Sorex araneus (shrew) 1 CGASYAQLMQTVGIHPTCAEEVAKLHISKRSGQDPTVTGCuG* 0 TXNRD2_dasNov Dasypus novemcinctus (armadillo) 1 CGASYAQMMRTVGIHPTCAEEVVKLRISKRSGLDPTVTGCuG* 0 TXNRD2_loxAfr Loxodonta africana (elephant) 1 CGATYSDLMKTVGIHPTCAEEVTKLKITKRSGLSAVITGCuG* 0 TXNRD2_monDom Monodelphis domestica (opossum) 1 CGATYPQMKKTVGIHPTSAEEVTKLWITKRSGLPATVTSCuG* 0 TXNRD2_macEug Macropus eugenii (wallaby) 1 CGATYSQMMKTVGIHPTCAEEVTKLHITKRSGLDPTVTGCuG* 0 TXNRD2_galGal Gallus gallus (chicken) 1 CGATYSQMMKTVGIHPTCAEEVTKLHITKRSGLDPTVTGCuG* 0 TXNRD2_anoCar Anolis carolinensis (lizard) 1 CGATYHQLMRTVGIHPTCAEEVTKLHITKRSGLDPTVTGCuG* 0 TXNRD2_xenTro Xenopus tropicalis (frog) 1 CGLTYEHLRNTVGIHPTCAEELTKLNITKRSGLDATVTGCuG* 0 TXNRD2_danRer Danio rerio (zebrafish) 1 CGATYSHLLQTVGIHPTCAEEVIKVNITKRSGLDATVTGCuG* 0 TXNRD2_tetNig Tetraodon nigroviridis (pufferfish) 1 CGATYSHLLQTVGIHPTCAEEVIKVNITKRSGLDATVTGCuG* 0 TXNRD2_takRub Takifugu rubripes (fugu) 1 CGATYSHLLQTVGIHPTCAEEVVKVNITKRSGLDPAVTGCuG* 0 TXNRD2_gasAcu Gasterosteus aculeatus (stickleback) 1 CGATYRQLMQTVGIHPTSAEELVKINITKRSGLDPTVTGCuG* 0 TXNRD2_oryLap Oryzias latipes (medaka) 1 CEATYSQLVATVGIHPTCAEDVNKLNITKRSRLDPTVTGCuG* 0 TXNRD2_calMil Callorhinchus milii (elephantfish) 1 CNATYSQLASTVGIHPTCAEDVNKLNITKRSGQDATVTGCuG* 0 TXNRD2_squAca Squalus acanthias (spiny_dogfish) CLRVQIFTMLAVTKRSGADIKQTGCLAVTKRSGADIKQTGCuG* 0 TXNRD2_petMar Petromyzon marinus (lamprey)* SGLTYQQLASSVGIHPTCAEEVVKMGITKRSGLDPTVTGCuG* 0 TXNRD2_braFlo Branchiostoma floridae (amphioxus)* *may not be orthologs: TXNRD2_braFlo has last 3 exons fused though correct phase 0 0 IYHAFYKPLEYYIPERDASQCYIKAVCKRVGDQEIVGLHFLGPNAGEVTQGFAVALRSGLTYQQLASSVGIHPTCAEEVVKMGITKRSGLDPTVTGCuG* 0 not available TXNRD2_ornAna Ornithorhynchus anatinus (platypus)
SEPP1 (SELP): 15 vertebrate sequences
>SEPP1_homSap Homo sapiens (human) chr5 4 exons 10 selenocysteines 0 MWRSLGLALALCLLPSGGTESQDQSSLCKQPPAWSIRDQDPMLNSNGSVTVVALLQASuYLCILQASK 2 1 LEDLRVKLKKEGYSNISYIVVNHQGISSRLKYTHLKNKVSEHIPVYQQEENQTDVWTLLNGSKDDFLIYDR 2 1 CGRLVYHLGLPFSFLTFPYVEEAIKIAYCEKKCGNCSLT 0 0 TLKDEDFCKRVSLATVDKTVETPSPHYHHEHHHNHGHQHLGSSELSENQQPGAPNAPTHPAPPGLHHHHKHKGQHRQGH PENRDMPASEDLQDLQKKLCRKRCINQLLCKLPTDSELAPRSuCCHCRHLIFEKTGSAITuQCKENLPSLCSuQGLRAEEN ITESCQuRLPPAAuQISQQLIPTEASASuRuKNQAKKuEuPSN* 0 >SEPP1_panTro Pan troglodytes (chimp) 0 MWRSLGLALALCLLPSGGTESQDQSSLCKQPPAWSIRDQDPMLNSSGSVTVVALLQASuYLCILQASK 2 1 LEDLRVKLKKEGYSNISYIVVNHQGISSRLKYTHLKNKVSEHIPVYQQEENQTDVWTLLNGSKDDFLIYDR 2 1 CGRLVYHLGLPFSFLTFPYVEEAIKIAYCEKKCGNCSLT 0 0 TLKDEDFCKRVSLATVDKTVETPSPHYHHEHHHNHGHQHLGSSELSENQQPGAPNAPTHPAPPGLHHHHKHKGQH RQGHPENQDMPGSEDLQDLQKKLCRKRCINQLLCKLPKDSELAPRSCCCHCRHLIFEKTGSAITuQCKENLPSLCSu QGLRAEENITESCQuRLPPAAuQISQQLIPTEASTSuCuKNQAKKuEuPSN* 0 >SEPP1_ponPyg Pongo pygmaeus (orang_sumatran) 0 MWRSLGLALALCLLPLGGTESQDQSSLCKQPPAWSIRDQGPMLNSNGSVTVVALLQASuYLCILQASK 2 1 LEDLRVKLKKEGYSNISHIVVNHQGISSRLKYTHLKNKVSEHIPVYQQEENQTDVWTLLNGSKDDFLIYDR 2 1 CGRLVYHLGLPFSFLTFPYVEEAVKIAYCEKKCGYCSLT 0 0 TLKDEDFCKSVSLATVDKTVEAPSPHYHHEHHHNHRHQHLGSSELSENQQPGAPDAPTHPAPPGLHHHHKHKGQH RQGHPENRDMPGSEDIQDLQKKLCRKRCINQLLCKLPKDSELAPRSCCCHCRHLIFEKTGSAITuQCKENLPSLCSu QGLRAEENITESCQuRLPPPAuQISQQLIPTEASTSuRuKNQAKKuEuPSN* 0 >SEPP1_macMul Macaca mulatta (rhesus) 0 MWRSLGLALALCLLPSGGTESQDQSSFCKQPPAWSIRDQDPMLDSNGSVTVVALLQASuYLCILQASK 2 1 LEELRVKLEKEGYSNISYIVVNHQGISSRLKYTHLKNKVSEHIPVYQQEENQTDVWTLLNGSKDDFLIYDR 2 1 CGLLVYHLGLPFSFLTLPYVEEAIKIVYCEKKCGNCSLT 0 0 TLKDEDFCKSVSLATVDETVEAPQAHYHHEHHHNQGHQSSELSENQQPGAPGAPTHPAPPGLHHHHKHKGQHRQGH PESuDMPGSEGLQHLQKKLuRKRCINQLLCKLPKDSELAPRSuCCHCRHLIFEKTGSAITuQCKENLPSLCSuQGLLAEET* 0 >SEPP1_musMus Mus musculus (mouse) 0 MWRSLGLALALCLLPYGGAESQGQSSACYKAPEWYIGDQNPMLNSEGKVTVVALLQASuYLCLLQASR 2 1 LEDLRIKLESQGYFNISYIVVNHQGSPSQLKHSHLKKQVSEHIAVYRQEEDGIDVWTLLNGNKDDFLIYDR 2 1 CGRLVYHLGLPYSFLTFPYVEEAIKIAYCEERCGNCNLT 0 0 SLEDEDFCKTVTSATANKTAEPSEAHSHHKHHNKHGQEHLGSSKPSENQQPGPSETTLPPSGLHHHHRHRGQHRQGH LESuDTTASEGLHLSLAQRKLuRRGCINQLLCKLSKESEAAPSSCCCHCRHLIFEKSGSAIAuQCuENLPSLCSuQGLF AEEKVTESCQCRSPPAAuQNQPMNPMEANPNuSuDNQTRKuKuHSN* 0 >SEPP1_ratNor Rattus norvegicus (rat) 0 MWRSLGLALALCLLPYGGAESQGQSPACKQAPPWNIGDQNPMLNSEGTVTVVALLQASuYLCLLQASR 2 1 LEDLRIKLENQGYFNISYIVVNHQGSPSQLKHAHLKKQVSDHIAVYRQDEHQTDVWTLLNGNKDDFLIYDR 2 1 CGRLVYHLGLPYSFLTFPYVEEAIKIAYCEKRCGNCSFT 0 0 SLEDEAFCKNVSSATASKTTEPSEEHNHHKHHDKHGHEHLGSSKPSENQQPGALDVETSLPPSGLHHHHHHHKHKGQH RQGHLESuDMGASEGLQLSLAQRKLuRRGCINQLLCKLSEESGAATSSCCCHCRHLIFEKSGSAITuQCAENLPSLCSuQ GLFAEEKVIESCQCRSPPAAuHSQHVSPTEASPNuSuNNKTKKuKuNLN* 0 >SEPP1_canFam Canis familiaris (dog) 0 MWRSLGLALALCLLPWGGAESQGQSSFCKQPPAWSIRDQNPMLNSSGSVTVVALLQASuYLCILQASR 2 1 LEDLRVKLEKEGFLNISYVVVNHQGLSSQLKYMYLKNKVSEHIPVYQQEENQTDVWTLLNGKKDDFLIYDR 2 1 CGRLVYHLGLPYSFLTFPYVEEAIKRAYCEEKCGNCSLT 0 0 VLEDEEVCKMVSSGTVESTTEAPQPHPHDHHLHHHHHHHHKHWHRLMPHGNDELSENQQPEEPDVSEHPAPQGLHRHHKHKDHQRQGH PDNuDMPAGSESLQLSVPQNQLuRKGCRNQLLCKLPRDSGLAPSSuCuHuRHLIFEKTGSAITuQCKETLPSLCSuQGLWAEENVIESuQ uRWPPAAuQASQQLRPTEASTNuSuKYKTKMuKuLTY* 0 >SEPP1_equCab Equus caballus (horse) 0 MWRSLGLALALCLLPWGGTESQGQSSFCKQPPAWSIRDQDPMLNSYGSVTVVALLQASuYLCLLQASR 2 1 LEDLRVKLEKEGYSNISYVVVNHQEISARLKYIHLKNKVSEYIAVYQQEENQTDIWTLLNGSKDDFLIYDR 2 1 CGRLVYHLGLPYSFLTFPYVEEAIKIAYCEKKCGNCSLM 0 0 TLEDEDFCKTLSLATVEKTTEDSQPHHHHHHQHHHKHGHQHVGNSQLSENQQPEATGAPEHPPPPGLHHHHKHKGQQ RRGHPENuDMPGGESLQLSLPQKKLuQKGCINQLLCKMPKDSKLAPSSuCuHCRHLIFENTRSAITuQCTENLPSLCSuQ GLWAEENVIESCQuRLPPAAuQPS-QQLKPTEASTNuSuKYKAAMuKuPSN* 0 >SEPP1_bosTau Bos taurus (cow) 12 sel 0 MWRGLGLALALCLLLTGGTESQGQSSYCKQPPPWSIKDQDPMLNSYGSVTVVALLQASuYLCILQASR 2 1 LEDLRVKLEKEGYSNISYVVVNHQGISSRLKYVHLKNKVSEHIPVYQQEENQPDVWTLLNGNKDDFLIYDR 2 1 CGRLVYHLGLPYSFLTFTYVEDSIKTVYCEDKCGNCSLS 0 0 RPQDEDVCKNVFLATKEKTAEASQRHHHPHPHSHPHPHPHPHPHPHPHPHHGHQLHENAHLSESPKPDTPDTPENPPTSGLHHHHHRHKGPQRQGH SDNCDTPLGSESLQPSLPQKKLuRKRCINQLLuQFPKYSESALSSCCCHCRHLVFEKTGSAITuQCTEKLPSLCSuQGLLAEENVIESuQuRLPPAAu QAAGQQLNPTEASTKuSuKNKAKMuKuPSN* 0 >SEPP1_choHof Choloepus hoffmanni (sloth) 0 MWKSLGLALALCLLPWGGTESQEQSPFCKRPPAWTIRGQDPMLNSNGSVTVVALLQASuYLCILQASR 2 1 LEDLRVKLEKEGYSNISYIIVNHHEITSQLKYIHLKNKVSEHISVYQQKENERDVWALLNGKKDDFFIYDR 2 1 CGHLVYHFRLPYSFLTFPYVEEGIKIAYCENKCGNCPPT 0 0 FINLFQALKDEDVCENVSLAITEKTTEVSQPHPHHNHHHKHPGNSQLPENQQTGAPDAPQQPPPPGLHHHHKHQGQQREGHPDNCDMPESEGLQLSPHQKKLuRKRCINQLFCKVSKDSE LAPRSCCuHCRHLVFEKTGSAITCQCKENLPSLCSuQGLWAEENVIESuQuRLPPAAuQISQQLLKPTEVSTNuEuNNKADMuKuPAN* 0 >SEPP1_ornAna Ornithorhynchus anatinus (platypus) 0 MWQGLGLALALCLLPGGGAESQSASSHCKEAPRWQIRDQDPMLNSLGTVTVVALLQASuYLCILQASR 2 1 LEDLRVKLENEGYSNISYIIVNHQGMPSQLNHKTLKEKVSEHIPVYQQDEKQTDVWSTLKGNKDDFLIYDR 2 1 CGRLVYHLSLPYTFLSFSYVEDSIKTTYCEQNCGNCSYT 0 0 MPEAEEFCTNTSSAAKEKATEAPLPHNDRPHHHHHHHHHHGHKPHPSGTEQAPADPDGPLRSPAPQGLHKRLRPAGQPRQGQ GGSREAAEGRGEELPSPRKKAuRKGNASCQNQLLuDWHKRSGPAPSSuCuHCRHLLFGSKATATALuQCRDALPALCSuQGRQS GEDVIESuQuRSPLPAuPPAAQLPSPSPTDPNAAuKuENTAGMuKuPTR* 0 >SEPP1_galGal Gallus gallus (chicken) reqd traces tga-sel taa-stop 0 MWAGLGLALVLCLLPGGGTESQRCQEPPEWHIGEESPMLNARGSVAVVALLQASuYLCLLQASR 2 1 LEDLRVKLENEGLVNISYVVVNHQSPHSQKKFHLLQESVSDHITVYQQDDHQADVWTTLNGNKDDFLIYDR 2 1 CGRLVYHLGLPYSFLSFQYVEEAIKIAYCENNCGNCSYT 0 0 EPDIDNICENITKKEDENLAGIEPEPEPSGQHSHHHHQLHRHRHHHHHREGGRHSKNQNHQAPSESQRRHPHNGRRHRVFNHNRHDQ IGSHEQVETLPPGEGVENLPRVTKLuKKGKTICKNQLTuNWQTASDSTTSSuCCHCRHLLFEELGNSITuQ CRGALPNSCRuHGQLLAEDITESuQuRLLTAAuESAAGGGSETSDTuLuERAGNuAuKTN* 0 >SEPP1_xenTro Xenopus tropicalis (frog) frag 0 MWKGFALALALCLLPWGGAESQGHRKRCKEPPEWSIGDQNPMMQSAGKVTVVALLQASuSLCLLQASR 2 1 LEDLRLKLEKEKLVGISYVVVNHQGRQSRAKYDLLKSKVSEHIPVYQQEEYQPDVWSLLKGDKDDFFVYDR 2 1 CGRLVQHLELPYSLLHFPYVEEAIRIAYCEDKCGECEHK 0 0 ILDADVCKKPEEQQEQDKPVEEKVERPRHHRNHHRHHRPKHSGHRHRHHHSEDGQVAELDVLRSSAQANNRAGSQNGQGQ SVVQVVPQSEVLFVPQREADIPVLARQPuKKAKSuKKQYLuDWREEGGKAFNSuCuHuRQLSFEVTENEVAuHuQEALPSSC SuQELLSDSLPESuQuRLSAAAuHSESTGLPETKLESEPNAPuAuPQEAENuQuKELuRFLM* 0 >SEPP1_xenTro Xenopus laevis (frog) 0 MWKGFALALALCLLPWGGAESQGHRSRCKQPPDWSIGDQNPMIQSAGKVTVVALLQASuYLCLLQASR 2 1 LEDLRLKLEKEKLVGISYVVVNHQGRHSRAKYDLLKSKVSEHIPVYQQEENQPDVWSLLKGDKDDFFIYDR 2 1 CGRLVQHLELPYSLLHFPYVEEAVRIAYCGDKCGECEHK 0 0 IPDADVCKKPEEQPELEKPVEEKVERPRPHRNHHRHHRPKHSGHRHRHHNNEGGQAAEVDAFQTNNRAGSHNGQGQSVVPQSEVVFVPQ READVPVLALQPuKKAKSuKKQYLuEWREDAGKAFNSuCuHuRQLSFEIAQNEVTuRCQEALPASuQuQELLSDSLSESuQuRLSAAAuHSHS TGLPELDTESETNAPuAuPQEAENuQuKELuRFLM* 0 >SEPP1_danRer Danio rerio (zebrafish) 17 sel 0 MWKALSLTLALCLLVGCSAESETEGARCKLPPEWKVGDVEPMKNALGQVTVVAYLQASuLFCLEQASK 2 1 LNDLLLKLENQGYPNIAYMVVNNREERSQRLHHLLQERLLNITLYAQDLSQPDAWQAVNAEKDDILVYDR 2 1 CGRLTYHLSLPYTILSHPHVEEAIKHTYCDRICGECSLES 0 0 SAQLEECKKATEEVNKPVEEEPRQDHGHHEHGHHEHQGEAERHRHGHHHPHHHHHHHRGQQQVDVDQQVLSQVDFGQVAVETPMMKRPuAKHSRuKVQYSuQQGADSPVASu CuHuRQLFGGEGNGRVAGLuHCDEPLPASuPuQGLKEQDNHIKETuQuRPAPPAEuELSQPTuVuPAGDATuGuRKK* 0 >SEPP1_torCal Torpedo californica (electric_ray) 1 tga-sel taa-stop 0 MQQGLGVALALCLLLVGQAESQGQCDKPSEWTIGDKNPMKDSLGKVTVLALFQASuHFCLEQAGS 2 1 LKNLKRKLDQDGLEHISYMVVNHQGNDSRKRYQELKRNVQGKLPVYQQEIDQPDVWNLLQGQKDDFFIYDR 2 1 CGQQTFHLELPFTILSQMYVEHAIKQTYCQEICTNCS 0 0 LAELPFGMQQHKMQKSQRRWEEDGSPVPKHRGSTMGHKTTPPQSPR*
SEPP2 (SELPB): 16 vertebrate sequences
>SEPP2_monDom Monodelphis domestica (opossum) chr2:9,451,774-9,455,977 1 selenocys tga stop taa 0 MPPPGLSLAVLLGLLGASLAFENRTRFCQPAPPWQVGGGRAPMEEALGNVTVVALLKASuHFCLKQAAS 2 1 MGSLQERLARMGAPDVRFIIVNEKSPQSRALHGELELHAPPGVPVYGQPELGPDIWSILGGAKDDFLIYDR 2 1 CGRLTFHIRLPFSFLHFPYVESAIRFTHRQDSCGNCSFYPAQ 0 0 VNSTDERKGESKHSPGLEGEGQEPLGEKPDSRGLTLGSSAPTAHAHDPVHGGMEKPSPSLPSPALEPSLHEGEAPG* 0 >SEPP2_macEug Macropus eugenii (wallaby) trace archive tga stop taa 0 MGLPGLAAALLLGLVGATLASENGTRICQPAPKWQVDRGASPMEEALGQVTVVALLKASuHFCLKQAAS 2 1 IGNLQERLARVGATGVRFIIINEKSPLSQALYGELELHAPSGVLVYNQQGPGPDIWSILGGAKDDFLIYDR 2 1 CGRLTFHLPLPLSFLYFPYVESAIRFTHQQDHCGNCSFYPAQ 0 0 VNNTDKGKAESLTQSPRLEGEGQESLAEGPSTHGPTLWPSTPHPAHARGPVHSPGSKRLSPSLPPPSLEWPPHEEANK* 0 >SEPP2_ornAna Ornithorhynchus anatinus (platypus) synteny last exon uncertain 0 MAGSGLLGPALTLATLLAAAGALPDLENGTRICQPAPRWTVNGVAPMEGTEGQVIVVALLKASuHFCLKQAAR 2 1 LAGLRERLAGHGAGNVSFLIVNQRDPTAQLLHTELERHAPPGVPVYAQDGPDPDVWSVLGGDKDDFFVYDR 2 1 CGRLTFHIQLPFSFLHFPYVEAAVRFTHRRDFCGNCSYYFPQVGT 0 0 VNDTTTQESELEKSPGAPGEEPEGSPVREPDRPQSQDPTGPFSGVLLQGKENKIIPWKTPLQAAPRKPSHPPGAHD* 0 >SEPP1_tacAcu Tachyglossus aculeatus (echidna) EUEMSW408ERBZB length=227 run=R_2007_08_22_12_11_10_ 59% 1 VPVYAQDGPDPDVWSILGGGKDDFLVYDR 2 1 CGRLTFHIRLPFSFLHFPYVEAAVLFSHRHDFCGNCS >SEPP2_galGal Gallus gallus (chicken) taa stop synteny Zswim5 HPDL MUTYH 0 MGSLLLALASCLGLAVASEGATNGSRLCHEAPAWRINGSSPMEGAAGQVTVVALLKASuHFCLLQARS 2 1 LGALRERLGQQGVSDVRYVIVNEQAPLSRAMFGELQRHAPPGVPVLQQQPHEPDVWQLLGGDKDDFLVYDR 2 1 CGRLAFHIQLPYSFLHLPYVESAIRFTHRKDFCGNCSLYPNSTQE 0 0 ANSTMEVPATLTPLPKQEEKESETPAHHQPNHLHPHHRAVGNGTAPEPSGDHRPAHAHHHHGAHGKLHPKGQTPEGRDP* 0 >SEPP2_taeGut Taeniopygia guttata (finch) 0 MGLLVLALATWLGLGLASASEETANSSRICQEAPAWTINGSSPMEGAAGQVTVVALLKASuQFCLRQAHS 2 1 LGGLRERLARQGMADVRYMIVNEKAPLSRAMLPELQRHAPPGVPVFQPEREDPDVWQVLGGDKDDFLVYDR 2 1 CGRLAFHIQLPFSFLHFPYVEAAIRSSHIKDFCGNCSLYPNTTQE 0 0 ANSTMEGPATPSSLPEHEGMESETPVHQHKPLHPHRHHEVNSERDTNPSEDHKPATHAHHHHGDHGQLHHKGRKQKEGDEH* 0 >SEPP2_anoCar Anolis carolinensis (lizard) scaffold_4:5,993,109-5,994,846 0 MDYSLATRILLLGLVVISATQAEVTENKTRICQPAPLWKINGTAPMAGMEGQVTVVALLKASuPFCLKQAAK 2 1 IGGLQKKLSNEGVANVSFLIVNEKTPLSRAMYWELKRNAPQGIPVYQQQILEPDVWQILDGDKDDFLIYDR 2 1 CSRLTFHIQLPYSFLHLPYVEAAVHYTHRKDYCGNCSRYYSE 0 0 * 0 >SEPP2_xenTro Xenopus tropicalis (frog) NM_001006907 misannotated, no selenocysteine 0 MHNLALTVSILMGLLGQVSSSEQTNSSICKPPPKWSIEGEVPMAEALGKVTVVALLQASCGFCLVQAAR 2 1 MGPLRYKLSLQGMTDIKYMIVNDQSLHSANMFPELKRWAPEGIPVYQQTPGQDDVWELLDGNKDDFLIYDR 2 1 CGRLTFHVRLPLSFLHFPYVEAAILSTYNESFCGNCSFTSNSTLIPM 0 0 NGTTVSPSGDDSSSPLQNKDEPVNKEPSPTLEKHNDQRKLDSELRLHDHSQHHPINSHKRQENQNNHPRNLIKNGKQN* 0 >SEPP2_danRer Danio rerio (zebrafish) 3 possible tga-sel taa-stop 0 MQALWPLLLSALPALLGASSLFVEKESNGSRICKPAPQWEIDGKTPMKELLGNVVVVALLKASuHFCLTQAAR 2 1 LGDLRDKLANGGLTNISFMVVNEQDSQSRAMYWELKRRTAQDIPVYQQSPLQNDVWEILEGDKDDFLVYDR 2 1 CGYLTFHIVLPFSFLHYPYIEAAIRATYHKNMCNCS 0 0 LNANFSISESSDSTKNEPAGENNQRPNSTEPVTAAHHHHHQQHEPHHHHNPYPNNHKKSGDSDVTGKPKEPTHHSHQEHVHNHRFYTVFSFIILFYIMFFFFVNIWSCSCVRTSYLPQRG VFGCLQCIFVLNEVYTLNLRQGSVCGCuTQESFIYTKLLISPLFGKMuLCuTKWLLGLLC* 0 >SEPP2_tetNig Tetraodon nigroviridis (pufferfish) from cDNA, genome misassembled 0 MSSPWLLWLQVALTGLLWASQGQSATSRICKAAPRWEVGGQAPMEALVGRVVVVALLKASuQFCHIQASK 2 1 IGPLREKLSRRNVTEVSFVIVNEQEPVSRALYWQLRRRAPPGVPVYQQAPLQDDVWEALDGDKDDFLIYDR 2 1 CGQLTFHIGLPYSSLRYTYVEAAVIATHQGNICNCS 0 0 ANFTSLSISNSSGSGGMPSQTNQTVTAETDGPHTTHHHPHPHRHHHHHQHLSPEQPTPTAMPGQATPTPA* >SEPP2_gasAcu Gasterosteus aculeatus (stickleback) tga-sel taa-stop 0 MQALWPLLYAALPGLLWASHVSLLVEGDNDASRICKPAPHWTIKERAPMQELLGNVVVVALLKASuQFCLTQASK 2 1 IGGLRDKLNRSNLTDVSFMIVNEREPHSRAMYWELKRRAPPGVPVYQQAALQSDVWEALDGDKDDFLVYDR 2 1 CGLLTFHIVLPYSFLHNVYVEAAIRATYLKNICNCT 0 0 VDSVVSSLNNSVMNNETDFNVSQTNATPRIQPDTNDPEGAGTPPPPTHHHSHHHHHHHQHHHPHHPHLNQPLHDTSHPLSVHHHHHPHHHHHQQQQPGNHSGTAVDH* 0 >SEPP2_oryLat Oryzias latipes (medaka) 0 MLRLCSALPALLWASLSVLSAEGDSNASKICKPAPYWDIEGHVPMQEHLGNVVVVALLKATuEFCLTQASK 2 1 IGNLRDKLNRNNITEVSFMIVNELEALSQTMHWKLKKKAPTGVPVYQQSSLQKDVWEILDGDKDDFLIYDR 2 1 CGLLTFHIVLPNSFLQNADVENAITATYTQDICNCS 0 0 GNSTLSGGGNNFTRNSSQSHSGVQRPADEPENVTTTLEASPNDSRHEHAHHHHPHHQHLHHHHPHHQHLHHHHPHHQHLHHHHPHLQQHSKHQNDDSY* 0 >SEPP2_cypCar Cyprinus carpio (carp) 0 MQALWALWLSAFPALLWASPLFLERESNASRICKPAPKWEIDGEIPMKNLLGNVVVVALLKASuQFCLTQAAR 2 1 LGKLRDKLALGKLTDISFLVVNEQDAQSRAMYWELKRRTTEGIPVYQQSPLQNDVWDILEGDKDDFLVYDR 2 1 CGHLTFHIVLPFSFLHYPYIEAAIRATYHKNICNCS 0 0 LHANSSISEGFKNNTNNANKQTEVKNQKTNNTEPVTVEHHHHRHEHHHQHVHYHYHQHVHHHHQNKNNSTGDSDVTSVPKEPIQHSHQEHS >SEPP2_oncMyk Oncorhynchus mykiss (trout) 0 MQGLLTLWLCAALPGLLCASPLLVEGDNDASKICKPAPHWEIKGHEAPMKELLGNVVVLALLKASuHFCRTQASK 2 1 LGGLRDKLLRSNLTDVSFLIVNEREAQSRAMYWELKRRAPPGIPVYQQAPLQDDVWEALDGDKDDFLVYDR 2 1 CGRLTFHIVLPYSFLHYPYIEAAVRATYHKDICGNCT 0 0 VDSNTTSSAGWNSTRRNETLSSSQMRVNETDSTVRSIDVDTVSNPVPSDGPQMSAEGGW* 0 >SEPP2_squAca Squalus acanthias (spiny_dogfish) 0 MGLEMPLLMILLGLTITVAVAKNETRICQVAPHWGIANQSPIEEQFGHVVVVALLKASuQFCLTQAAK 2 1 LGSLRDKLARQGLKDVRYMIVNEKTPRSRAMLWELKRHAPKDVPVYQQSPFQPDVWRILQGNKDDFLVYDR 2 1 CGKLTFHIVLPYSYLHFRYIEAAIRATYNKDICGNCS 0 0 VSDITLERERNITTTYKVTQSNTTSL >SEPP2_leuEri Leucoraja erinacea (skate) potential 2 terminal sel uRYFKIGEuWKKFPSQIRAKCQY* 0 MGLQRSLLIILVGAAITLAAANNDTRICQVAPHWEIGNQSPMEQLSGQLVVVALLKASuQFCLTQAAK 2 1 LGILRDKLSLQGLKNIHYIIVNEKTLESRAMFWKLKLKTPKNITVYQQSAFQPDVWRILRGNKDDFLIYDR 2 1 CGKLTFHITSPYSYLNFRYVEAAIMATYNTDYCGNCM 0 0 GSSTTLEATSYISTGPNWEYSQHHSRSLK* 0 >SEPP2_calMil Callorhinchus milii (elephantfish) 1 LGSLQDKLARQGLKDVHYMIVNEKAPESRAMLWELKRHVPNNVSVYQQSPIQPDVWHSLQGGKDDFLIYDR 2 1 CGRLTFHVVLPYSSLQYPYIEAAIRATHKRDICGECT 0
SELU1: 13 vertebrate sequences
>SELU1_homSap Homo sapiens (human) chr10 processed pseudogenes chr8 and chr12 0 MSFLQDPSFFTMGMWSIGAGALGAAALALLLANTDVFLSKPQKAALEYLEDIDLKTLEK 1 2 EPRTFKAKELWEKNGAVIMAVRRPGcFLCREE 0 0 AADLSSLKSMLDQLGVPLYAVVKEHIRTEVKDFQPYFKGEIFLDEK 0 0 KKFYGPQRRKMMFMGFIRLGVWYNFFRAWNGGFSGNLEGEGFILGGVFVVGSGKQ 0 0 GILLEHRENEFGDKVNLLSVLEAAKMIKPQTLASEKK* 0 >SELU1_monDom Monodelphis domestica (opossum) tgt-cys 0 MSFLDLNFFSMSMWSLGAGALGAAALSLILANTDLFLTKSVDATLEFLEEIQLKTLDN 1 2 ESPKTFKARELWEHRGAVIMAVRRPGCFLCREV 0 0 AADLSALKPQLDLLGVPLYAVVKEKIGSEVENFQPYFKGKIFLDER 0 0 KKFYGPQKRKMMFMGFVRLGVWQNFFRARSKGFSGNLEGEGFVLGGVYVIGPGKQ 0 0 GILLEHREKEFGDKVNPASVLEAAKKIKPHTSTSEGK* 0 >SELU1_macEug Macropus eugenii (wallaby) EX196548 full 0 MSFLDLSFLSMGMWSLGAGALGAAVLSLILANTDVFLTKSVTATLEFLEDIELKTLDN 1 2 KTFKARELWEHRGAVIMAVRRPGCFLCREE 0 0 AADLSALKPQLDQLGIPLYAVVKEKIGSEVEDFQPYFKGKIFLDER 0 0 KKFYGPQKRKMMFMGFVRLGVWQNFFRARSKGFSGNLEGEGFILGGVYVIGPRKQ 0 0 GILLDHREKELGDKVNPASVLEACKKIKLHA* 0 >SELU1_triVul Trichosurus vulpecula (opossum) EC360881 0 MSFLDLSFFSMGMWSLGAGALGAAVLSLILANTNLFLTKSVTATLEFLEEIELKTLDN 1 2 ESPKTFKARELWEHRGAVIMAVRRPGCFLCREE 0 0 AAELSALKPQLDQLGIPLYAVVKEKIGSEVENFQPYFKGKIFLDER 0 0 KKFYGPQKRKMMFMGFVRLGVWQNFFRARSKGFSGNLEGEGFILGGVYVIGPGKQ 0 0 GILLEHREKEFGDKVDPASVLEAA * 0 >SELU1_ornAna Ornithorhynchus anatinus (platypus) taa early stop full 0 MPLPPDLGLFNLGMWSVGVGALGAAAVGLLLANTDLLLTKPEKATLEYLEDTELKTLGK 1 2 EPRTFKARELWQRNGAVIMAVRRPGuFLCREE 0 0 AAELSSLKPQLDRLGVPLYAVVKEKIGTEVEDFQPYFKGEIFLDER 0 0 KKFYGPHKRKMLFLGFIRLGVWQNFLRARNRGFSGNLEGEGLILGGVYVLGAGKQ 0 0 GILLEHREREFGDKVSPASVLEAAQRIKPQPL* 0 >SELU1_tacAcu Tachyglossus aculeatus (echidna) 454:EUEMSW405C31QQ (74%) tSASEKK terminus? frag 0 1 2 EPRTFKARELWQRNGAVIMAVRRPGuFLCREE 0 0 AAELSSLKPQLDQLGVPLYAVVKENIGTEVEDFQPYFKGEIFLDER 0 0 KRFYGPHKRKMLFLGLIRLGVWQNFIRARNKGFPPVTWEGEG 0 0 GVLLEHREREFGDKVSPASVLEAAQKIKPQ* 0 >SELU1_galGal Gallus gallus (chicken) 0 MSFLPDFGIFTMGMWSVGLGAVGAAITGIVLANTDLFLSKPEKATLEFLEAIELKTLGS 1 2 EPRTFKASELWKKNGAVIMAVRRPGuFLCREE 0 0 ASELSSLKPQLSKLGVPLYAVVKEKIGTEVEDFQHYFQGEIFLDEK 0 0 RSFYGPRKRKMMLSGFFRXGVWQNFFRAWKNGYSGNLEGEGFTLGGVYVIGAGRQ 0 0 GVLLEHREKEFGDKVSLPSVLEAAEKIKPQAS* 0 >SELU1_taeGut Taeniopygia guttata (finch) 0 msflpdfgiFTMGMWSVGLGAIGAAVTGIVLANTDLFLSKPEKATLEFLEEIELKTLGS 1 2 EKRTFKAGELWKQNGAVIMAVRRPGuFLCREE 0 0 ASELSSLKPQLSKLGVPLYAVVKENIGTEVEDFQHYFKGEIFLDEK 0 0 KGFYGPRRRKMMLSGFFRLGVWQNFVRAWRSGYSGNLEGEGFTLGGVYVIGAGRQ 0 0 GVLLEHREKEFGDKVSLPSVLEAAEKIKPQAS* 0 >SELU1_anoCar Anolis carolinensis (lizard) 0 MWTIGLGAIGAAVTGIILANTDLFLSKAEQASLDFLEAIDLKTLGE 1 2 NQRTFKAEELWKKNGAVIMAVRRPGuFLCREV 0 0 AAELSSLKPQLDKLGVPLYAVVKENLGTEVMDFQPYFKGEIFLDEK 0. 0 KQFYGPQKRKMLFMGFIRCSVWRNFFRAWKSGYTGNIDGEGFVLGGVFVVGPGKQ 0 0 GVLLEHREKEFGDKVSLDAVLEAVKNIQPQPSEKDK* 0 >SELU1_takRub Takifugu rubripes (fugu) 0 MGLLAKLLAAVGGFVTAVMNSVTDAFLTPPLRATLEHLEETDLKTLSG 1 2 ALVIRLIPTRTETKTFKAKSLWENSGAVVMAVRRPGuFLCRE 0 0 EAAELSSLKPRLDQLGVPLYAVVKEDVGTEIQNFRPYFQGEIFLDEK 0 0 RRFYGPRERKMGLLGFLRVGVWMNGLRAFRSGFMGNVLGEGFVLGGVFVIGREQQ 0 0 GILLEHREREFGDKVNIEDVIQAVDRIAQELMPVTQN* 0 >SELU1_calMil Callorhinchus milii (elephantfish) frag 2 ENRTFRASELWAGRGAVIMAVRRPGuFLCRE 0 0 AAALSSLRPSLAQLGVPL 0 GHLLEHREKEFGDAVNLTAVMEAAGKISPRQSAE* 0 >SELU1_squAca Squalus acanthias (spiny_dogfish) also selenocysteine 0 MVVVVEDFHMGLWTLGLGALGAAITGVILANTDLLLPKAETASLAYLSGAELRTLDR 1 2 EERTLKAGDLWSRSGAVIMVVRRPGuFLCREE 0 0 AAEISSLRPQLDELGVPLYGVIKENINNELKNFQPFFKGEIFLDVE 0 0 MRFYGPKPRTMGLMGFMRLGVWKNFVRAWQKGFSGNTDGEGFILgGVFVIGAGQQ 0 0 GVLLEHREKEFGDVVNISSVLEARRKIETQRTEP* 0 >SELU1_braFlo Branchiostoma floridae (amphioxus) 8.9 m traces somewhat different intronation 0 MGWRLLVA 00 TMGWVVNAVGVAVVGMVAVNLNFWLPKHIPAQLVHIA 1 2 DAALEELDGNKKKLQAQELWKNHGAVIMAVRRPGuQLCREE 0 0 ALGLSSLKPQLDRAGVPLYAVVHERKGVPEFQPYFQGKVFLDLE 0 0 RRFYGPHERWMSLAGLLRVNFWLNIRRVEKKVEGNYEGEGRLLGGVFVVGSGNQGILMQHAEQEFGHHANLTDIMKAVGEIQHINAKSKI* 0
SELU2: 1 vertebrate sequence
>SELU2_homSap Homo sapiens (human) 7 exons chr1p36.32 36% id NM_152371 0 MSTVDLARVGACILKHAVTGE 0 0 AVELRSLWREHACVVAGLRRFGCVVCRWIAQDLSSLAGLLDQHGVRLVGVGPEALGLQEFLDGDYFAG 1 2 ELYLDESKQLYKELGFKR 2 1 YNSLSILPAALGKPVRDVAAK 0 0 AKAVGIQGNLSGDLLQSGGLLVVSK 1 2 GGDKVLLHFVQKSPGDYVPKEHILQVLGISAEVCASDPPQ 0 0 CDREV* 0 >SELU2_strPur Strongylocentrotus purpuratus (sea_urchin) 0 MAFDLSKIASNLVKNFATGE 0 0 MVAISSLWEKQACVIHFMRRFGuSACRLGASELDSLRPQLDEADVRLVGIGLEDLGAQEFLDGGFWKG 1 2 DLFIDQQQKSYQGLGFKKYSLMSIMKAVFSAKTRAALSR 0 0 ASDKKIKANLKGDKLQIGGTLIISK 1 2 GGEKVLVDFKQEAPGDHIPLQTVLDAFGIKGETPKTEGAEGGTVVCNDDVCQMK* 0
SELU3: 3 eukaryote sequences
>SELU3_homSap Homo sapiens (human) 6 exons chr9q22.32 25% id processed pseudogene chrX
0 MAAPAPVTRQVSGAAALVPAPSGPDSGQPLAAAVAELPVLDARGQRVPFGALFRERRAVVVFVR 0
0 HFLCYICKEYVEDLAKIPRSFLQ 0
0 EANVTLIVIGQSSYHHIE 0
0 PFCKLTGYSHEIYVDPEREIYKRLGMKRGEEIASS 1
2 GQSPHIKSNLLSGSLQSLWRAVTGPLFDFQGDPAQQGGTLILGP 1
2 GNNIHFIHRDRNRLDHKPINSVLQLVGVQHVNFTNRPSVIHV* 0
>SELU3_petMar Petromyzon marinus (lamprey) cDNA EE741479 ti|1430987375 tga-sel taa-stop frag
VRPDPYIYKALELKMGEDVDKIY 1
2 KSPHVHCGLWWGVARGLWRAMWSESFDFQGDPKQQGGALVLGP 1
2 GGRVLFSHRDEAVLDHTPINRLLAVAGIPPVDFTHKHMVKHVuPRPLTPTG* 0
>SELU3_monBre Monosiga brevicollis (choanoflagellate) tga-sel
MATGAGVGASWEDVAKMTVYDHQQEPCSLGSLCSPGQMTAIGTLARDNSARVPPPLITHHATLPPRVCRLVFVR
HFLuFVCKDYVTDLARVPDEHWA
GARVVVIGCANPVFID
RFAADTGFPASNVYCDPKREVYQALQLPSVLAAINA
SKSIHIHSSALGGTMNSIYRGLRAMRAQGNVKQQGGAFVVNG
QGQLIYAHRDQRPEDHCPINTLLEQVGGRPFF*
SELV: 19 placental mammal sequences
>SELV_homSap Homo sapiens (human) chr 19 uc002oly.1 66 pro + 32 thr +21 ser first exon 0 MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTLTPVLTPSPAGTSPLVLTPAPAQIPTLVPTPALARIPRLVPPPAPAWIPTPVPT PVPVRNPTPVPTPARTLTPPVRVPAPAPAQLLAGIRAALPVLDSYLAPALPLDPPPEPAPELPLLPEEDPEPAPSLKLIPSVSSEAGPAPGPLPTRTPL AANSPGPTLDFTFRADPSAIGLADPPIPSPVPSPILGTIPSAISLQNCTETFPSSSENFALDKRVLIRVTYC 2 1 GLuSYSLR 0 0 YILLKKSLEQQFPNHLLF 0 0 EEDRAAQATGEFEVFVNGRLVHSKK 0 0 RGDGFVNESRLQKIVSVIDEEIKKR* 0 >SELV_panTro Pan troglodytes (chimp) essentially identical to humna MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTLTPVLTPSPAGTPPLVLTPAPAQIPTLVPTPALARIPRLVPPPAPAWIPTPVPTPV PVRNPTPVPTPARTLTPPVRVPAPAPAQLLAGIRAALPVLDSYLAPALPLDPPPEPAPELPLLPEEDPEPAPSLKLIPSVSSEAGPAPGPLPTRTPLAANS PGPTLDFTFRADPSATGLADPPIPSPVPSPILGTIPSAISLQNSTETFPSSSENFALDKRVLIRVTYC 2 1 GLuSYSLR 0 0 YILLKKSLEQQFPNHLLF 0 0 EEDRAAQATGEFEVFVNGRLVHSKK 0 0 RGDGFVNESRLQKIVSVIDEEIKKR* 0 >SELV_ponPyg Pongo abelii (orang_borneo) 0 MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTLTPVLTPSPAGTPPLVLTPAPAQIPTLVPTPALARIPRLVPPPAPAWIPTPVPTPVPVRNPTPVPTPARTLTTPV RVPAPAPAPPPAQVLAGIRAALPILDSYLAPALRLHPPPEPAPELPLSPEEDPEPAPSLKLIPSVSSEAGPAPGPLPARTPQAANSPGPTLDFTFRADLSATGLADPPIPSPVPSPILGT TSSAISLQNSTENFASSSENFALDKRVLIRVTYc 2 1 GLuSYSLR 0 0 YILLKKSLEQQFPNHLLF 0 0 EEDRAAQATGEFEVFVNGRLVHSKK 0 0 RGDGFVNESTLQKIVSVIDEEIKKR* 0 >SELV_macMul Macaca mulatta (rhesus) distal seq from M. nemestrina transcript 0 MNNQARTPAPSSARTSTSVRASTPTRTPTPLRTPTPVRTRTPIRTQTPVLTPSPARLPALVQPPAPAHIPTQVPTPALARIPGLVPPPARAWIPTPVPTPARVRNPTPVPTPARTLTPPV RVPAPAPDPAQVLAGIRTALPALDSYPAPALSLDSPPEPAQEPPLSPEEDPEPAPSLKLIPSVSSEAGPALGPLPAHTPLAAKSSGPTLDFTFRADPSATGLADPHIPSPVPAPILGTIP SALSLQNFTETLVSTSENFALDKRVLIRVTYC 2 1 GLuSYSLR 0 0 YILLKNSLEQQFPSHLLF 0 0 EEDRAAQATGEFEVFVNGRLVHSKK 0 >SELV_calJac Callithrix jacchus (marmoset) 0 MNNQARTPAPSSARTSTSVRASTPTRTPTPVRTPTAVRIRTPIRTLTPSLAGTPALVPTPTPARISRLVPTPAPARTPTPIPTLVRTLTPVPLPAPARIPAPAPAPAPAPAPAPSPALVP AGIRATLPVLDSYPALALPWDPPPEPVPEPLVSVSSEEDPEPAPSLKLVPSVSGETGPAPGPLPACTPLATNPPEPTLDFTADSSATELAVPTIPGSVPAPILGTIPLAASLLNSTESFL SASENFALDKRVPIRVTYc 2 1 GLuSYSLR 0 0 YILLKKSLEQRFPDCLLF 0 0 EEDRAAQATGDFEVFVDGRLVHSKK 0 0 RGDGFVDEASLQKIVSVIDEEIKKR* 0 >SELV_otoGar Otolemur garnettii (bushbaby) 0 MNSQARASVHPSRTSTAVRASIPARVHPRARTTPVQPRTLITQDRIPAPVRVPP 1 GLuSYGLQ 0 0 YILLRQNLEHHFPNRLLF 0 0 EEGRAAQATGEFEVFVDGKLVHSKK 0 0 NGDGFVDEIRLQKIVNIIDEEIKKRQ 0 >SELV_musMus Mus musculus (mouse) AV279316 58 Pro, 25 Thr, 25 Ser syntenic chr 7 46% hsa! MNNKARVPAPSSVRANTPARTPAPIRTATPVRAPNPAHNSTPVRTSIRVRAPAQVPNPVPIRFPTPAPVPAPTLTPAPTPAPVRHAAPVRTPAPVRAPNLGRV FPKISPGRFFPSLASPTAQPLSSRAASALLKDPTLAQNQKPSIHSLAEAIQGPLPVLTPSSSKTQGSIPDTASPIDSLASTAMASSTLGPIPGPNPTLEFLAS PLKETPGLGKLSTISPAPSFGSTKEIPSTSEDVPTPNRILIRVMYC 2 1 GLuSYGLR 0 0 YIILKRTLEHQFPNLLEF 0 0 EEERATQVTGEFEVFVDGKLIHSKK 0 0 KGDGFVDESGLKKLVGAIDEEIKKR* 0 >SELV_ratNor Rattus norvegicus (rat) chr 1 synteny 83% mmu MNNKARNPAPSSVRANTPSRTPTPVRTATPVRASTPAHNRSPVRTSIRVRTPANPVPIRFPTPAPAPAPTPTPAPTPAPVPAAAPVRTPAPVRASIQGRSFPTIS PVRFLRNLALPAAQPLSSGGAGSLSKDLTLAQKQKPSIHSLAEAIQGPFPVLTPSASSETHGSIPDPAPPTDSLASTAMASSTLDPIPGPKPTLEFLASPLKETP DLGKLSTISPAQNFVSTKEVPSTSEDVPTANRILIRVMYC 2 1 GLuSYGLR 0 0 YILLKKTLEHQFPNLLEF 0 0 EEERATQVTGEFEVFVDGKLIHSKK 0 0 KGDGFVDETSLKKLVGAIDEEIKKR* 0 >SELV_cavPor Cavia porcellus (guinea_pig) 1 GLuSYGFQ 0 0 YTLLKMSLKQQFPNLLRF 0 0 EDERATQVTGEFEVFVEGKLVHSKK 0 0 QGDGFVDDNRMQTIVNAINEEIKRR* 0 >SELV_oryCun Oryctolagus cuniculus (rabbit) 0 MNNQARTPAPAQARTSSVVRASAPTRVSTQIRTPATGWTPTPVQASTPVRTQTPVRTPTLVQASTPVRTPTLGQASTPVRTPTPVQALTPVRTPTPVSGPDSGRTPTPVGTPVGTL 1 GLuSYGLR 0 0 YILLKKNLEELFPDCLLF 0 0 EEERATQASGEFEVFVNGKLVHSKK 0 0 KGDGFVDEVKLRKIVTAINEEIK... 0 >SELV_canFam Canis familiaris (dog) CO599097 0 MNNQARAPPRTSARVLAWVRASTPVRTSIPVRTPTPARIPTSSRAPTSVQTPAPARTPNPVQIPTPVQTSTSARIPNPARTLTPVQTPASAWTPNPVQMLTPARTPTPVPTP VPTPIPARTPTPARTPTPVEAAAAAPASESFGSSALPLEPPPEPASEPTTSPHQDLSPTPSVKPLPSVTNGFGSTQEPLPDLTPPATDFLGPTLGSTSRADSSATKLTDSSESVR VPIPGTPSATALATSTNTFAPVGESCSVKIAVRVIYC 1 GLuSYGLR 0 0 YILLKKSLEQQFPNCLLF 0 0 EEERAAQATGEFEVFVDGKLVHSKK 0 0 KGDGFVDEARLQKIMNVIEEEVRKR* 0 >SELV_felCat Felis catus (cat) 0 MNNQARAPNPSPARVLALVRASTPVRSSIPVRIPTPARIPARTRNPQTPVRAPNPVQIPNPVQISARASNPARAPRPVQIPARTPNPPQTLTPGRAPNPVPTPVQTPTLVQTP TPVQTPTPVWTETPVQTPTAVGTPTRVQTLTPVIPRVRTPTPIRPLWPDPSPNLIPSGPDHPAPESSLRNSAPSFWINSPDSLPAPILETPSTAFASTFENLPEDSKILIRVIYC 1 GLuSYSLR 0 0 YILLRKSLELQFPNCLRF 0 0 EEERSAQATGEFEVFVDGKLVHSKK 0 0 KGDGFVDEARLQKIVSTIDEEIRKR* 0 >SELV_bosTau Bos taurus (cow) DT829759 0 MNNQPRTPAPTPARASTPVRGSTLHRTSIQVRTPTPGPDSGPTRITTLLRTPALIRTLTPIRTPTPVRTPTPVPPSTPVRSPIPVRTPTPVPPSTPVRTPTPVPLSTPVRPPTPVRPPTPVRPPTPVRPPTP IGTPTLIRSPTPVQIPIPEPIPTPIPSRVLIPPLESFPDSALPSGPPLELEPTLTVSPAKNLEPSPAKNLEPSPRVKQVSSAANGFPPIQEPLPALTPLATDLRSPSLGSPLRTDTSTTNLIASSSGHVPGTPILGAIQAILPVPATALASISGNLKEENKIMIRVVYC 1 GLuSYSLR 0 0 YILLRKSLEQQFPNSLIF 0 0 EEEISAQATGEFEVFVDGKLIHSKK 0 0 NGDGFVDEVKLQTIVNLLNEEFKKR* 0 >SELV_susScr Sus scrofa (pig) ti|851198642 CX061656 0 MNNPARAPAPSPVRPSASLRPLASVRASTPARGSTLARTSLLVRTPRTPNLVPASGPGPIRTPTPVRTPTPVRTPTPVRTLTPVRTPTPVRTPTPVRTPTPVRTPTPVRTPTPVQVPTPVRVPTTVGIPTP TLSQVLVPALESLPNPALPSLDPPLEPDPELTLSPDEDPAPTPRAKHLPLVANGFVPVQEPLPALSPLATNLLESTPGAGTDSSTTKLTDSTSGHVPGTPILATIPLAVALPVSTNALASTSENIQVEQQILIRVVYC 2 1 GL*SYGLR 0 0 YILLKKSLEQQFPNCLVF 0 0 EEDISAQATGAFEVFVDGKLVHSKK 0 0 KGDGFVDETKLQKIVSHINEEIKTKVAGSC* 0 >SELV_equCab Equus caballus (horse) 1 GLuSYGLR 0 0 YILLRKSLDQQFPNRLVF 0 0 EEDVGAQASGEFEVFVEGKLVHSKK 0 0 KGDGFVDEARLRKIVSAINEEIKRR* 0 >SELV_myoLuc Myotis lucifugus (microbat) 0 MNHQARASHPFPARTPASVRASIPVRPSNQTPAPSSAPTRTSIPVPASTAVRTPTLVRTPIPFQAVSPVRTSTRFSASVPVQFPTPVSASTPVQTPTRVPAPVRTSTRFPASV PVQFPTPVPASTPVRTSTPVPASTPVQTPTRVPAPAQVRTSIPVPAPAPARTSTPVPAPAPVRTSTTVPAPVPFRTPTPVPAPAPVRTSTPVPAPAPVRTSTPVPAPGPNPDSSPCPGPG 1 GLuSYNLR 0 0 YILLKKSLEQQFPNCLYF 0 0 EEDISEQATGEFEVFVDGKLVHSKK 0 0 KGDGFVDEVKMQKIVNIIDEEIRKN* 0 >SELV_pteVam Pteropus vampyrus (macrobat) 0 MNNKARASAPSLPGTSALVRASAPVRPSTPVRAPTLIRTPTPVRTPNPVPASTPARVPTPVRTLTPVRTPTPIRTPTPVRTQIPFRAANPVRAANPVRTPTPLRTPTPVRTSIPVRTL APVSTPTPIPPPTPPAVPAVVPAAVPGLEFFPSPPPEPVPEPALSPDKDPAPTQSVKHVPSVANEFGLTQEPLSALAPLATDLLGPTRASIRRADPEATKLTGSTAEPSPASILELISSAHIGIASEYLQVDTGSVIRVILL 1 GLuSYNFR 0 0 YILLRKNLEQQFPHGLLF 0 0 EEEVSAEGTGEFEVLVDGKLIHSKK 0 0 RGDGFVDEARLQKIVNVINE IRKR* 0 >SELV_dasNov Dasypus novemcinctus (armadillo) ti|593909023 1 GLuSYALR 0 0 YILLKKSLEMQFPNRLIF 0 0 EEARSSQAVGEFEVFVDGALVHSKK 0 0 RGDGFVDDSKMEKIVSSIEEAVKKT* 0 >SELV_loxAfr Loxodonta africana (elephant) 0 2 1 GLuSYGLR 0 0 YILLRKSLEQQFPNHLIF 0 0 EEDISSQATGEFEVFVDGKLVHSKK 0 0 KGDGFVDDTRLQKIVNSINETIKKE* 0 >SELV_proCap Procavia capensis (hyrax) 0 2 1 GLuSYSLR 0 0 YILLRKNLEQQFPNHLLF 0 0 EVDISSQATGEFEVFVDGKLVHSKK 0 0 KGDGFVDDAQLQKIVNSINETIQRR* 0
SEPW1: 29 vertebrate sequences
>SEPW1_homSap Homo sapiens (human) Selenoprotein W chr19 87 aa uc002phn.1 has retroprocessed pseudogene 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_panTro Pan troglodytes (chimp) 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 RGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_ponPyg Pongo pygmaeus (orang_sumatran) CR926472 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_macMul Macaca mulatta (rhesus) 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_macFas Macaca fascicularis (cynomolgus_monkey) AB169486 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_papAnu Papio anubis (baboon) EY285690 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQATGFFEVMVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_calJac Callithrix jacchus (marmoset) 0 MALTVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 SGEGTPQATGFFEVTVAGKLIHSKK 0 0 KGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_micMur Microcebus murinus (mouse_lemur) 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCLDI 0 0 CGEGTPQATGFFEVMVAGKLVHSKK 0 0 GDGYVDTESKFLKLV >SEPW1_musMus Mus musculus (mouse) 0 MALAVRVVYC 2 1 GAuGYKPK 0 0 YLQLKEKLEHEFPGCLDI 0 0 CGEGTPQVTGFFEVTVAGKLVHSKK 0 0 RGDGYVDTESKFRKLVTAIKAALAQCQ* 0 >SEPW1_ratNor Rattus norvegicus (rat) BC087625 0 MALAVRVVYC 2 1 GAuGYKPK 0 0 YLQLKEKLEHEFPGCLDI 0 0 CGEGTPQVTGFFEVTVAGKLVHSKK 0 0 RGDGYVDTESKFRKLVTAIKAALAQCQ* 0 >SEPW1_cavPor Cavia porcellus (guinea_pig) 0 MALAVRVVYC 2 1 GAuGYKPK 0 0 YLQLKEKLEDEFPGCLDI 0 0 CGEGTPQTTGFFEVTVAGKLVHSKK 0 0 GGDGFVDTEGKFRKLVAAIKAALAQG* 0 >SEPW1_oryCun Oryctolagus cuniculus (rabbit) 0 MALAVRVVYC 2 1 GAuGYKPK 0 0 YLQLKKKLEDEFPGCLDI 0 0 CGEGTPQVTGFFEVTVAGKLVHSKK 0 0 RGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_ochPri Ochotona princeps (pika) 0 MALSVRVVYW 2 1 GAuGYKPK 0 0 YLQLKKRLEDEFPGCLDI 0 0 GEGTPQVTGFFEVMVAGKLVHSKK 0 0 SGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_canFam Canis familiaris (dog) 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCLDI 0 0 RGEGTPQATGFFEVTVAGKLVHSKK 0 0 RGDGYVDTESKFLRLVAAIKTALAQG* 0 >SEPW1_felCat Felis catus (cat) 0 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCLDI 0 0 RGEGTPQATGFFEVMVGGKLVHSKK 0 0 RGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_bosTau Bos taurus (cow) 0 MAVVVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPSRLDI 0 0 RGEGTPQVTGFFEVFVAGKLVHSKK 0 0 GGDGYVDTESKFLKLVAAIKAALAQA* 0 >SEPW1_oviAri Ovis aries (sheep) 0 MAVVVRVVYC 2 1 GAuGYKPK 0 0 YLQLKKKLEDEFPSRLDI 0 0 CGEGTPQVTGFFEVFVAGKLVHSKK 0 0 GGDGYVDTESKFLKLVAAIKAALAQA* 0 >SEPW1_susScr Sus scrofa (pig) AF380118 0 MGVAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGRLDI 0 0 CGEGTPQVTGFFEVLVAGKLVHSKK 0 0 GGDGYVDTESKFLKLVAAIKAALAQG* 0 >SEPW1_eriEur Erinaceus europaeus (hedgehog) 0 MALAVRVVYC 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCLDI 0 0 RGEGTPQGTGFFEVLVAGKLVHSKK 0 0 KGDGYVDTETKFLKLVTAIKAALAQG* 0 >SEPW1_sorAra Sorex araneus (shrew) 0 2 1 GAuGYKSK 0 0 YLQLKKKLEDEFPGCVDV 0 0 CGEGTPQVTGFFEVMVAGKLVHSKK 0 0 RGDGYVDSESKYVRLVTAIKTALAQA* 0 >SEPW1_choHof Choloepus hoffmanni (sloth) 0 MALAVRVVYW 2 1 GAuGYKPK 0 0 YVQLKKKLEDEFPGCLDI 0 0 SGEGTPQTTGFFEVMVAGKLVHSKK 0 0 QKGDGFVDTESKFLRLVAAIKAALAQG* 0 >SEPW1_monDom Monodelphis domestica (opossum) diverged 0 MAIQVRVVYW 2 1 GAuGYKPK 0 0 YLLLKKKLEDEYPGLLRH 0 0 NGEGTPEVTGFFEVTVAGKLVHSKK 0 0 AGHGFVDTADKYLQIVAEIKAALA* 0 >SEPW1_ornAna Ornithorhynchus anatinus (platypus) 0 MASLEAFPRGVVPVHVVYC 2 1 GAuGYKPK 0 0 FLQLKKKLENEFPGQVEI 0 0 SGEGTPQVTGWFEVTVAGKLVHSKK 0 0 EGDGFVDSESKFAKIRMAIKAALVPGY* 0 >SEPW1_galGal Gallus gallus (chicken) tga confirmed 0 MPLRVTVLYC 2 1 GAuGYKPK 0 0 YERLRAELEKRFPGALEM 0 0 RGQGTQEVTGWFEVTVGSRLVHSKK 0 0 NGDGFVDTNAKLQRIVAAIQAALP* 0 >SEPW1_anoCar Anolis carolinensis (lizard) 0 2 1 GAuGYSPK 0 0 YQQLKRGLEKEFPGKLEI 0 0 TGEGTPQVTGWFEVTVAGKLVHSKK 0 0 NGDGFVDNDTKLHKILMAIKAALA* 0 >SEPW1_xenTro Xenopus tropicalis (frog) tga confirmed 0 MPDTMVKVNVVYC 2 1 GAuGYLSK 0 0 FRRLKKELEQRFPGKLSI 0 0 DGEGTERMTGWFEVSINGKLVHSKK 0 0 NGDGYVDNDAKLQKIILAIEAALKQ* 0 >SEPW1_danRer Danio rerio (zebrafish) tga confirmed 0 MTVKVHVVYC 2 1 GGuGYRPK 0 0 FIKLKTLLEDEFPNELEI 0 0 TGEGTPSTTGWLEVEVNGKLVHSKK 0 0 NGDGFVDSDSKMQKIVTAIEQAMGK* 0 >SEPW1_takRub Takifugu rubripes (fugu) 0 MGVTIRVEYC 2 1 GGuGYGPR 0 0 YEELARVVRAEFPDADVSGFVGRM 1 2 GSFEIQINEQLIFSKLETGGFPYEDD 0 >SEPW1_calMil Callorhinchus milii (elephantfish) 1 GAuGYEPRYQKLAIVIKDEFPDADVSGKVGRT 1 2 GSFEIEINGQLIFSKLETGGFPYEND 0 0 ISEAVQKANNGEELQKIENSRPPCVIL* 0 >SEPW1_squAca Squalus acanthias (spiny_dogfish) EE722256 0 MGVKIHFEYC 2 1 GAuGYKPR 0 0 YQELANTIMGTFPDAAISGDVGRT 1 2 GSFEIEINGQLVFSKLETSGFPYEDD 0 0 IMDAVQKASAGDDVQKIVKSRPPCVIL* 0 >SEPW1_torCal Torpedo californica (electric_ray) 0 MSVKVHVEYC 2 1 GAuGYGNR 0 0 YQDLANNILKTFPDADISGDVGRK 1 2 GSFEVEINGQLVFSKLETKGFPFEND 0 0 IITAVQNASNGVEMQQITNSRASCVIL* 0 >SEPW1_petMar Petromyzon marinus (lamprey) cDNA FD700531 tga-sel tag-stop full 0 MPLKIHIVYC 2 1 GAuGYRSR 0 0 FHRLKDELETEFPGELEI 0 0 TGEGTPTQTGFLEVQIVGGKLLHSKA 0 0 NGDGFVDSDEKLQKIFSGVEKALKK* 0 >SEPW1_eptBur Eptatretus burgeri (hagfish) cdna BJ662449 tgn-sel tag-stop 0 MPLKIHVVYC 2 1 GSuGYASK 0 0 FRALKVKLDHEFPGKLEI 0 0 TSEGTPGLTGKFEVQVGEKLVHSKK 0 0 NGDGFVDSPEKLQKIFKAVENALKGQ* 0 >SEPW1_strPur Strongylocentrotus purpuratus (sea_urchin) tga confirmed 0 MVVIVKVIYC 2 1 GGuGYGPR 0 0 YRRLEQELKDEFGDDVDM 0 0 VGESTPGVTGWLEVEVNGKLIHSKK 0 0 NGDGYIDSEGKLKKIVNAVSAAM* 0
SEPWb: 2 vertebrate sequences
>SEPWb_squAca Squalus acanthias (spiny_dogfish) 0 MPVEVAIEYC 2 1 SGuGFLTR 00 YLSLANEIRERVPGTNVTGFIGQA 1 2 GSFEVTINNKLVFSKLESNGFPYQED 0 0 VIEAVKTAKEGSAVERIISEEEIITQKRGCIII* 0 >SEPWb_danRer AAO86697 0 MVVKVKIEYC 2 1 GAuGYEPR 00 FQELKREICGNCPDAEVSGFVGRR 1 2 GCFEIQINDFLVFSKLESGGFPYSED 0 0 IIEAVVKAKDGKPEKIIRNRKECIIL* 0
RDX12 (C17orf37, SEPW2A): 49 deuterostome sequences
>RDX12_homSap C17orf37 NM_032339 0 MSGEPGQTSVAPPPEEVEPGSGVRIVVEYC 2 1 EPCGFEATYLELASAVKEQYPGIEIESRLGGT 1 2 GAFEIEINGQLVFSKLENGGFPYEKD 0 0 LIEAIRRASNGETLEKITNSRPPCVIL* 0 >RDX12_panTro MSGEPGQTSVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_gorGor MSGEPGQTSVAPPPEEVEPGSGVRMVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_ponAbe MSGESGQTFVAPPPEEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_rheMac MSGEPGQTSVAPPPEEVEPGSGVRIMVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASKGEPLEKITNSRPPCVIL* >RDX12_calJac MSGEPGQTSVAPLPGEVEPGSGVHIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASTGEPLEKITNSRPPCIIL* >RDX12_otoGar MSGEPGHSSVAPPPGEVEPGSGVRIMVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCIIL* >RDX12_tupBel MSGESGETSVAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGT-------------------------------------------TNSRPPCVIL* >RDX12_musMus MSGEPAPVSVVPPPGEVEAGSGVHIVVEYCKPCGFEATYLELASAVKEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASNGEPVEKITNSRPPCVIL* >RDX12_ratNor MSGEPGQVSVVPPPGEVEAGSGVHIVVEYCKPCGFEATYLELASSLEEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_dipOrd MSGEPGEVSLAPPPGEAEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_cavPor MSGEPGELSVARPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIQIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_oryCun MSGEPGPTSAAPSPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCTIL* >RDX12_ochPri MSGEPGPTSAAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELANAVKEEYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_vicPac MSGETGPTSVAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_turTru MSGETGPASSAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_bosTau MSGDTGTTSVAPPPGETEPGHGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_equCab MSGEPGLTSVAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_canFam MSGEPGPTSEAPPPGEIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_myoLuc MSGDAGPVSAAPHPGELEPGSGVRIVVEYCEPCGFEATYLELASAVKEQFPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_pteVam MSGDLGPTSAAPHPREIEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCIIL* >RDX12_eriEur MSGEPGPTAVAPPPGEVEPGSGVRIVVEYCEPCGFEATYQELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_sorAra MSG----LAVAPPPGEGEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_loxAfr MNGEPGPISLAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_proCap MNGEPGPVSLAPPPGEVEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITNSRPPCVIL* >RDX12_echTel MSGEPGPVSLAPPPGEAEPGSGVRIVVEYCEPCGFEATYLELASAVKEQYPGIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASNGEPLEKITSSRPPCAIL* >RDX12_monDom MSGESGAELVAPLPGEALPGSGVRIVVEYCEPCGFESTYLELASAVKEEYPGIKIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASSGEPLEKITNSCPPCVIL* >RDX12_macEug MSGEPGADLEEPLP.....GSGVRIVVEYCEPCGFESTYLELASAVKEEYPGIQIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRASSGEPLQKITNSRPPCVIL* >RDX12_ornAna ---------------------------------------------------------------AFEIEINGQLVFSKLENGGFPYEKDLMEAIRRASLGEPLEKITNSRPPCSIL* >RDX12_galGal MSSSGGNGAAAVGTESEAGDGDGFGSDSGSERRVHIMVEYCEPCGFGATYEELASAVREEYPDIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCTIL* >RDX12_melGal MSSSGGGNGAAAAGTETEAGNGADGFVSGSGSERRVHIMVEYCEPCGFGATYEELASAVREEYPDIEIESRLGGTGAFEIEINGQLIFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCVIL* >RDX12_taeGut MSGGTGDGTGDGNGAERRVRIVVEYCEPCGFEATYQELASAVRDEYPDIEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRRARNGEPLEKITNSRPPCVIL* >RDX12_anoCar MSDGSGEPAAEAPPATEGGVRIVVEYCKPCGFESAYLELANAVKEEYPDVEIESRLGGTGAFEIEINGQLVFSKLENGGFPYEKDLIEAIRKAINGEPLEKITKSRPPCVIL* >RDX12_xenTro MSVSIVVEYCEPCGFKSHYEELASAVLEEFPDVTIDSRPGGTGAFEIEINGQLVFSKLELGGFPYAKDLIEAIRKASNGEPVEKITNSQAPCVIL* >RDX12_ambMex MAAVTIVVEYCKHCGFESHYLELESAVKEEFPDVVIESRCGETGTFEIKINGQIVFSKLELGGFPYEKDLMEAIKRASNGEPVERITNSRAPCVIL* >RDX12_tetNig MAVKIIVEFCGGuGYGPRYEELARVVKAEFLDADVSGFVGRLGSFEIVINEQLVFSKLETGGFPYEDDVLQVIQCAYDGKPVEKLTKSRPPCVIM* >RDX12_fugRub MGVTIRVEYCGGuGYGPRYEELARVVRAEFPDADVSGFVGRMGSFEIQINEQLIFSKLETGGFPYEDDVMHAIQCVSDGKPVEKITKSRPPCVIM* >RDX12_gasAcu MGVTMRVEYCGNuGYEPRYQELRTAVKQDFPDADVTGFVGRRGSFEIVLNGQLIFSRLESWGFPHVEDVLDAVKKAADGKPVDKITISRAPCVIM* >RDX12_oryLat MGVKIDVEYCGRuGYEPRYQDLASTVKDEFPEAEVSGFVGRSGSFEIQINGQLVFSKLELGGFPYEDDVLNAVQNAHDGKPVQKITKSRAPCVIM* >RDX12_danRer MGVQIKVEYCGGuGYEPRYQELKRVVTAEFTDADVSGFVGRQGSFEIEINGQLIFSKLETSGFPYEDDIMGVIQRAYDGQPVEKITKSQPPCVIL* >RDX12_oreMas MGVKVRVEYCGGuGYEPRYRELARVVKGEFSDADVTGVVGRTGSFEIEINGQLVFSKLETGGFPYEDDVMDAIHNAYDGKPLQKITKSRAPCVIM* >RDX12_calMil ..........GAuGYEPRYQKLAIVIKDEFPDADVSGKVGRTGSFEIEINGQLIFSKLETGGFPYENDISEAVQKANNGEELQKIENSRPPCVIL* >RDX12_squAca MGVKIHFEYCGAuGYKPRYQELANTIMGTFPDAAISGDVGRTGSFEIEINGQLVFSKLETSGFPYEDDIMDAVQKASAGDDVQKIVKSRPPCVIL* >RDX12_torCal MSVKVHVEYCGAuGYGNRYQDLANNILKTFPDADISGDVGRKGSFEVEINGQLVFSKLETKGFPFENDIITAVQNASNGVEMQQITNSRASCVIL* >RDX12_petMar MAVNVNVEYCGGuGYEPRYQELAANILKQAPGVEVIGQVGRSGSFEVTINGELIFSKLECGGFPFAEDIIAEVKKVQGGEKVGKVTKSQAPCVIL* >RDX12_cioInt MADSNKVKVEIEYCGSuGYYGRFMDLKNDLESGCPNALVSGFVGREGSFEVSINGKQIFSKLETFGFPYPNDLIEAMKTAQNGEEVAAINNSQSPCTIL* >RDX12_braFlo MNYSRSKMAPKVEVEYCGGuGYAPRYWELANQIKTAVPDAEVTGVVGRSSSFEIIVDGQLLFSKLESGGFPQEQEILEALSNYKEGEKVEQVTNIQFPWCTIL* >RDX12_strPur MAQKKVTIEYCGSuGYYPRYRELHEMIESAIKGVDVSGKRGRPSSFEVKLNGQVLYSKLKNGGFPDLDAIVEAIEDYKGEGTVTPVEKKAASCIIL* >RDX12_astPec MATRKVEIEYCGGuGFYPRFMDLHNDIKSGVPGVAVDGRVGRSTSFEVTLNGKLLFSKLKTGGFPINKEIVTAIKEYKGEGDVTPVTNTASNCVLL*
DIO1: 50 vertebrate iodothyronine deiodinases
>DIO1_homSap Homo sapiens (human) iodothyronine deiodinase type I 4 exons chr1 uc001cwb.1
0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWEFMQ 1
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLVIYIEEAHAS 1
2 DGWAFKNNMDIRNHQNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYIIQEGRILYK 0
0 GKSGPWNYNPEEVRAVLEKLHS* 0
>DIO1_panTro Pan troglodytes (chimp)
0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVHLSGQRCNIWEFMQ 1
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLVVYIEEAHAS 1
2 DGWAFKNNMDIRNHQNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYVIQEGRILYK 0
0 GKSGPWNYNPEEVRAVLEKLHS* 0
>DIO1_ponPyg Pongo pygmaeus (orang_sumatran)
0 MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTETGGLAPNCPVVRLSGQRCNIWEFMQ 1
>DIO1_macMul Macaca mulatta (rhesus)
0 MGLPQSGLWVKKLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWDFMQ 1
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLIIYIEEAHAS 1
2 DGWAFKNNMDIRNHRNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYVIQEGRILYK 0
0 GKSGPWNYNPEEVRAVLEKLYS* 0
>DIO1_macFas Macaca fascicularis (crab_macaque)
0 MGLPQSGLWVKKLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWDFMQ 1
>DIO1_macNem Macaca nemestrina (pigtailed_macaque)
0 MVLPQSGLWVKKLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGPSCNIWDFMQ 1
>DIO1_calJac Callithrix jacchus (marmoset)
0 MGLPGPGLWLKRLWVLLEVAVHVAVGKVLLTLFPDRVKKNILAMGDKTGMTRNPNFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVHLSGQRCNIWDFMQ 1
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLTEDFSSVADFLIIYIEEAHAS 1
2 DGWAFKNNVDIRNHQNLQDRLQAAHLLLARSPQCPVVVDTMQNQSSELYAALPERLYIIQEGRILYK 0
0 GKSGPWNYNPEEVRDVLEKLHS* 0
>DIO1_otoGar Otolemur garnettii (bushbaby)
0 MGLPRPGLWLKRLWVFLEVAVHVAVGKMLLILFPDRVKSQILAMGQQTVMAKNPHFSHDNWIPTFLQHPVFLVCPEGPLQRLEDMTERGGLAPNCPVSASSGQKCNIWDFMQ 1
2 GNRPLVLNFGSCTuPSFLFKFDQFKRLVEDFSSVADFLIIYIEEAHAS 1
2 DGWAFKNNVDIRTHRNLQDRLRAAHLLLARSPQCPVVVDTMENQSSQLYAALPERLYVLQEGRILYK 0
0 GKSGPWNYQPEEVRAVLEKFDN* 0
>DIO1_micMur Microcebus murinus (mouse_lemur)
0 MGLPCPGLWLKRLWVLLQVAVHVAVGKVLLTLFPERVMQHILSIGQKTGMARNPHFTPDNWVPTFFSTQYFWFVLKVRWQQLEDVTERGGLAPNCPVVRLSGQRCNIWDFMQ 1
>DIO1_tupBel Tupaia belangeri (tree_shrew)
0 RAGLWLKRLWVFLQLTVQVAVGKVLLTLFPERVKQHILALGQKTGIARNPNFAHDNWIPTFFSTQYFWFVLKVYWQRLEDTTEPGGLAPNGPVVHLSGQRCDIWDFMQ 1
2 GDRPLVLNFGSCTuPSFLFKFDQFKRLIEDFSSIADFLIIYIEEAHAS 1
2 DGWAFKNNVNIRNHQNLQDRLQAAHLLLDRSPQCPVVVDTMQNQSSQLYAALPERLYVLQEGRILYK 0
0 GKPGPWNYDPEEVRAVLEKLRS* 0
>DIO1_musMus Mus musculus (mouse)
0 MGLPQLWLWLKRLVIFLQVALEVAVGKVLMTLFPGRVKQSILAMGQKTGMARNPRFAPDNWVPTFFSIQYFWFVLKVRWQRLEDRAEFGGLAPNCTVVCLSGQKCNIWDFIQ 1
2 GSRPLVLNFGSCTuPSFLLKFDQFKRLVDDFASTADFLIIYIEEAHAT 1
2 DGWAFKNNVDIRQHRSLQERVRAARMLLARSPQCPVVVDTMQNQSSQLYAALPERLYVIQEGRICYK 0
0 GKAGPWNYNPEEVRAVLEKLCTPPRHVPQL* 0
>DIO1_ratNor Rattus norvegicus (rat)
0 MGLSQLWLWLKRLVIFLQVALEVATGKVLMTLFPERVKQNILAMGQKTGMTRNPRFAPDNWVPTFFSIQYFWFVLKVRWQRLEDRAEYGGLAPNCTVVRLSGQKCNVWDFIQ 1
2 GSRPLVLNFGSCTuPSFLLKFDQFKRLVDDFASTADFLIIYIEEAHAT 1
2 DGWAFKNNVDIRQHRSLQDRLRAAHLLLARSPQCPVVVDTMQNQSSQLYAALPERLYVIQEGRICYK 1
0 GKPGPWNYNPEEVRAVLEKLCIPPGHMPQF* 0
>DIO1_speTri Spermophilus tridecemlineatus (squirrel)
0 MGLLRPGLWLKKLWILLLVIVEVAMGKVLMTLFPERATQNILAMGQKTGMTRNPQFSPDNWVPTFFSIQYFWFVLKVRWQRLEDKAELGGLAPNCPVIRLSGEKCNIWDFIQ 1
2 GNRPLVLNFGSCTuPSFLFKFDQFKRLIEDFSPIADFLIIYIEEAHAS 1
2 1
0 * 0
>DIO1_dipOrd Dipodomys ordii (kangaroo_rat)
0 MELSRLGLWLKRLWIFLQVVVEVAMGKMLMILFPERVKKHILAMGQKTGMTRNPRFSPDNWVPTFFSTQYFWFVLKVRWQRLEDKAMYGGLAPNCSVISLSGQRCSIWDFMQ 1
2 GNRPLVLNFGSCTuTSFLFKFDQFKRLVEDFDSTADFLIIYIEEAHAS 1
2 DGWAFKNNVNISHHRNLQDRLQAAQLLLDQKPQCPVVVDTMENQSSQLYAALPERLYVLQEGRILYK 1
0 GKPGPWNYNPEEVRAVLEKLCT* 0
>DIO1_ochPri Ochotona princeps (pika)
0 MAWPQVRLWLRRLWVLVQVAVEVAVGKVLMTLFPERVKQSILAMGQKTGLAQNPLFTHDNWIPTFFSIQYFWFILKVRWQRLEDATQPGGLAPNCPVVCLSGQECHIWDFMQ 1
2 GNRPLVLNFGSCTuPSFLSKFNQFKRLIEDFSSIADFLIIYIEEAHAS 1
2 DGWAFKNNVDIRNHRNLQERLQAAHLLLPRSPQCPVVVDTMQNQSSQHYAALPERLYVLQQGRILYK 1
0 GQPGPWNYDPEEVRAVLLELHS* 0
>DIO1_cavPor Cavia porcellus (guinea_pig)
0 MGLTWPGLWLKRLWVLVQVAVEVAMGKVLMTLFPERIKKSILAMGEKTGMTRNPQFSHDNWIPTFFSTQYFWFILKVRWQRLEETAELGGLAPDCSVVCLSGEKRTIWDFMH 1
2 GNRPLVLNFGSCTuPSFMFKFDQFKRLIEDFSSIADFLVIYIEEAHAS 1
2 DGWAFKNNVDIRQHQNLQDRMRAAHLLLAKSPQCPVVVDTMQNESSQLYAALPERLYVQEGRILYK 1
2 GKSGPWNYNPEEVRGVLEKLHT* 0
>DIO1_oryCun Oryctolagus cuniculus (rabbit)
0 MGLPRPGLWLKRLWVLVQVAVEVAVGKVLMTLFPERVKQNILAMGQKTGIAQNPNFAQDSWIPTFFSTQYFWFVLKVRWQRLEDATEPGGLAPNCSVVRLSGQQCSIWDFMR 1
2 GNRPLVLNFGSCTuPSFLSKFDQFKRLIQDFSSIADFLIIYIEEAHAS 1
2 DGWAFKNNVDIKNHRNLQDRLRAASLLLARSPQCPVVVDTMQNQSSQLYAALPERLYVLRQGRILYK 0
0 GESGPWNYNPEEVRAVLEELHS* 0
>DIO1_canFam Canis familiaris (dog)
0 MGLPRPVLWLRRLWVLLQVAVQVAVGKVFLKLFPARVKQHIVAMNGNKPHFSYDNWAPTLYSMQYFWFVLKVQWQRLEDRTEPGGLAPNCPVVRLSGQRCNIWDFMQ 1
2 GNRPLVLNFGSCTuPSFLFKFDQFKRLIEDFCSTADFLIIYIEEAHAS 1
2 DGWAFKNNVNIRTHQTLQDRLQAARLLLDRAPPCPVVVDTMRNQSSQFYAALPERLFVLQEGRILYK 0
0 GKPGPWNYHPEEVRAVLEKLHS* 0
>DIO1_ursArc Ursus arctos (bear) AM748338
0 VAMHVAVGKVLLILFPERVKQQVLAMNKKNPHFSYDNWLPTFYSMQYFWFVLKVRWQRLEDRTEPGGLAPNCPVVRLS
>DIO1_melUrs Melursus ursinus (bear) AM748341
AMHVAVGKVLLILFPERVKQQVLAMNQKNPHFSYDNWLPTFYSMQYFWFVLKVRWQRLEDRTEPGGLAPNCPVVRLSG
>DIO1_ursAme Ursus americanus (bear) AM748339
AMHVAVGKVLLILFPERVKQQVLAMNQKNPHFSYDNWLPTFYSMQYFWFVLKVRWQRLEDRTEPGGLAPNCPVVRLSG
>DIO1_ursMar Ursus maritimus (bear) AM748337
AMHVAVGKVLLILFPERVKQQVLAMNQKNPHFSYDNWLPTFYSMQYFWFVLKVRWQRLEDRTEPGGLAPNCPVVRLSG
>DIO1_ailMel Ailuropoda melanoleuca (bear) AM748344
AMHVAVGKVFLILFPERVKQQVLAMNQKNPHFSYDNWLPTFYGMQYFWFVLKVRWQRLEDRTEPGGLAPNCPVVQLSG
>DIO1_treOrn Tremarctos ornatus (bear) AM748343
AMHVAVGKVLLILFPERVKQQVLAMNQKNPHFSYDNWLPTFYSMQYFWFVLKVRWQRLEDRTEPGGLAPNCPVVQLSG
>DIO1_helMal Helarctos malayanus (bear) AM748340
GKVLLILFPERVKQQVLAMNQKNPHFSYDNWLPTFYSMQYFWFVLKVRWQRLEDRTEPGGLAPNCP
>DIO1_ursthi Ursus thibetanus (bear) AM748342
AMHVAVGKVLLILFPERVKQQVLAMNQKNPHFSYDNWLPTFYSMQYFWFVLKVRWQRLEDRTEPGGLAP
>DIO1_felCat Felis catus (cat)
0 MGLSQLGLWLRRLWVLFQVALQVAVGKVFLILFPSRVKQHIVAMNRKNPHFSYDNWAPTLYSVQYFWFVLKVRWQRLEDRTEPGGLAPNCPVVRLSGQRCSIWDFMK 1
2 GNRPLVLNFGSCTuPSFLFKFDQFKRLIEDFCSIADFLIIYIEEAHAS 1
2 DGWAFKNNVNIRNHRNLQDRLQAACLLLDRSPRCPVVVDTMKNQSSRLYAALPERLYVLQAGRILYK 0
0 GKPGPWNYHPEEVRAVLEKLHS* 0
>DIO1_equCab Equus caballus (horse)
0 MGLPRAGLWLKRLWVLLQVALQVAVGKVLLTLFPDRVKQHIVAMNQKNPHFSYDNWVPTLYSTQYFWFVLKVHWQRLEDTTKRGGLAPNSPVVRLSGQRCNIWDFMQ 1
2 GNRPLVLNFGSCTuPSFLFKFDQFKRLIEDFSSIADFLIIYIEEAHAS 1
2 DGWAFKNNVVIRNHRNLQDRLRAARLLLDRSPPCPVVVDSMENRSSQLYAALPDRLYVLQARRILYK 0
0 GKPGPWNYHPEEVRAVLEKLHS* 0
>DIO1_myoLuc Myotis lucifugus (microbat)
0 MGLPQPGLWLKRLWILLQVALHVTLGKVQLKLFPRRVKQHILAMNRKNPHFSYDNWAPTLFSTPYFWFILKVRWQRLEDKTEEGSLAPNCPVVRLSGQRCHIWDFMQ 1
2 GNRPLVLNFGSCTuPSFLFKFDQFKRLIEDFSAIADFLVIYIEEAHAS 1
2 DGWAFKNNVVIKNHRNLQDRLQAAHLLLDRSPRCPVVVDTMKNQSSQLYAALPDRLYVLQEGRILYK 0
0 GKPGPWNYHPEEVRAVLEKLRS* 0
>DIO1_pteVam Pteropus vampyrus (macrobat)
0 MELPWPGRWLKRLWVLLQVALHVAVGKVQLTLFPRRVKQNIVAMNRKNPHFSFDNWLPTLFSTQYFWFVLKVRWQQLEDTTKEGGLAPNCPVVCLSGQRCNIWDFMQ 1
2 GKRPLVLNFGSCTuPSFLLKFDQFKKLIEDFSSIADFLVIYIEEAHAS 1
2 ERLQAARMLLDRSPPVPVVVDTMKNQSSHLYAALHERLYVLQEGRILYK 0
0 GKPGPWNYHPEEVHAVLEKLHS* 0
>DIO1_bosTau Bos taurus (cow)
0 MGLPSPGLWLKRLWVLFQVALHVAIGKVLLTLFPRRVKQNILAMGEKTGMTRNPHFSHENWIPTFFSTQYFWFILKVRWQRLEDMTEQGGLAPNCPVVRLSGERCSIWDFMQ 1
2 GNRPLVLNFGSCTuPSFIFKFDQFKRLIEDFGSVADFLIIYIEEAHAS 1
2 DGWAFKNNVDIKNHRNLQDRLRAAHLLLDRSPPCPVVVDTMTNQSSSCYAALPERLYVLQEGRVLYK 0
0 GKPGPWNYHPEEVRAVLEKLHS* 0
>DIO1_turTru Tursiops truncatus (dolphin)
0 MGLPLPGLWLKRLWVLFQVGLHVAMGKVLLTLFPRRVKQNILAMSEKTGMAKNPHFSYENWIPTFFSAQYFWFILKVRWQRLEDMTEQGGRAPNCPVVRLSGQRCNIWDFMQG 1
2 GNRPLVLNFGSCTuPSFIFKFDQFKRLIEDFSSIADFLIIYIEEAHAS 1
2 DGWAFKNNVDIKNHQHLQDRLRAARLLLDRSPQCPVVVDTMKNQSSQLYAALPERLYVLQDGRILYK 0
0 GKPGPWNYRPEEVRAVLEKLHS* 0
>DIO1_susScr Sus scrofa (pig)
0 MELPLPGLWLKRLWVLFQVALHVAMGKVLMTLFPGRVKQDILAMSQKTGMAKNPHFSHENWIPTFFSAQYFWFVLKVRWQRLEDKTEEGGLAPNCPVVSLSGQRCHIWDFMQ 1
2 GNRPLVLNFGSCTuPSFIFKFDQFKRLIEDFSSIADFLIIYIEEAHAS 1
2 DGWAFKNNVDIKNHQNLQDRLRAAHLLLDRSPQCPVVVDTMKNQSSRLYAALPERLYVLQAGRILYK 0
0 GKPGPWNYHPEEVRAVLEKLHS* 0
>DIO1_vicVic Vicugna vicugna (vicugna)
0 MGLSLPGLWLKRLWVLFQVVLHVALGKVLLTLFPGRVKQDILAMSQRTGMAQNPHFSYENWIPTFFSTQYFWFILKVRWQRLEDTTEQGGLAPDCPVVCLSDRVHIWDFMQ 1
2 FDQFKRLIEDFNSIADFLIIYIEEAHAS 1
2 DGWAFKNNVDIKNHRNLQDRLRAAHLLLDRSPQCPVVVDTMKNQSSQLYAALPERLYVLQKGRILk 0
0 GKPGPWNYHPEEVRAVLEKLHS* 0
>DIO1_eriEur Erinaceus europaeus (hedgehog)
0 MGLPSPGLWLKRLWVLFQVALHVAIGKVLLTLFPRRVKQNILAMGEKTGMTRNPHFSHENWIPTFFSTQYFWFILKVRWQRLEDMTEQGGLAPNCPVVRLSGERCSIWDFMQ 1
2 1
2 DRWAFKNNVDIRTHRNLQDRMRAALLLLDRDPQCPVVVDTMENQSSQLYAALPERLYVLQEGRILYK 0
0 GKPGPWDYQPQEVRAVLEKLRGKCGQTLPKL* 0
>DIO1_sunMur Suncus murinus (shrew)
0 MGLPGLGLLLKRFGVLVRVALKVAVGKVLLTLWPSAIRPHLLAMSEKTGMAKNPRFTYEDWAPTFFSTQYFWFVLKVNWQQLEDRTKQGDIAPDSPVVHLSGQRARLWDFMQ 1
2 GNRPLVLNFGSCSuPSFLFKFDQFKRLVEDFSSVADFLTVYIEEAHAS 1
2 DGWAFKNNVDIRRHRDLQERLQAARLLLDRNPGCPVVVDTMENRSSQLYAALPERLYVLQEGRILYK 0
0 GGPGPWNYHPEEVHAVLEQLCRSSAQSPRL* 0
>DIO1_loxAfr Loxodonta africana (elephant)
0 MGLPQPGLWLKRLWIFLKVALHVAMGKVLLILFPGRVKKNILAQNPHFAYDMWGSTLFSIPYFWFILKVYWQRLEDKTEEGGPAPDCPVVCLSGQRCNISDFMQ 1
2 GIRPLVLNFGSCTuPSFLSKLDQFKRLVEDFSSMADFLIIYIEEAHAT 1
2 DGWAFKNNVAIRNHRNLQDRLQAAHLLLDRSPQCPVVVDTMQNVSSQLYAALPERLYVLQEGRILYK 0
0 GKPGPWNYHPEEVRAVLEKLNS* 0
>DIO1_proCap Procavia capensis (hyrax)
0 MGLPQPVLWLKRLWVLLRVALHVAMGKVLLALFPGRVKKNILAQNPHFAYDMWCSTLFSVPYFWFVLKVYWQRLEDKTEEGGLAPNCPVVHLSGQRRNIWDFMQ 1
2 GIRPLVLNFGSCTuPSFLSKLDQFKRLVEDFSSMADFLIIYIEEAHAT 1
2 DGWAFKNNVAIRTHRNLQDRLQAARLLLDRSPQCPVVVDTMQNVSSQFYAALPERLYVLQEGRILYK 0
0 GKPGPWNYHPEEVRAFLEKFGS* 0
>DIO1_echTel Echinops telfairi (tenrec)
0 1
2 GTRPLVLNFGSCTuPSFLLKFDQFKRLMDDFHATADFLI 1
2 DGWAFKNNVTIRTHQNLEDRLRAARLLLDRGPQCPVVVDTMENESGRLYAALPERLYVLQEGRILYK 0
0 GKPGPWNYRPEEVRAVLEKLDS* 0
>DIO1_choHof Choloepus hoffmanni (sloth)
0 MGLSWPGLWLKRLWVLLQVALHVAMGKILLTLFPGRVKQNILAMSRRANNTKDPQFPYDNWGPTFFNTQYFWFVLKVRWQRLEDKTEQGGLAPNCPVVHLSRQRCNIWDFMQG 1
2 GNRPLVLNFGSCTuPSFLFKLDQFKRLVDDFSSIADFLIIYIEEAHAS 1
2 RRRAARLLLDRSPQCPVVVDTMQNQSSQLYAALPERLYMLQEGRILYK 0
0* 0
>DIO1_dasNov Dasypus novemcinctus (armadillo)
0 MGLSQPGLWLKRLWILFQVALHVAVGKTLLTLFPGRVKQNILAKSQKSHKAENPHFPYDNWGPTFFNTQYFWFLLKIGWQRLEDKTEQGGLAPNCPVVHLSGQRCNIWDFMQ 1
2 GNRPLVLNFGSCTuPSFLFKFDQFKRLVEDFSSIADFLIIYIEEAHAS 1
2 DGWAFKNNVDIRNHRNLQDRQRAACLLLDRSPQCPVVVDTMQNQSSQLYAALPDRLYVLQEGRILYK 0
0 GKPGPWDYQPEEVRAVLETLNG* 0
>DIO1_monDom Monodelphis domestica (opossum)
0 MAELLRLWLWLWLRLQRLWVLLQVVGHVLMGKLMKMLSPDRMKQHILGMGQKSSIFQNPNFKYENWGPTFFTLPYFLFVLRVRWQRLEDQALQGGPAPDCPVVSLRGQPRRLWDFMH 1
2 ANRPLVLSFGSCTuPSFIFKFDQFHRIMEDFSSVADFLIIYIEEAHAT 1
2 DGWAFANNIDIKQHRTLQDRMEAARLLMDRKPLCPIVVDTMDNLATRKYAALPERFYILLEGHILYK 0
0 GGPGPWSYNPEEVRAVLETLSR* 0
>DIO1_triVul Trichosurus vulpecula (possum)
0 MAGPLLWVRRFWALLQVAFHVVVGKLLKTLFPNMMKKHILSLGQRSSISQNTQFAYENWGPTFFSIQYFFFVLKVRWQRLEDQALQGGLAPNPPVVTLKGESRHIWDFMH 1
2 GNRPLVLNFGSCTuPSFILKFDQFRKLIEDFSSIADFLIIYIEEAHAA 1
2 DGWAFENNIDIKQHRTLQDRRETAQLLMDRKPLCPIVLDTMDNLTSKKYAALPERFYVLLEGRILFK 0
0 GDPGPWNYHPEEVQAVLQ* 0
>DIO1_ornAna Ornithorhynchus anatinus (platypus)
0 1
2 GSRPLVLTFGSCTuPSFLFKFDQFNQLVQDFNSIADFLIIYIEEAHPT 1
2 DGWAFANNVDIPSHRSLQERQEAARRLLARGPRCPVVVDTMDNASSRQYAALPERLYLLREGKVVYK 0
0 GGPGPWNYNPGEVRAVLEKLS 0*
>DIO1_galGal Gallus gallus (chicken)
0 MLSIRVLLHKLLILLQVTLSVVVGKTMMILFPDTTKRYILKLGEKSRMNQNPKFSYENWGPTFFSFQYLLFVLKVKWRRLEDEAHEGRPAPNTPVVALNGEMQHLFSFMR 1
2 DNRPLILNFGSCTuPSFMLKFDEFNKLVKDFSSIADFLIIYIEEAHAV 1
2 DGWAFRNNVVIKNHRSLEDRKTAAQFLQQKNPLCPVVLDTMENLSSSKYAALPERLYILQAGNVIYK 0
0 GGVGPWNYHPQEIRAVLEKLK* 0
>DIO1_anoCar Anolis carolinensis (lizard)
0 MFKAGRLVLKTWLLLQVCLSTAVGKLFMILFPATAKRYILKQSERSSMGRNPNFVYENWGPTFFSFQYLLFVLKVKWKRLEDKALQGCPAPNTPVVDFDGKIHHILDFMQ 1
2 DNRPLVLAFGSCTuPSFMFRFGEFKKLIEDFSFAADFLVIYIEEAHAS 1
2 DGWAFKNNIVIKSHRTLHDRMQAAEILLKQYPLCSVVMDTMENLSSSTYAALPERLYVLQGGNIVYK 0
0 GGVGPWNYNPQEVREVLEKL* 0
>DIO1_xenTro Xenopus laevis (frog) tga confirmed
0 MLRYIQKALILFFLFLYVVVGKVLMFLFPQTMASVLKSRFEISGVHDPKFQYEDWGPTFFTYKFLRSVLEIMWMRLEDEAFVGHSAPNTPVVDLSGELHHIWDYLQ 1
2 GTRPLVLSFGSCTuPPFLFRLGEFNKLVNEFNSIADFLIIYIDEAHAA 1
2 DEWALKNNLHIKKHRSLQDRLAAAKRLMEESPSCPVVLDTMSNLCSAKYAALPERLYILQEGKIIYK 0
0 GKMGPWGYKPEEVCSVLEKKK* 0
>DIO1_danRer Danio rerio (zebrafish)
0 MGSAVGFALRKLFVYISAVLMVCAAILQMSMLKLLSFISPGRMRKIHMKMGERSTMTQNPKFRYEDWGPAFFSLAFIKTLFFVNWCSLGLEAFEGHAAPDSALITLDRQKTSVHRFLK 1
2 GNRPLVLSFGSCTuPPFLYKLDEFKQLVKDFSNVADFLIVYLAEAHAT 1
2 DAWAFKNNVDISVHKNLEERLAAARTLLKEDPPCPVVVDEMNNITASKYGALPERLYVIQSGKVIY 0
0 QASDLGGQA* 0
>DIO1_takRub Takifugu rubripes (fugu) genome glitches
0 MLLQKLAMYLSTAGLFCFMITLNVVLWILNIVAPALAKKIALKMGEKATMTQDPLFKYEDWGLTFASTALVKTASRHMWLSLGQEAFAGLEAPDSPVVTMERKRSSIGEFMK 1
2 TNRPLVLNFGSCTuPPFMFKLEEFKQLVRDFSDVADFLVVYIAEAHST 1
2 DGWAFKNNFDINQHRNLEDRLSAAQILVQKDPLCPVVVDDMNNSCAIKYGALPERLYVLQAGKVLYK 0
0 GAVGPWGYDPREVRSYLEKMK* 0
>DIO1_calMil Callorhinchus milii (elephantfish) weak match but introns ok
0 MWRRVVVYMQTVLLLVCICVRVAVGRVMLTLFPATTRRLELRNGLKTTMTLNPRFRFEDWGPSMFSLSSLRAVTTSIIANSGDRAFPGQPAPDTTLIDLDNTAHTIRSFIR 1
2 1
2 0
0 SGLGPWGYKPEEVRGVLHTLE* 0
>DIO1_braFlo Branchiostoma floridae (amphioxus) fused terminal exons frag
0 FTSTSFWTMWESEVMKDLQCETRVGGKAPDAPLLHMDGLTQARLMDFAMK 1
2 PGRPLVVNFGSCTuPPFMAHLAKFNDMVAEFADRADFLTVYIAEAHAS 1
2 RPHSSVTFTVFRKHQILEERLVAAKMLVDSNVSGGPLLVDTMDDVANQKYGAWPERLYILLDGRVMYK 00 GMPGPHGYHVHQVRDWLEKYLRNN* 0
DIO2: 27 vertebrate iodothyronine deiodinases
>DIO2_homSap Homo sapiens (human) iodothyronine deiodinase type II 2 exons chr14 uc001xuu.1 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGLRCVWKSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSSAEGGDNSGNGTQEKIAEGATCHLLDFASPERPLVVNFGSATuPPFTSQLPAFRKLVEEFSSVADFLLVYIDEAHPSDG WAIPGDSSLSFEVKKHQNQEDRCAAAQQLLERFSLPPQCRVVADRMDNNANIAYGVAFERVCIVQRQKIAYLGGKGPFSYNLQEVRHWLEKNFSKRuKKTRLAG* 0 >DIO2_macMul Macaca mulatta (rhesus) 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGLRCVWKSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSSAEGGDNSGHGTQEKIAEGAACHLLDFASPERPLVVNFGSATuPPFTSQLPAFRKLVEEFSSVADFLLVYIDEAHPSDG WAIPGDSSLSFEVKKHQNQEDRCAAAQQLLERFSLPPQCRVVADRMDNNANIAYGVAFERVCIVQRQKIAYLGGKGPFSYNLQEVRHWLEKNFSKRuKKTRLAG* 0 >DIO2_otoGar Otolemur garnettii (bushbaby) 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGLRCVWKSFLLDAYKQ 0 0 VKLGEEAPNSSVVHVSNPEKGDSSGNGAPENAAAECHLLDFASPERPLVVNFGSATuPPFTSQLPAFRKLVEEFSAVADFLLVYIDEAHPSDG WAVPGDSSLSFEVKKHRNQEDRCAAAHQLLERFSLPPQCRVVADRMDNNANVAYGVAFERVCIVQRQKIAYLGGKGPFFYNLQEVRRWLEKNFSKRuNRLAG* 0 >DIO2_tupBel Tupaia belangeri (tree_shrew) 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGLRCVWKSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSNLEGGHNGGNGTQEKTADGAECHLLDFANSERPLVVNFGSATuPPFTSQLPAFSKLLEESSVADFLVVYIDEAHPSDG WAVPGDSSLSFEVKKHRNQEDRCAAAHQLLERFSLPPQCRVVADRMDNNANVAYGVAFERVCIVQRQKIAYLGGKGPFYYNLQEVRRWLEKNFSKRuNSLAG* 0 >DIO2_ratNor Rattus norvegicus (rat) 0 MGLLSVDLLITLQILPVFFSNCLFLALYDSVILLKHVALLLSRSKSTRGEWRRMLTSEGLRCVWNSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSNPEAGNNCASEKTADGAECHLLDFACAERPLVVNFGSATuPPFTRQLPAFRQLVEEFSSVADFLLVYIDEAHPSDG WAVPGDSSMSFEVKKHRNQEDRCAAAHQLLERFSLPPQCQVVADRMDNNANVAYGVAFERVCIVQRRKIAYLGGKGPFSYNLQEVRSWLEKNFSKRuILD* 0 >DIO2_canFam Canis familiaris (dog) 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGMRCIWKSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSNSEGGDNSRNGAQVKIVDGAECHLLDFASPERPLVVNFGSATuPPFTSQLPAFSKLVEEFSSVADFLLVYIDEAHPSDGWAVPGD SSLSFEVKKHRNQEDRCAAAHQLLERFSLPPQCRVVADRMDNNANVAYGVAFERVCIVQRQKIAYLGGKGPFYYNLQEVRRWLEKNFSKRuNRLAG* 0 >DIO2_bosTau Bos taurus (cow) 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGQWRRMLTSEGMRCIWKSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSSPEGGDTSGNGAQEKTVDGTECHLLDFASPERPLVVNFGSATuPPFTNQLPAFSKLVEEFSSVADFLLVYIDEAHPSDGWAVPGD SSLFFEVKKHRNQEDRCAAAHQLLERFSLPPQCRVVADRMDNNANVAYGVAFERVCIVQRQKIAYLGGKGPFFYNLQEVRRWLEKNFSKRuKRLAG* 0 >DIO2_susScr Sus scrofa (pig) tga tag 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGMRCIWKSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSNPEGSNNHGHGTQEKTVDGAECHLLDFANPERPLVVNFGSATuPPFTSQLPAFSKLVEEFSSVADFLLVYIDEAHPSDG WAVPGDSSLSFEVKKHQNQEDRCAAAHQLLERFSLPPQCRVVADRMDNNANVAYGVAFERVCIVQRQKIAYLGGKGPFYYNLQEVRRWLEKNFSKRuKLD* 0 >DIO2_equCab Equus caballus (horse) 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGMRCIWKSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSNPDRGDNRGNGAQGKTVDGTECHLLDFASSERPLVVNFGSATuPPFISQLPAFSKLVEEFSSVADFLLVYIDEAHPSDG WAVPGDSSLSFEVKKHQNQEDRCAAAHQLLERFSLPPQCRVVADRMDNNANVAYGVAFERVCIVQRQKIAYLGGKGPFYYNLQEVRRWLEKNFSERuNKLG* 0 >DIO2_sorAra Sorex araneus (shrew) 0 MGNLSVHLLITLQILPVFFSNCLFLALYDSVILLKHVVLMLSRSKSTRGEWRRMLTSEGMRCIWKSFLLDAYKQ 0 0 VKLGEDAPNSSVVHVSSSEDGSRNGAHEKTVDGAECHLLDFASPERPLVVNFGSATuPPFTSQLPAFSKLVEEFSSVADFLLVYIDEAHPSDG WAVPRDSSLSFEVKKHRSQEDRCAAAHQLLERFSLPPQCRVVADRMDNNANVAYGVAFERVCIVQRQKIAYLGGKGPFCYNLQEVRDWLEKNFSKRuNSL* 0 >DIO2_echTel Echinops telfairi (tenrec) 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVLLLKHVMLLLSRSKSTCGEWRRLLTFEGLRCVWKSFLLDAYKQ 0 0 VKLGEDAPNSGVVHVSNFEGGGNRGPGAQEKTADGAQCHLLDFARAERPLVVNFGSATuPPFTSQLPAFSQLVEEFSSVADFLLVYIDEAHPSDG WAVPGDSSSSFEVKKHRNQEDRCAAAHRLLERFSLPPQCLVVADRMDNNANVAYGVAFERVCIVQQKIAYLGGKGPFCYNLQEVRCWLEKNFSKRuTRVAGQ* 0 >DIO2_dasNov Dasypus novemcinctus (armadillo) 0 MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGLRCVWKSFLLDAYKQ 0 0 VKLGEDAPNSGVVHVSNPEGGDNSGNSAQEKTEDGAECHLLDFASPERPLVINFGSATuPPFTSQLPAFSKLIEEFSSVADFLLVYIDEAHPSDG WAVPGGSSLSFEVKKHRNQEDRCAAVHKLLDRFSLPPQCHVVADRMDNNANIAYGVAFERVCIVQRQKIVYLGGKGPFCYNLQEVQHWLEKNFSKRuNRLAG* 0 >DIO2_monDom Monodelphis domestica (opossum) tga tag 0 MGLLSIDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGLRCVWKSFLLDAYKQ 0 0 VKLGGDAPNSSVIHITSAEVGSGQQNNSRWKRFDGAECHLLDFANPERPLVVNFGSATuPPFTSQLPAFSKLVEEFSTVADFLLVYIDEAHPSDG WAVPGNSSVSFEVKKHRNQEDRCAAAHQLLERFSLPPQCQVVADCMDNNANIAYGVSFERVCIVQRQKIAYLGGKGPFFYNLQEVRLWLEKNFSKRuNPG* 0 >DIO2_ornAna Ornithorhynchus anatinus (platypus) tga tga then tag 0 MGLLSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGEWRRMLTSEGLRCVWNSFLMDAYKQ 0 0 VKLGGDAPNSSVVHVANANGETSGGNSPKWKNFSGRYGAECHLLDFASSERPLVVNFGSATuPPFISQLPAFSKLVEEFSAVADFLLVYIDEAHPSDG WAVPGEFSLPFEVRKHQNQEDRCAAAHQLLERFSLPPQCQVVADCMDNNANVAYGVSFERVCIVQRQKIAYLGGKGPFFYNLQEVRLWLEQNFSKRuNPD* 0 >DIO2_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01A7SZJ 454 GAECHLLDFASSERPLVINFGSATuPPQPAAGLQQAGEEFSTVADFLLVYI >DIO2_galGal Gallus gallus (chicken) 2 exons 0 MGLLSADLLITLQILPVFFSNCLFLALYDSVILLKHMVLFLSRSKSARGEWRRMLTSEGLRCVWNSFLLDAYKQ 0 0 VKLGGEAPNSSVIHIAKGNDGSNSSWKSVGGKCGTKCHLLDFANSERPLVVNFGSATuPPFTSQLSAFSKLVEEFSGVADFLLVYIDEAHPSDG WAAPGISPSSFEVKKHRNQEDRCAAAHQLLERFSLPPQCQVVADCMDNNANVAYGVSFERVCIVQRQKIAYLGGKGPFFYNLQEVRLWLEQNFSKRuNPLSTEDLSTDVSL* 0 >DIO2_anoCar Anolis carolinensis (lizard) 0 MGLLSVDLLITLQILPVFFSNCLFLALYDSVILLKHMVLFLSRSKSARGEWRRMLTSEGLRCVWNSFLLDAYKQ 0 0 VKLGGEAPNSSVIHIAKGNDGSNSSWKSVGGKCGTKCHLLDFANSERPLVVNFGSATuPPFTSQLSAFSKLVEEFSGVADFLLVYIDEAHPSDG WAAPGISPSSFEVKKHRNQEDRCAAAHQLLERFSLPPQCQVVADCMDNNANVAYGVSFERVCIVQRQKIAYLGGKGPPFFYNLQEVRLWLEQNFRKRuNPNLPQEDMLNISPLK* 0 >DIO2_xenTro Xenopus tropicalis (frog) tga tag 0 MGLLSVDLLITLQILPGFFSNCLFLALYDSVVLVKHVLLQLSRSKSSQSQWRRMLTPEGLRCVWNSFLLDAYKQ 0 0 VKLGQDAPNSNVIQVSNKICKSVQRKLVGKCHLLDFASSERPLVVNFGSATuPPFISQLPAFSKLVEEFSSVADFVLVYIDEAHPSDG WAAPGTTSYEVKKHRNQEERCAAASKLLEHFSIPPQCQVVADCMDNNANVAYGVSFERVCIVQKQKIVYLGGKGPFFYNIQEIRRWLELSFGKRuT* 0 >DIO2_neoFor Neoceratodus forsteri (lungfish) exons unknown 0 MGLLSVDLLITLQILPWFFSNCLFLALYDSVVLLKHVILLLSCSKSSRGEWRRMLTSEGLRTVWNSFLLDAYKQ 0 0 VKLGGDAPNSKVVRVTSGCCRRRSFSGKGESECHLLDFASSNRPLVVNFGSATuPPFISQLPTFRKLVEEFSDVADFLLVYIDEAHPADG WAAPGVATKSFEVKKHRSQEERCVAAHKLLEHFSLPPQCQVVADCMDNNTNVAYGVSFERVCIVQRQKIAYLGGKGPFFYNLKEVRHWLEQTYRKRuVPTCELIM* 0 >DIO2_gasAcu Gasterosteus aculeatus (stickleback) cDNA taa stop 0 MGMASGGLRVTLQILPGFFSNCLFLALYDSVVLLKAVVSLLSCSRAAGRGARRRMLTSAGLRSVWRSFLLDAYKQ 0 0 VKVGLEAPNSKVVKVPGSPQRSSNLSNLISLPPGAMIANEVECHLLDFESSDRPLVVNFGSATuPPFISHLPAFRQLVEDFSDVANFLLVYIDEAHPSDG WVAPALGSCSFDVPKHQNLEERLGAARKLIEHFSLPPQCQLVADCMDNNANVAYGVSNERVCIVKQRKIAYLGGQGPFFNLKDVQHWLEQNFGKRFSRTSAEKDMSHISKKGIIHQ* 0 >DIO2_oryLap Oryzias latipes (medaka) tga stop 0 MGSAGAELLVTLQILPGFFSNCLFMALYDSVVLLKRVVSLLSCSRSVSCGEWRRMLTSAGLRAIWNSFLLDAYKQ 0 0 VKVPESPRWSSSIKSMTSVPRGARAQTGDECRLLDFESSDRPLVVNFGSATuPPFISHLPAFRQLVEDFSDVANFLLVYIDEAHPSDG WVAPQMGPCSFSFRKHQNLEERMGAARQLTEHFSLPPQCQLVADCMDNNANVAYGVSNERVCIVHQRKIAYLGGKGPFFYNLKEVRQWLEQLRQTVGPNTEE* 0 >DIO2_danRer Danio rerio (zebrafish) cDNA tga taa 0 MGLLSVDLLVTLQILPGFFSNCLFFVLYDSIVLVKRVVSLLSCSGSTGEWQRMLTTAGVRSIWNSFLLDAYKQ 0 0 VKLGEAAPNSKVVKVTGINRCWSISGKTHNQCHLLDFESPDRPLVVNFGSATuPPFISQLPVFRRMVEEFSDVADFLLVYIDEAHPSDG WVGPPMENFSFEVRKHRNLEERMFAARTLLEHFSLPPQCQLVADCMDNNANIAYGVSYERVCIVQKNKIAYLGGKGPFFYNLKDVRRWLEKCYGKu* 0 >DIO2_tetNig Tetraodon nigroviridis (pufferfish) tag 0 MGMASEDLLITLQILPGFFSNCLFLALYDSVVLVRRVVSRLSCSRSAGAKEWRPMLTSAGLRSIWNSFLLDAYKQ 0 0 VKVPNGPRWSSISNMLPGANLRNGIECHLLDFESSNRPLVVNFGSATuPPFISHLPAFRQLVEDFSDVADFLLVYIDEAHPSDG WEAPPMGPCSFNVRKHQNLEERLGAARKLIEHFSLPPQCQLVADCMDNNANVAYGVSNERVCIVQQRKIAYLGGKGPFFYNLKEVRQWLEHSYGKR* 0 >DIO2_takRub Takifugu rubripes (fugu) taa 0 MGMASEDLLITLQILPGFFSNCLFLALYDSVVLVRRVVSRLSCSRSAGPKEWRPMLTSAGLRSIWNSFLLDAYKQ 0 0 VKLGCEAPNSKLVKVPDGARWSSINNITNMLPGASLCNGIECHLLDFESSNRPLVVNFGSATuPPFISHLPAFRQLVEDFSDVADFLLVYIDEAHPSD GWKAPPMGPISFNVRKHQNLEERIGAAQKLIEHFSLPPQCQLVADCMDNNANVAYGVSNERVCIVQQRKITYLGGKGPFFYNLKEVRQWLEQSYGPHLI* 0 >DIO2_funHet Fundulus heteroclitis (mummichog) tag tag 0 MGSASEDLLVTLQILPGFFSNCLFLALYDSVVLVKRVVALLSRSRSAGCGEWRRMLTSEGLRSIWNSFLLDAHKQ 0 0 VKLGCEAPNSKVVKVPDGPRWSSTVVPCGSRIQTGGECRLLDFESSDRPLVVNFGSATuPPFISHLPAFRQLVEDFSDVADFLLVYIDEAHPSDG WVAPQMGACSFSFRKHQNLEERIGAARKLIEHFSLPPQCQLVADCMDNNANVAYGVANERVCIVHQRKIAYLGGKGPFFYSLKDVRQWLELSYGRR* 0 >DIO2_calMil Callorhinchus milii (elephantfish) no intron tga stop many extra aa subsequent 0 MGLLSMDLIMKLQILPGFSNCLFLAAYDSFVLLRQAVSLLSCSGLGPDPQHRMLTAEGMQVVWQSFLLDALKQ 0 0 VKVGLEAPNSAVARLDGGAPCRLLDFASRDRPLVVNFGSATuPPFVSRLPAFRQMVERYAEVADFLLVYVDEAHPSDGWALRSRFQLRRHRSQEER CSAAGLLAREFGLPAACGVVADLMDNNANRAYGVAFERLCVVQSQKIAYLGGKGPFFYNLNGVREWLERHSGQRWG* 0 >DIO2a_petMar Petromyzon marinus (lamprey) 0 MTAELNVSVFVALRILPGFFTNCLFLGLRDVLALLARSTRALFARHVPASCCPCPPEACRRVLTRAGMRAVWRSFLLDARRE 0 0 AQRAGDPAPNPRVALLADARSSSPAAAAAIPCRLQELSREGRPLVINMGSASuPPFVGRLPEFRRVVDDFSHAADFLLVYVEEAHPSDGWAVPGALQVSRARSLENNNSMA >DIO2b_petMar Petromyzon marinus (lamprey) frag 0 SRPVVDSLLILPGVFSNCLFLALYDAVSFLRRALQASLTHSAKGDAQHPRMLAGQGMLSVWRSYVLDAHKK VRLGGEAPNSSV RPSPPQPPPPQLRQAPPCRLLDFARAHRPLVVNFGSASuPPFVEQLGEFCDLVRDFADVADFLVVYIEEAHPSDAWPAPGGLEVPRHLALGDRCVAASQLRGLMPPLGRCPVVADAMDNNANIDYGVSYERLYVIQDG RIRYLGGKGPFFYRVREVKSFLESVKASR* 0
DIO3: 37 vertebrate iodothyronine deiodinases
>DIO3_homSap Homo sapiens (human) 1 exon chr14 0 MLRSLLLHSLRLCAQTASCLVLFPRFLGTAFMLWLLDFLCIRKHFLGRRRRGQPEPEVELNSEGEEVPPDDPPICVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVLPDGFQSQHILDYAQGNRPLVLNFGS CTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYIIPQHRSLEDRVSAARVLQQGAPGCALVLDTMANSSSSAYGAYFERLYVIQSGTIMYQGGRGPDGYQVSELRTWLERYDEQLHGARPRRV* 0 >DIO3_macMul Macaca mulatta (rhesus) 0 MLRSLLLHSLRLCAQTASCLVLFPRFLGTAFMLWLLDFLCIRKHFLGRRRRGQPEPEVELNSEGEEVPPDDPPICVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVLPDGFQSQRILDYAQGNRPLVLNFGS CTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYIIPQHRSLEDRVSAARVLQQGAPGCALVLDTMANSSSSAYGAYFERLYVIQSGTIMYQGGRGPDGYQVSELRTWLERYDEQLHGARPRRV* 0 >DIO3_musMus Mus musculus (mouse) 0 MLRSLLLHSLRLCAQTASCLVLFPRFLGTAFMLWLLDFLCIRKHFLRRRHPDHPEPEVELNSEGEEMPPDDPPICVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVRPDGFQSQRILDYAQGTRPLVLNFGS CTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYVIPQHRSLEDRVSAARVLQQGAPGCALVLDTMANSSSSAYGAYFERLYVIQSGTIMYQGGRGPDGYQVSELRTWLERYDEQLHGTRPHRF* 0 >DIO3_ratNor Rattus norvegicus (rat) 0 MLRSLLLHSLRLCAQTASCLVLFPRFLGTAFMLWLLDFLCIRKHFLRRRHPDHPEPEVELNSEGEEMPPDDPPICVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVRPDGFQSQRILDYAQGTRPLVLNFGS CTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYVIPQHRSLEDRVSAARVLQQGAPGCALVLDTMANSSSSAYGAYFERLYVIQSGTIMYQGGRGPDGYQVSELRTWLERYDEQLHGTRPRRL* 0 >DIO3_cavPor Cavia porcellus (guinea_pig) 0 MLRSLLLHSLRLCAQIASCLVLFPRFLGTAFMLWLLDFLCIRKHFLRRRGLGQPEPEAELNSEGEEVPPDDPPICVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVLPDGFQSQRILDYEHGNRPLVLNFGS CTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYIIPQHRSLEDRVSAARVLQQGAPGCALVLDTMANSSSSAYGAYFERLYVIQSGTIMYQGGRGPDGYQVSELRTWLERYDEQLHGPRHGRF* 0 >DIO3_canFam Canis familiaris (dog) 0 MLRSLLLHSLRLCAQTASCLVLFPRFLGTAFMLWLLDFLCIRKHFLGRRRRGQPEPAVELDSDGEELPPDDPPVCVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVLPNGFQNQHILDYARGNRPLVLNFGS CTuPPFMARMSAFHRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYSIPQHRSLEDRVSAAQVLQQGAPSCSLVLDTMANSSSSAYGAYFERLYVIQNGTVMYQGGRGPDGYQVSELRTWLERYDEQLHGAQPRRV* 0 >DIO3_felCat Felis catus (cat) 0 MLRSLLLHSLRLCAQTASCLVLFPRFLGTAFMLWLLDFLCIRKHFLGRRRRGQPEPAAELDSDGEEVPPDDPPVCVSDDNRLCTLASLKAVWHGQKuDIFKQAHEGGPAPNSEVVLPNGFQNQHILDYARGNRQLVLNFGR CTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYSIPQHRSLEDRVSAAEVLQQGAPSCSLVLDTMANSSSSAYGAYFERLYVVQNGTIMYQGGRGPDGYQVSELRTWLERYDAQLRGGQPRRV* 0 >DIO3_bosTau Bos taurus (cow) 0 MLRSLLLHSLRLCSQTASCLVLFPRFLGTAFMLWLLDFLCIRKHLLGRRRRGQPEIEVELNSDGEEVPPDDPPVCVSDDNRLCTLASLRAVWHGQKLDFFKQAHEGGPAPNSEVVLPDGFQNQHILDYARGNRPLVLNFGS CTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYSIPQHRSLEDRVSAARVLQQGAPECALVLDTMTNSSSSAYGAYFERLYIIQSGTIMYQGGRGPDGYQVSEVRTWLERYDEQLHGPQPRRV* 0 >DIO3_susScr Sus scrofa (pig) 0 MLHSLLLHSLRLCAQTASCLVLFPRFLGTACMLWLLDFLCIRKHLLGRRRRGEPETEVELNSDGDEVPPDDPPICVSDDNRLCTLASLRAVWHGQKLDFFKQAHEGGPAPNSEVVLPDGFQNQHILDYARGNRPLVLNFGS CTuPPFMARMSAFQRLVTKYQRDVDFLIIYIEEAHPSDGWVTTDSPYSIPQHRSLEDRVSAARVLQQGAPECSLVLDTMANSSSSAYGAYFERLYVIQSGTIMYQGGRGPDGYQVSELRTWLERYDQQLHGPQPRRV* 0 >DIO3_loxAfr Loxodonta africana (elephant) 0 MLRCMLLHSLRLCAQTASCIVLFPRFLGTAFMLWLLDFLCIRKHFLGRRRRGQPETEVELNSDGEEVPPDDPPISVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVLPDGFQSQRILDYVRGNRPLVLNFGS CTuPPFMARMSAFQRLVTKYRRDVDFLIIYIEEAHPSDGWVTTDSPYSIPQHRSLEDRVSAARVLQQGAPGCALVLDTMANSSSSAYGAYFERLYVIQNGTIVYQGGRGPDGYQVAKLRTWLERYDEQLHSAQTRRV* 0 >DIO3_echTel Echinops telfairi (tenrec) 0 MLRSPVLHSLVLCAQDAYSIVLFPCLLGTAFMQRQDFLCIRKHFLGLRRRGQPEPEVELNSDGEEVPPDDPPICVSDDNRLCTLASLKAVWHGQKLDFFKQAHEGGPAPNSEVVLPDGFQSQHILDYARGSRPLVLNFGS CTuPPFMARMSAFQRLVSKYRGEVDFLIIYIEEAHPSDGWVTTDSPYSIPQHRSLEDRVSAARVLQQGAPDCALVLDTMTNSSSSAYGAYFERLYVIQNGTVMFQGGRGPDGYQVAELRTWLQQYSEQQHRAQPRRV* 0 >DIO3_monDom Monodelphis domestica (opossum) tga taa 0 MRSRREGDTEEPRKAGKGGGGGMGDEEDGAAVIQSPGLPPDDPPVWVSDSNRLCTLESLKAVWHGQKMDFFKTARKGSLAPNPEVIQLDGLKRLRILDFARGSRPLVLNFGS CTuPPFMARLRAFQRLASTFLDIADFLLVYIEEAHPSDGWVSSDAAYDIPKHQCLQDRLRAARLLQEGVPGCLLAVDTMDNASSAAYGAYFERLYIIQDARVMYQGGRGPEGYKISELRHWLDQYKDRLQSPEASVVLPV* 0 >DIO3_ornAna Ornithorhynchus anatinus (platypus) DCLLVYIEEAHPSDGWVSSDAPYDIPKHRCLQDRLRAARLMQRGAPGCRLAVDTMDNAASAAYGAYFERLYVVQDARVVYQGGRGPEGYKISELRLWLEQYRLRLRGPGPATAAAAAVLDV* 0 >DIO3_galGal Gallus gallus (chicken) 0 MLHSLGAHTLQLLTQAAACILLFPRFLLTAVMLWLLDFLCIRKKMLTMPTAEEAAGAGEGPPPDDPPVCVSDSNRMFTLESLKAVWHGQKLDFFKSAHVGSPAPNPEVIQLDGQKRLRILDFARGKRPLILNFGS CTuPPFMARLRSFRRLAADFVDIADFLLVYIEEAHPSDGWVSSDAAYSIPKHQCLQDRLRAAQLMREGAPDCPLAVDTMDNASSAAYGAYFERLYVIQEEKVMYQGGRGPEGYKISELRTWLDQYKTRLQSPGAVVIQV* 0 >DIO3_xenTro Xenopus laevis (frog) 0 MLHCAGPHTGKLVKQVAACCLLLPRFLLTGLMLWLLDFQCIRRRVLLTAREESTAEHEDPPLCVSDSNRMCTVESLRAVWHGQKLDYFKSAHLGCSAPNTEVVMLEGRRLCKILDFSQGKRPLVVNFGS CTuPPFMARLQAYRRLAAQHVGIADFLLVYIEEAHPSDGWLSTDASYQIPQHQCLQDRLAAAQLMLQGAPGCRVVVDTMDNSSNAAYGAYFERLYIVLEGKVVYQGGRGPEGYKISELRMWLEQYQQGLMGTKGSGQVVIQV* 0 >DIO3_ranCat Rana catesbiana (bullfrog) 0 MLPAPHTCCRLLQQLLACCLLLPRFLLTVLLLWLLDFPCVRRRVIRGAKEEDPGAPEREDPPLCVSDTNRMCTLESLKAVWYGQKLDFFKSAHLGGGAPNTEVVTLEGQRLCRILDFSKGHRPLVLNFGS CTuPPFMARLQAYQRLAAQRLDFADFLLVYIEEAHPCDGWLSTDAAYQIPTHQCLQDRLRAAQLMLQGAPGCRVVADTMTNASNAAYGAYFERLYVILDGKVVYQGGRGPEGYKIGELRNWLDQYQTRATGNGALVIQV* 0 >DIO3_neoFor Neoceratodus forsteri (lungfish) 1 exon 0 MYQSSGVHTMNEVLKQAFACFILLPRFLVTALMLWLLDFLCVRRRVLLHMSRRQEASDLPDEPELCVSDSNRMFTLKSLRAVWHDQKLDFFKAAHIGLVAPNTEVIKLEGQRKAKILEFGGGKRPLILNFGS CTuPPFMARLKAFRGVATQYKDVADFLLIYIEEAHPSDGWVSTDAPYQIPKHQCLEDRLKAAQLMNLEIPGCLVVVDTMDNASNAAYGAFFERLYIVQQERVVYQGGRGPEGYKISELKNWLDQYKSQLQNSSAVVIQV* 0 >DIO3a_danRer Danio rerio (zebrafish) MEMLQGSAGVQSALKNAAVCVLLLPRFLLAALMLCLLDFLCIRRKLLLKMQEGAFSSPDDPPLRVSDSNKMFTLESLRAVWYGQKLDFFKSARLGGAAPNTEVFPLDGDARAAERILDYARGRRPLILNFGS CSuPPFMTRLSAFQRVARQYADIADSLLVYIEEAHPSDGWVSSDAPVQIPRHRCLEDRLRAAQMLHREAAGSAVVVDSMQNSCNAAYGAYFERLYIVKDATVVYQGGRGPEGYRIAELRDWLERYRSGLQESAVLHV* 0 >DIO3bb_danRer Danio rerio (zebrafish) 0 MNTGRALKNALVCLLILPRFLVAAFMLWCLDFLCVRKRVLVHLQERAEEEEEDAEEEEEPLCISDSSRMFSWESLKAVFHGHKLDYMKSARLGHAAPDSEVFPLAEPRRGRVLEFARGHRPLVLSFGS CSuPPFMRRLKAFRRLVLRYADVADALLIYIEEAHPSDGWRSSDAPHQIRRHRSLEERLSAARLMEREAPGCAVVADGMENAANSAYGAYFDRLYIVQDGRVVYQGGRGPEGFRISELRHWLDRYRERLRDAVIPV* 0 >DIO3bbb_danRer Danio rerio (zebrafish) bizarre ... matches opossum yet in both genomes VHTLGFLTQVAACCFLGPRFLATAVTLWLLDFLCIRKKVLMRHRREGDAEDPDDPPIWVSDSNRFCTIESLKAVWHGQKMDFFKTAHKGSPAPNPEVIQLDGLKRLRILDFARGSRPLVLNFGS CTuPPFTARLRAFQHLASTFLDIADFLLVYIEEAHPSDGWVTSDAAYDIPKHQCLQDRLRAARLLQEGAPGCLLAVDTMDNASSAAYGAYSERLYIIQEARVMYQGGRGPEGYKISELRHWLDQYKDRLQSPPASVVLPV* 0 >DIO3b_danRer Danio rerio (zebrafish) 0 GALKNALVCLLILTRFLVAAFMLWCLDFLCVRKRVLVHLQERAYAEQEEEPLCISDSSRMFSWESLKAVFHGHKLDYMKSARLGHAAPDSEVFPLAEPRRGRVLEFARGHRPLVLSFGS CSuPPFMRRLKAFRRLVLRYADVADALLIYIEEAHPSDGWRSSDAPHQIRRHRSLEERLSAARLMEREAPGCAVVADGMENAANSAYGAYFDRLYIVQDGRVVYQ* 0 >DIO3a_takRub Takifugu rubripes (fugu) 1 exon 0 MLDSGGVQMATALKHAALCLMLLPRFLLTAVMLWLLDFLCIRRKVLLQMGQRQKSPDDPPVCVSDSNRMFTLESLGAVWYGQKLDFFKSAHLGSAAPNTEVMLVQERRQVRILDCMKGKRPLILNFGS CSuPPFMTRLAAFQRVVRQYADIADFLVVYIEEAHPSDGWVSTDAPYQiPKHRCLEDRLRAAQLMLAEVPESNVVVDNMDNSSNAAYGAYFERLYIVRDERVVYQGGRGPEGYRISELRSWLEQYRNDVARSQTAVLHV* 0 >DIO3a_tetNig Tetraodon nigroviridis (pufferfish) 0 MLLPRFLLTAVALWLLDFLCIRRKVLLQMGQRQKSADDPPVCVSDSNKMFTLESLRAVWYGQKLDFLKSAHLGSAAPNTEVMLVQERRQVRILDCMKGKRPLVLNFGS CSuPPFMTRLAAFQRIVRQYADIADFLVVYIEEAHPSDGWVSTDAPHQIPKHRCLEDRLRAAQLMLAEVPESNVVVDNMDNSSNAAYGAYFERLYILRDERVVYQGGRGPEGYRISELRTWLEQYRNDVAESQTAVLHV * 0 >DIO3a_gasAcu Gasterosteus aculeatus (stickleback) 0 MHDSGGGVRTARALKHAALCLMLLPRFLLAAVMLWLLDFLCIRKVLLKMGERQDSPDDPPLCVSDSNRMFTLESLRAVWYGQKLDCLKAAHLGRAAPNTEVMLVQQRRRVPILDCTKGKRPLILNFGS CSuPPFMTRLAAFQRVVSQYADIADFLLVYIEEAHPSDGWVSSDAPYQIPKHRCLEDRLRAARLMLAEVPGSDVVVDNMDNSSSAAYGAYFERLYVVRDERVVYQGGRGPEGYRISELRAWLDEYRSELARSQTAVLHV* 0 >DIO3a_oryLap Oryzias latipes (medaka) 0 MDDSSGVQMARALKQAALCLMLLPRFLLAAVMLWLLDFLCIRRKLLLKMGERQDSPDDPPLCVSDSNKMFTLESLRAVWHGQKLDILKTAHLGQTAPNTEVVLVQERRQVRILDCMKGNRPLILNFGS CSuPPFMMRLAAFQRVVSQYADIADFLVVYIEEAHPSDGWVSSDAPHQIPKHRCLEDRLRAAALMLTEVPGSKVVVDNMDNSSNAAYGAYFERLYVVRDETVVYQGGRGPEEYRISELKTWLQQYRKELLHSQNAVLHV* 0 >DIO3a_pimPro Pimephales promelas (minnow) 0 MEMLHGSAGVQTAKALKNAAVCLMLLPRFLLAALTLWLLDFLCIRRKLLMKMRESDIASPDDPPMCVSDSNKMFTLESLRAVWYGQKLDFFKTARLGGAAPNTEVVPLDSTRLRGTRRILDYARGRRPLIVNFGS CSuPPFMTRLSAFQRVARQYADIADSLLVYIEEAHPSDGWVSSDAPYQIPRHRCLEDRLRAAELMNQKVPECAIVVDTMENSSNSAYGAYFERLYILKDEKVVYQGGRGPEGYRISELRDWLERYRNELEAS* 0 >DIO3a_ictPun Ictalurus punctatus (catfish) 0 MPDVQDFVKALKNALVSLMLLPRFLLTAVLLWFLDFVCIRRKVLVKMREREGSSPDDPSVTVSDSNRMFTVESLRAVWYSQKLDFCKTAHLGLTAPNSEVVPLGERKRARILDYARGSRPLILNFGS CSuPPFMTRLAAFRRVADQYADIADSLLIYIEEAHPSDGWVSTDAPYQIPRHRCIKDRLRAARLMTATVPGSIVVVDTMDNSSNAAYGAYFERLYVVKDEKVVYQGGRGPEGYRIFEL NWLEPYRQKARGSRPIVGHV* 0 >DIO3a_spaAur Sparus aurata (seabream) 0 MHDSGGVQMARALKHAALCLMLLPRFLLAAVMLWLLDFLCIRKKVLLKMGERQDGPDDPPVCVSDSNKMFTLESLRAVWYGQKLDFLKSAHLGRTAPNTEVMLVQERRQVRILDCMKGKRPLILNFGS CSuPPFMTRLAAFQRVVSQYADIADFLVVYIEEAHPSDGWVSSDAPYQIPKHRCLEDRLRAAQLMLAEVPGSNVVVDNMDNSSNAAYGAYFERLYIVRDERVVYQGGRGPEGYRISELRNWLEQYRNGLVNSQTAVLHV* 0 >DIO3a_parOli Paralichthys olivaceus (halibut) mRNA MVHDSGGVQMARALKHAALCLMLLPRFLLAAVMLWLLDFLCIRKKVLLKMGERQDGPDDPPVCVSDSNKMFTLESLRAVWYGQRLDFFKSAHLGRAAPNTEVVLVQEGRQVRILDCMKGKRPLILNFGS CSuPPFMTRLAAFQRVVSQYADIADFLVVYIEEAHPSDGWVSSDAPFQIPKHRCLEDRLRAAQLMLSEVPGGNVVVDNMDNSSNAAYGAYFERLYIVRDERVVYQGGRGPEGYQISGLRDWLEQYRSDLVNSKTPVLHV* 0 >DIO3b_takRub Takifugu rubripes (fugu) 1 exon N's 0 MNTIKSIKNALVFFVLLPRFLVAAVMFWLLDFLCIRKRVFFRMKEQEDDAVDPPLCISDSNRLFTVESLKAVWHGHKLDFLKAAHLGQGAPNTEVVQLEDQRRSRILDYAKDKRPLILNFGS CTuPPFMARLKAFRGSWSKTQTSRLCSCVIEEAHPSDGWVSTDAPYQIPKHRCLADRLGAAQLMRLEVPGCLIVVDSMENSSNVAYGAYFDRLYILQEGKIVYQGGRGPEGYRITELRDWLVEYRESLKSSNDLVIHL* 0 >DIO3b_tetNig Tetraodon nigroviridis (pufferfish) 0 MNTMKSIKNALVFFVLLPRFLMAAVMFWLLDFLCIRKRVFFRMKEQEDDAVDPPLCISDSNRLFTVESLKAVWHGHKLDFLKAAHLGQGAPNTEVVQLEDQRRSRILDYAKDKRPLILNFGS CTuPPFMARLKAFQGVVEQNADIADTVVVYIEEAHPSDGWVSTDAPYQIPKHRCLEDRLSAAQLMRLEVPGCLVVVDSMENSSNAAYGAYFDRLYILQEGKIVYQGGRGPEGYRITELRDWLEKYRKSLETSNNLVIHL* 0 >DIO3b_gasAcu Gasterosteus aculeatus (stickleback) 0 MNTIKAIKNAIVCLALLPRFLMAAFMFWLLDFLCIRRRVFFRMEQQEGDAIDPPLCMSDSNRLFSLESLKAVWHGHKLDFLKAAHLGHGAPNTEVVHLGDQRRGHILDYAKEKRPLVLNFGS CTuPPFMARLKAFQGVVQQNADIADSVVVYIEEAHPSDGWMSTDAPYQIPKHRCLEDRLNAAQLMHLEVPGCPLVVDSMENSSNAAYGAYFDRLYILQEGTIVYQGGRGPEGYRVTELRDWLDRYRQTLGKSSQLVMSV** 0 >DIO3b_oryLap Oryzias latipes (medaka) 0 MNTMKALKNAIVCLLLLPRFLLAAVVFWLLDFSCIRKRVFLRMKEQGGDATDPPLCISDSNRLFSVESLKAVWHGHKLDFLKAAHLGRGAPNTEVVHLEDQRCSRILDYAKDKRPLILNFGS CTuPPFMARLKAFQGVVQQNADIADSLVVYIEEAHPSDGWMSTDAPYQIPKHRRLEDRLNAAQLMHLEVPGCLVVVDSMENSSNAAYGAFFDRLYILQEGKVVYQGGRGPEGYRISELRDWLDQYRKGMEKSNNLVINM* 0 >DIO3_calMil Callorhinchus milii (elephantfish) note ATU not CTU 0 MKLLKEAMAVLVLLPRFIVTALTLWLLDILCIRKRLLCKLRARYEE AAELLGPGAGPPAPD HRMLTAEGMQVVWQSFLLDALKQVKVGLEAPNSAVARLDGGAPCRLLDFASRDRPLVVNFGS ATuPPFVSRLPAFRQMVERYAEVADFLLVYVDEAHPSDGWALRSRFQLRRHRSQEERCSAAGLLAREFGLPAACGVVADLMDNNANRAYGVAFERLCVVQSQKIAYLGGKGPFFYNLNGVREWLERHSGQRWG* 0 >DIO3_squAca Squalus acanthias (spiny_dogfish) note CTU 0 MNLLKEAVAVLILLPRFLVTALMLWLLDILCIRKRLLSKSREQTEGNADDPPLCVSDTNRMFTLESLKAIWHGQKLDFFKSAHVGSPAPNPEVVQLQEQRKVRLLDYSRGARPLILNFGS CTuPPFMARLKAFQRVAIQYADIADFLLVYIEEAHPSDGWVSTDAPYDIPRHRCLEDRLKAARLMHKENPSCLVVADTM >DIO3_chiPun Chiloscyllium punctatum (bambooshark) EU275162 835-1324 0 MDLLKEIVAILILLPRFFITAFMLWLLDILCIRKRLLPKRREQTEGSADDPPLCVSDTNRMFTLESVKAIWYGQKLDFFKSAHVGSPAPNTEVVQLQDQRKVRLLDYSRGARPLVLNFGS CTuPPFMARLKSFQRVATQYADIADFLLIYIEEAHPSDGWVSTDAPYNIPRHRSLEDRLKAASLIDKESPGCLVVADTMDNSSNSAYGAYFERLYVLRDQKVVYQGGRGPEGYKISELRLWLEQYKSQSHNSSTVLIEV* 0 >DIO3_braFlo Branchiostoma floridae (amphioxus) taa 1 exon closer match to DIO3 note YTU not CTU 10x trace coverage 0 MGCGHHSSLSPHSVERPNAGMLCEAVRPLKALAAPILFVVLCFAFFLLKTASIVLTFVAPAKKEDLLAKLLGREEQEQTSSFQYDIDEFLTWNSLKGLVYSQMVGIMKRARHGKPAPDPTLVLLDSVTEKTLLSFAVADRPLFVNFGS YTuPPFVPDLAVFDEIVAEFGERVDFLLVYIEEAHPTDGWSFRAGPDITSHQSIGQRCTAARAMLANRTVLYNVAVDSMTNSANYQYGAFPDRVYIIRHGQVVYEGGKGPFKYSLQEARTWLQENIGQQKN* 0
SELM1 (SEP15): 6 vertebrate sequences
>SELM1_homSap Homo sapiens (human) 0 MAAGPSGCLVPAFGLRLLLATVLQA 0 0 VSAFGAEFSSEACRELGFSSNLLCSSCDLLGQFNLLQLDPDCRGCCQEEAQFETKK 0 0 LYAGAILEVCGuKLGRFPQVQ 1 2 AFVRSDKPKLFRGLQIKYVRGSDPVLKLLDDNGNIAEELSILKWNTDSVEEFLSEKLERI* 0 >SELM1_susScr Sus scrofa (pig) 0 MAAEPGGWLGPALGLRLLLATALQM 0 0 VSAFGAEFSSESCRELGFSSNLLCGSCDLLGQFDLLQLDPDCRGCCQEEAQFETKK 0 0 LYAGAILEVCGuKLGRFPQVQ 1 2 AFVRSDKPKLFRGLQIKYVRGSDPVLKLLDDNGNIAEELSILKWNTDSVEEFLSEKLQR* 0 >SELM1_ornAna Ornithorhynchus anatinus (platypus) EG339469 0 MAAAEAAVMAPWLRLFLAATLQA 0 0 ISAYGTELSSEACRDLGFSSNLLCSSCDLLGQFSLTQLDPVCRGCCQEEAQFESRK 0 0 LYAGAILEVCGuKLGRFPQVQ 1 2 AFVRSDKPKLFRGLQIKYVRGSDPVLKLLDESGNIAEELSILKWNTDSVEEFLSEKLERI* 0 >SELM1_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01BKAWJ EUEMSW404CBHS5 454 0 LGFSSNLLCSSCDLLGQFSLTQLDPSCRGCCQEEAQFESRK 0 0 LYAGAILEVCGuKLGRFPQVQ 1 2 AFVRSDKPKLFRGLQIKYVHGSDPVLKLLDESGNIAEELSILKWNTDSVEEFLSEKLERI* 0 >SELM1_petMar Petromyzon marinus (lamprey) cDNA EG333213 tga-sel taa-stop full 0 MAAVRPGLLLLLLKAVS 0 0 VYATELTSEACRDLGFSSNLLCSSCDLLSQFGLDQLDPGCKRCCQLEVEESALK 0 0 LYPGAVLEVCGuKLGRFPQVQ 1 2 AFVRSNKPSTFKGLTIKYVRGSDPVLKLLDESGNVAEELSITKWNTDSVEEFLSEKLERL* 0 >SELM1_eptBur Eptatretus burgeri (hagfish) BJ647169 tga-sel taa-stop 0 FGTRLGAGSFLALCLVS 0 0 VSAAELSSEVCRDRGFSSGLVCSSCDLLAQFDLHRLDPDCRSCCQPEVEEQEIKR 0 0 YAGAVLEVCGuKLGRFPQVQ 1 2 AFVKSNKPSTFKGLTTK 0 0 YVRGADPVLKLLDQDGNVAEELSITKWNTDSVEEFLSENLERL* 0
SELM2: 22 vertebrate sequences
>SELM2_homSap Homo sapiens (human) NM_080430 Selenoprotein M (uc003ajq.1) chr22 0 MSLLLPPLALLLLLAALVAPATAATAYRPDWNRLSGLTRARVE 0 0 TCGGuQLNRLKE 0 0 VKAFVTQDIPFY 2 1 HNLVMKHLPGADPELVLLGRRYEELE 0 0 RIPLSEMTREEINALVQELGFYRKAAPDAQVPPEYVWAPAKPPEETSDHADL* 0 >SELM2_musMus Mus musculus (mouse) 0 MSILLSPPSLLLLLAALVAPATSTTNYRPDWNRLRGLARGRVE 0 0 TCGGuQLNRLKE 0 0 VKAFVTEDIQLY 2 1 HNLVMKHLPGADPELVLLSRNYQELE 0 0 RIPLSQMTRDEINALVQELGFYRKSAPEAQVPPEYLWAPAKPPEEASEHDDL* 0 >SELM2_ratNor Rattus norvegicus (rat) 0 MNILLSPPPLLLLLAALVAPATSITTYRPDWNRLRGLARGRVE 0 0 TCGGuQLNRLKE 0 0 VKAFVTQDIQLY 2 1 HNLVMKHLPGADPELVLLSRNYQELE 0 0 RIPLSQMTRDEINALVQELGFYRKSAPEAKVPPEYLWAPAKPPEDASDRADL* 0 >SELM2_canFam Canis familiaris (dog) 0 MRLPLPPPPLLLLLAALAAAVTTFRPDWNRLHGLARARVE 0 0 TCGGuQLNRLKE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHLPGADPELVLLGHHYEELE 0 0 RIPLSEMTREEINELVQELGFYRKAAPDEAVPPEYLRAPARPAEGAPDRADL* 0 >SELM2_bosTau Bos taurus (cow) 0 MHLPLPPPPLLLLLAAVAAATTTFRPDWNRLQGLARARVE 0 0 TCGGuQLNRLKE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHLPGADPELVLLGHRFEELE 0 0 RIPLSDMTREEINALVQELGFYRKASPDEPVPPEYLRAPARPAGDAPDHADL* 0 >SELM2_susScr Sus scrofa (pig) 0 MHLPPLSLPLLLLLAALAAATTTFRPDWNRLQGLARARVE 0 0 TCGGuQLNRLKE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHLPGADPELVLLGHRFEELE 0 0 RIPLSDMTREEINALVQELGFYRKAAPDDPVPPEYMRAPARPAEGAPDRADL* 0 >SELM2_ornAna Ornithorhynchus anatinus (platypus) 0 MLVCPERRSWVLIPPLSLLLLLPGLLAAFQPDWSRLQGLARGKVE 0 0 TCGGuQLNRLKE 0 0 VKAFVTEDIPLY 2 1 HNLVMKHLPGADPELVLLNFRYEELE 0 0 RIPLSHMTRAEINQLVQDLGFYRKAERDAPVPPEFQQAPAKTSDLREKVQPQETPKSEEQNHPDL* 0 >SELM2_galGal Gallus gallus (chicken) 0 MRRAALAALLLLLAAAAGIERRPPRGLARGKVE 0 0 TCGGuRLSRLPE 0 0 VKAFVSQDIPLY 2 1 HNLEMKHLPGADPELVLLSFRYEELE 0 0 RIPLSDMTREEINQLVQELGFYRKETPEAPVPEEFQFAPAKPLPTLTPRRAPAADGKTLSEQDKKDHPDL* 0 >SELM2_botIns Bothrops insularis (snake) PVPDAFQMA ..PLLWLPLLLLGLLSAVAPLRAVQLDRSRLQWLARGKVE 0 0 SCGGuRLNRLPE 0 0 VKAFLNEDLPLY 2 1 HNMDLKYLAGADPELILLNIKFEELQ 0 0 RIPLSDMSREEINQLMQELGFYRKDTPDSLFRCFPNGAC* 0 >SELM2_xenTro Xenopus laevis (frog) 0 MWLPLPLLLGLLQLQPILSYQIDWNKLERINRGKVE 0 0 SCGGuQLNRLKE 0 0 VKGFVTEDLPLY 2 1 HNLEMKHIPGADPELVLITSRYEELE 0 0 RIPLSDMKRDEINQLLKDLGFYRKSSPDAPVPAEFKMAPARASGDTKEDL* 0 >SELM2_danRer Danio rerio (zebrafish) 0 MWPLIFTALLPSVILTYEVNIEKLSGLARARVE 0 0 TCGGuQLNRMRE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHIPGADPELVLLNHYYEELD 0 0 RIPLSEMTRAEINKLLAELGFYKKDHPEDQVPEEFRFSPAKDSPFEGRQSSTAAPETTEPSDSQHTDL* 0 >SELM2_ictPun Ictalurus punctatus (catfish) 0 MFSTWPLLWAAFLPCISLAYEVDWKKLDGLARAKVE 0 0 SCGGuQLNRLRE 0 0 VKAFVTQDIPFY 2 1 HNLVMKHIPGADPELVLLNHYYEELD 0 0 RIPLSHMTRTDINGLLEELGFYKKARAEDDVPEEFRFSPAKDSPFKEHHTRRAPANSDLAQEPQPENESPHKDL* 0 >SELM2_oncMyk Oncorhynchus mykiss (trout) 0 MWFFFFVSLLNCVSAYDVDLKKLDGLAKAKVE 0 0 SQSCGGuQLNRLRE 0 0 VKAFVTQDIPLY 2 1 HNLVMKHIPGADPELVLLNHYYEELD 0 0 RIALSDMTRSEINELLEKLGFYKKAQAEDQVPEEFRFSPAKDSPFKATPADNASSDSDAEAKHSDL* 0 >SELM2_strPur Strongylocentrotus purpuratus (sea_urchin) tga confirmed bad assembly 0 MAKERLFAAILGIVFLAGIHVSAEGDGQVMSARVE 0 0 SCTGuRLNKLPEVKQFINEDIPYY 2 1 HNVNFKPVPGANPDLLLLDESGNIVR 0 0 RIDLSKMKRAELNAMMTEELGFYKKGNLAEDVPDEHSTGPYSSKKEL* 0
MSRB1: 17 vertebrate sequences
>MSRB1_homSap Homo sapiens (human) SEPX1 (uc002cng.1) SELX SELR human chr16 0 MSFCSFFGGEVFQNHFEP 1 2 GVYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPEHNRSEALK 0 0 VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK 1 2 GKETSASQGH* 0 >MSRB1_macMul Macaca mulatta (rhesus) 0 MSFCSFFGGEVFQNHFEP 1 2 GVYACAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPEHNRPGALK 0 0 VSCGKCGNGLGHEFLNDGPKPGQSRFuIFSSSLKFVPK 1 2 GKETSTSQGH* 0 >MSRB1_musMus Mus musculus (mouse) exon break just at stop codon NP_038787 0 MSFCSFFGGEVFQNHFEP 1 2 GVYVCAKCSYELFSSHSKYAHSSPWPAFTETIHPDSVTKCPEKNRPEALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFVPK 1 2 GKEAAASQGH* 0 >MSRB1_ratNor Rattus norvegicus (rat) 0 MSFCSFFGGEVFQNHFEP 1 2 GVYVCAKCGYELFSSRSKYAHSSPWPAFTETIHEDSVAKCPEKNRPEALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 2 GKEAPASQGD* 0 >MSRB1_cavPor Cavia porcellus (guinea_pig) 0 MSFCSFFGGEVFQNHFES 1 2 GIYVCAKCGYELFSSRSKYAHSSPWPAFTDTIHADSVAKCPEHNRPGALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 2 DKETSASQGH* 0 >MSRB1_canFam Canis familiaris (dog) frag 0 MSFCSFFGGEVFQNHFEP 1 2 GVYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPERNSPEALK 0 0 VSCGKCGNGLGHEFLNDGPKPGKSRFuIFSSSLKFVPK 1 2 GKGTSGSQEA* 0 >MSRB1_bosTau Bos taurus (cow) 0 MSFCSFFGGEIFQNHFEP 1 2 GIYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPEHNRPGAIK 0 0 VSCGRCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 2 AEETSASQGQ* 0 >MSRB1_equCab Equus caballus (horse) 0 MSFCSFFGGEIFQNHFEP 1 2 GIYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVAKRPEHNRPEALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFVPK 1 2 GKESSASQGQ* 0 >MSRB1_loxAfr Loxodonta africana (elephant) 0 MSFCSFFRSEVFQNHFEP 1 2 GVYVCAKCGYELFSSRSKYAHSSPWPAFTETIHADSVGKHPEHNRPEALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 2 GKETSASQGK* 0 >MSRB1_monDom Monodelphis domestica (opossum) frag 2 GVYVCAKCGYELFSSRSKYHHSSPWPAFTETIHADSVSKRPESGRSEALK 0 0 VSCGKCGNGLGHEFINDGPKKGQSRFuIFSSSLKFVPKG 1 >MSRB1_ornAna Ornithorhynchus anatinus (platypus) 0 1 2 GTYVCARCGYELFSSRSKYEHSSPWPAFTETIHPDSVAKREEPGRPNAFK 0 0 VSCGKCGNGLGHEFLNDGPRRGQSRFuIFSSLKFIPK 1 2 GKDSQAAQDK* 0 >MSRB1_galGal Gallus gallus (chicken) 0 MSFCSFFGGEVFKDHFEP 1 2 GVYVCARCGYELFSSRAKYEHSSPWPAFTETIHEDSVAKRKERPGALK 0 0 VSCGKCGNGLGHEFLNDGPKRGQSRFuIFSSSLKFIPK 1 0 GKSPQEN* 0 >MSRB1_anoCar Anolis carolinensis (lizard) 0 MSFCAFSGGEIYQGHFEA 1 2 GMYVCSKCGFELFSSKSKYAHSSPWPAFTETIHDDSITKYLERPNAFK 0 0 VLCGKCGNGLGHEFINDGPKKGQSRFZIFSSSLKFVPK 1 >MSRB1_xenTro Xenopus tropicalis (frog) 0 MSFCSFFGGEVYKDHFKS 1 2 GIYVCSECNYELFSSRSKYQHSSPWPAFTETVHKDSISKYLERPNAYK 0 0 VSCGKCGNGLGHEFINDGPKKGQSRFuIFSSSLKFIPK 0 0 DKVDGEVQRE* 0 >MSRB1_danRer Danio rerio (zebrafish) tga confirmed 0 MSFCSFSGGEIYKDHFES 1 2 GMYVCAQCGYELFSSRSKYEHSSPWPAFTETIHEDSVSKQEERWGAYK 0 0 VRCGKCGNGLGHEFVNDGPKHGLSRFuIFSSSLKFI 0 PKVKNEQQ* 0 >MSRB1_ictFur Ictalurus furcatus (fish) 0 MAFCSFKGGEIFKDHYEP 1 2 GIYVCVKCGYELFSSTSKYKLSSPWPAFTTTIHEDSVSKQEERPGALK 0 0 IRCGKCNNGLGHEFLNDGPKHGLSRFuIFSSSLKFV 0 0 PKDKGGQ* 0 >MSRB1_oncMyk Oncorhynchus mykiss (trout) 0 MSFCSFFGGEVFKDHFKT 1 2 GLYMCAQCGHQLFSSRSKYEHSSPWPAFTETILQDSVSKHEERPGAFK 0 0 VRCGKCGNGLGHEFVGDGPKKGLSRFuIFSSSLKFV 0 0 PKDKVDGQ* 0 >MSRB1_salSal Salmo salar (salmon) 0 MSFCSFFGGEVFKDHFKT 1 2 GLYVCAQCGHQLFSSRSKYEHSSPWPAFTETVLQDSVSKHEERPGAFK 0 0 VRCGKCGNGLGHEFVGDGPKKGLSRFuIFSSSLKFV 0 0 PKDKVDGQ* 0
MSRB2: 6 vertebrate sequences
>MSRB2_homSap Homo sapiens (human) CBS-1 PILB 5 exons chr10 0 MARLLWLLRGLTLGTAPRRAVRGQAGGGGPGTGPGLGEA 1 2 GSLATCELPLAKSEWQKKLTPEQFYVTREKGTEP 0 0 PFSGIYLNNKEAGMYHCVCCDSPLFS 2 1 SEKKYCSGTGWPSFSEAHGTSGSDESHTGILRRLDTSLGSARTEVVCKQ 0 0 CEAHLGHVFPDGPGPNGQRFCINSVALKFKPRKH* 0 >MSRB2_musMus Mus musculus (mouse) 0 MARLLRALRGLPLLQAPGRLARGCAGS 1 2 GSKDTGSLTKSKRSLSEADWQKKLTPEQFYVTREKGTEA 0 0 PFSGMYLNNKETGMYHCVCCDSPLFSSEKKYCSGTGWPSF 2 1 SEAYGSKGSDESHTGILRRLDTSLGCPRMEVVCKQ 0 0 CEAHLGHVFPDGPKPTGQRFCINSVALKFKPSKP* 0 >MSRB2_bosTau Bos taurus (cow) 0 MARLLRALRGLTLREAPGWAVRGRADCGGFRAGA 1 2 GSPQAAHPDTFPFHRGSKSEWQKKLTPEQFHVTREKGTEP 0 0 PFSGIYLNNKEPGMYHCVCCDSPLFS 2 1 SEKKFCSGTGWPSFSEAHGTSGSDESNTGILRRADTSLGPARTEVVCKQ 0 0 CEAHLGHVFPDGPGPAGQRFCINSVALRFKPRKH* 0 >MSRB2_canFam Canis familiaris (dog) 0 MGDSSTTLRFLIRKRHSPYDFVVKIDKSKPKEERSLTKHEL 0 0 PLTKSEWQKKLTPEQFYVTREKGTEP 0 0 PFSGVYLNNKESGMYHCVCCDSPLFS 2 1 SEKKYCSGTGWPAFSEAHGTSGSDERDTGILRRVDTSLGLTRTEVVCKQ 0 0 CEAHLGHVFPDGPGPSGQRFCINSVALKFKPRKH* 0 >MSRB2_ornAna Ornithorhynchus anatinus (platypus) 0 0 0 DWKKKLTPEQYCVTRGKGTEP 0 0 PFSGIHLNNTERGMYHCLCCDMPLFS 2 1 SEKKFCSGTGWPSFLEAHGTQGTDESQTGILRRPDHSLGLARTEVVCKR 0 0 CEAHLGHVFDDGPPPTGQRFCINSVALTFKPS* 0 >MSRB2_xenTro Xenopus laevis (frog) 0 MSRLLTGFRLLLRVREISFPSVTAQRLQAWSRVRMAQTGADL 1 2 GSLTRYDDSAVSTDWQKKLTPEQYYVTREKGTEL 0 0 PFSGIYLNNTEKGMYHCVCCSAPLFS 2 1 SEKKYNSGTGWPSFSEAYGAQGADESNTNVLRRLDNSLGSTGTEVICKE 0 0 CDAHLGHVFEDGPPPYGQRFCINSVALTFAPSSM* 0
MSRB3: 5 vertebrate sequences
>MSRB3_homSap Homo sapiens (human) ER retention signal KAEL* AY358229 0 MSPRRTLPRPLSLCLSLCLCLCLAAALGSAQS 1 2 GSCRDKKNCKVVFSQQELRKRLTPLQYHVTQEKGTES 2 1 AFEGEYTHHKDPGIYKCVVCGTPLFK 2 1 SETKFDSGS 1 2 GWPSFHDVINSEAITFTDDFSYGMHRVETSCSQ 0 0 CGAHLGHIFDDGPRPTGKRYCINSAALSFTPADSSGTAEGGSGVASPAQADKAEL* 0 >MSRB3_musMus Mus musculus (mouse) 0 MSAFNLLHLVTKSQPVAPRACGLPS 1 2 GSCRDKKNCKVVFSQQELRKRLTPLQYHVTQEKGTES 2 1 AFEGEYTHHKDPGIYKCVVCGTPLFK 2 1 SETKFDSGS 1 2 GWPAFHDVISSEAIEFTDDFSYGMHRVETSCSQ 0 0 CGAHLGHIFDDGPRPTGKRYCINSASLSFTPADSSEAEGSGIKESGSPAAADRAEL* >MSRB3_canFam Canis familiaris (dog) 0 MSAFNLLHLVTKSQPVALRACGLPS 1 2 GSCRDKKNCKVVFSQQELRKRLTPLQYHVTQEKGTES 2 1 AFEGEYTHHKDPGIYKCVVCGTPLFR 2 1 SESKFDSGS 1 2 GWPSFHDVISSDAITFTDDFSYGMHRVETSCSQ 0 0 CGAHLGHIFDDGPRPTGKRYCINSASLAFTPAGGGTQGSSGPGGPAAGGRAEL* >MSRB3_ornAna Ornithorhynchus anatinus (platypus) (uc001ssm.1 0 MSAFNLLHLVTKSQPGSLQACGLPS 1 2 GSRRDKRSCKVVFSQEELRKRLTPLQYHVTQEKGTES 2 1 AFEGEFTHHKAPGTYKCVVCGTPLFKSETKFDSGS 1 1 1 2 GWPSFYDVIGSDAITLTDDFSYGMHRVETSCSQ 0 0 CGAHLGHIFEDGPRPTGKRYCINSASLSFAAVEENIDKENGSGDSPIQPGKTEL* >MSRB3_petMar Petromyzon marinus (lamprey) cDNA FD728382 tga-sel taa-stop full 0 MSWAARLLGCAAVRAAGRAFGSVHARRHIALSTTLV 1 2 ASCKDTRTCRVNFAEEELRKKLSPLEFHVTQEKGTEP 1 2 AFTGKFAEHKGKGTYGCVVCQAPLFA 1 2 GWPSFYNIIRANAVSHSKDRSHGMHRTEVTCGQ 0 0 CGAHLGHVFDDGPPPTGKRFCINSASLNFNPSAEEEGGAEAVEEAPVKPELuGIGGIV* 0
GPX3: 33 chordate sequences
>GPX3_homSap Homo sapiens (human) chr5 5 exons plasma glutathione peroxidase 3 0 MARLLQASCLLSLLLAGFVSQSRGQEKSK 0 0 MDCHGGISGTIYEYGALTIDGEEYIPFKQYAGKYVLFVNVASYuGLTGQYI 1 2 ELNALQEELAPFGLVILGFPCNQFGKQEPGENSEILPTLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELLGTSDRLFWEPMKVHDIRWNFEKFLVGPDGIPIMRWHHRTTVSNVKMDILSYMRRQAALGVKRK* 0 >GPX3_panTro Pan troglodytes (chimp) 0 MARLLQASCLLSLLLAGFVSQSRGQEKSK 0 0 MDCHGGISGTIYEYGALTIDGEEYIPFKQYAGKYVLFVNVASYuGLTGQYI 1 2 ELNALQEELAPFGLVILGFPCNQFGKQEPGENSEILPTLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELLGTSDRLFWEPMKVHDIRWNFEKFLVGPDGIPIMRWHHRTTVSNVKMDILSYMRRQAALGVKRK* 0 >GPX3_macMul Macaca mulatta (rhesus) 0 MARLLQASCLLSLLLAGFLPQSRGQDKSK 0 0 MDCHGGVSGTIYEYGALTIDGEEYIPFKQYIGKYVLFVNVASYuGLTGQYI 1 2 ELNALQEELAPFGLVLLGFPCNQFGKQEPGENSEILPSLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELLGTSDRLFWEPMKVHDIRWNFEKFLVGPDGIPVMRWHHRTTISNVKMDILSYMRRQAALGVKRK* 0 >GPX3_otoGar Otolemur garnettii (bushbaby) 0 MARLLRASCLLSLLLAGFVPPGRGQEKSK 0 0 TDCHGGMSGTVYEYGALTIDGEEYIPFKQYTGKYILFVNVASYuGLTGQYV 1 2 ELNALQEELAPFGLVILGFPSNQFGKQEPGENSEILATLK 2 1 HVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELLGSPGRLFWEPMKVHDIRWNFEKFLVGPDGIPVMRWHHRTTVSNVKMDILSYMRRHAAQGGTGK* 0 >GPX3_tupBel Tupaia belangeri (tree_shrew) 0 MARLLRASCLLSLLLAGFVPPGRGQEKSK 0 0 TDCHGGVSGTIYEYGALTLDGEEYIPFKQYAGKYILFVNVASYuGLTGQYI 1 2 ELNALQEELAPFGLVILGFPCNQFGKQEPGENSEILPTLK 2 1 YVRPGGGFVPNFQLFEKGDVNGDKEQKFYTFLK 0 0 NSCPPTSELLGTPSRLFWEPMKIHDIRWNFEKFLVGPDGIPIMRWYHRTTISNVKMDILAYMRRQAALGLRGK* 0 >GPX3_musMus Mus musculus (mouse) 0 MARILRASCLLSLLLAGFVPPGRGQEKSK 0 0 TDCHGGMSGTIYEYGALTIDGEEYIPFKQYAGKYILFVNVASYuGLTDQYL 1 2 ELNALQEELGPFGLVILGFPSNQFGKQEPGENSEILPSLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTAELLGSPGRLFWEPMKIHDIRWNFEKFLVGPDGIPVMRWYHRTTVSNVKMDILSYMRRQAALSARGK* 0 >GPX3_ratNor Rattus norvegicus (rat) 0 MARILRASCLLSLLLAGFVPPGRGQEKSK 0 0 TDCHGGMSGTIYEYGALTIDGEEYIPFKQYAGKYILFVNVASYuGLTDQYL 1 2 ELNALQEELGPFGLVILGFPCNQFGKQEPGENSEILPSLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTAELLGSPGRLFWEPMKIHDIRWNFEKFLVGPDGIPIMRWYHRTTVSNVKMDILSYMRRQAALGARGK* 0 >GPX3_cavPor Cavia porcellus (guinea_pig) 0 MARLLRASCLLPLLLAGLAPPGRGQEKAK 0 0 TDCHSAVSGTIYEYGALTIDGEEYIPFNKYRGKYILFVNVASYuGLTDQYL 1 2 ELNALQEELAPFGLVILGFPCNQFGKQEPGENSEILPSLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELVGSPNRLFWEPMKVHDIRWNFEKFLVGPDGRPIMRWYHRTTVSNVKMDILACMRRQASLMALGN* 0 >GPX3_oryCun Oryctolagus cuniculus (rabbit) 0 MARLLRASCLLSLLLAGFVPPGRGQEKSK 0 0 TDCHDGVSSTIYEYGALTIDEKEYIPFKQYADKYVLFVNVATYuGLTGQYV 1 2 ELNALQEELAPFGLVILGFPCNQFGKQEPGENSEILPALK 2 1 YVRPGGGFVPNFQLFEKGDVNGDKEQKVYTFLK 0 0 NSCPPTSELLGSPNRLFWEPMKMHDVRWNFEKFLVGPDGVPIMRWYHRATVSNVKMDILAYMRRQAAMGAKGK* 0 >GPX3_ochPri Ochotona princeps (pika) 3rd exon again ends in potential PALKu 0 0 0 1 tDCHGGMSGTIYDYGALTIDGEEYIPFKQYAGKYVLFVNVATYuGLTGQYV 1 2 ELNALQEEFAPFGLVILGFPCNQFGKQEPGTNSEILPALK 0 0 HVRPGGGFVPNFQLFEKGDVNGDKEQKVYTFLK 0 0 NSCPPTSELLGSHSSLFWDPMKNHDIRWNFEKFLVGPDGVPIMRWHHRTTISNVKMDILTYMRRQAALGVRGK* >GPX3_canFam Canis familiaris (dog) 0 MARLLRASCLLSLLLAGFVPPSRGQEKSK 0 0 TDCHGGVSGTIYEYGALTINGEEYIPFKQYAGKYILFVNVASYuGLTGQYL 1 2 ELNALQEELAPFGLVILGFPCNQFGKQEPGENSEILPSLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELLGSPGRLFWEPMKVHDIRWNFEKFLVGPDGIPIMRWYHRTTVSTVKMDILAYMRRQAALAIKGK* 0 >GPX3_felCat Felis catus (cat) 0 MARLLRASCLLSLLLAGFVPPSRGQEKSK 0 0 TDCHAGGSGTIYEYGALTIDGEEYIPFKQYAGKYILFVNVASYuGLTGQYV 1 2 ELNALQEELAPFGLVILGFPCNQFGKQEPGENSEILPSLK 2 1 HVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELLGSPNRLFWEPMKVHDIRWNFEKFLVGPDGVPLMRWYHRTTVSTVKMDILAYMRRQAALTVNGK* 0 >GPX3_equCab Equus caballus (horse) 0 MARLLRASCLLSLLLAGFVPPSRGQEKSK 0 0 TDCHAGVSGTIYEYGALTIDGEEYIPFKQYAGKYILFVNVASYuGLTGQYV 1 2 ELNALQEEFAPFGLVILGFPSNQFGKQEPGENSEIPLTLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NACPPTSELLGSPDRLFWEPMKIHDIRWNFEKFLVGPDGKPIMRWYHRTTVNTVKMDILAYMRREAALAARGK* 0 >GPX3_myoLuc Myotis lucifugus (microbat) frag 0 0 0 TDCHAGVSGTIYEYGALTLNGEEYIPFKQYAGKYVLFVNVASFuGLTSQYV 1 2 ELNALQEELQPFGLVILGFPCNQFGKQEPGENSEILPILK 2 1 VRPGGGFVPNFQLFEKGDVNGEKEQKVYTFLK 0 1 * 0 >GPX3_pteVam Pteropus vampyrus (macrobat) 0 VDCHAGMSGTIYDYGALTIDGEEYIPFKQYAGKYVLFVNVATYuGLTGQYI 1 >GPX3_bosTau Bos taurus (cow) 0 MARLFRASCLLSLLLAGFIPPSQGQEKSK 0 0 TDCHAGVGGTIYEYGALTIDGEEYIPFKQYAGKYILFVNVASYuGLTGQYV 1 2 ELNALQEELEPFGLVILGFPCNQFGKQEPGENSEILATLK 2 1 YVRPGGGFTPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELLGSPDRLFWEPMKVHDIRWNFEKFLVGPDGIPIMRWYHRTTVNSVKMDILTYMRRRAVWEAKGK* 0 >GPX3_oviAri Ovis aries (sheep) 0 MARLLRASCLLSLLLAGFIPPSQGQEKSK 0 0 MDCHAGVGGTIYEYGALTIDGEEYIPFKRYAGKYILFVNVASYuGLTGQYV 1 2 ELNALQEELEPFGLVILGFPCNQFGKQEPGENSEILATLK 1 2 YVRPGGGFTPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELLGSPGRLFWEPMKVHDIRWNFEKFLVGPDGIPIMRWYHRTTVNSVKMDILTYMRRRAVLEARGSN* 0 >GPX3_eriEur Erinaceus europaeus (hedgehog) 0 MARLLRVSCLLPLLLAGLVSPGQGQEKSK 0 0 TNCHPDVSGTIYDYGALTIDGEEYIPFKQYAGKYVLFVNVASYuGLTGQYI 1 2 ELNALQEELGPFGLVILGFPSNQFGKQEPGENSEILATLK 2 1 HVRPGGGFVPKFQLFEKGDVNGDKEQKVFTFLK 0 0 NSCPPTSELLGTSGRLFWEPMVHDIPSNSVNVLVGPKGINIIYLYHRITFTNVEWDILCYMNPQPSMGVKGQL* 0 >GPX3_dasNov Dasypus novemcinctus (armadillo) 0 MALRAPCLLLLLLAGLVSPSQQEKSR 0 0 TDCHGGTSGSIYEYGALTLNGEEYIPFKQYAGKYILFVNVASFuGLTSQYL 1 2 ELNALQEELAPFGLVVLGFPCNQFGQQEPGENSEILPTLR 2 1 YVRPGQGFVPNFQLFEKWDVNADKEHKIYTFLN 0 1 NSCPPTSELLGTTDRLFWEPMKVHDIRWNFEKFLVGPDGKPIMRWYHRTMVSTVKMDILAHMRKQAALGVERK * 0 >GPX3_choHof Choloepus hoffmanni (sloth) 0 MARLLRASALLSLLLDGIVPPSLGQEKSR 0 0 1 2 ELNALQEEFAPFGLVVLGFPCNQFGHQEPGENSEILPILK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 1 * 0 >GPX3_loxAfr Loxodonta africana (elephant) 0 MARLLRASCLLSLLLAGFVPPSWGQEKSK 0 0 TDCHGGMSGTIYEYGALTIDGEEYIPFKQYAGKYILFVNVASYuGLTGQYI 1 2 ELNALQEELAPFGLVILGFPCNQFGKQEPGENSEILASLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLKV 0 1 NSCPPTSELMGSPGRLFWDPMKVHDIRWNFEKFLVGPDGLPVMRWYHRTTVNTVKIDILNYMRRQVALGAKK* 0 >GPX3_echTel Echinops telfairi (tenrec) 0 MAGQLGASFLLPLFLAGFVQPSPKPEKMK 0 0 TDCHGGISGTIYEYGALTLDGEEYVPFKKYAGKYILFVNVASYuGLTGQYV 1 2 ELNALQEELEPFGLVILGFPSNQFGKQEPGENSEILASLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELVGSPGRLFWEPMKIHDIRWNFEKFLVGPDGLPIMRWYHRTTVNTVKLDILHYMRQRAELRAQKK* 0 >GPX3_proCap Procavia capensis (hyrax) 0 MARLLRASCLLSLLLAGFVPPSWGQEKSK 0 0 TDCHGGMSGTIYDYGALTIDGEEYIPFKQYAGKYILFVNVATYuGLTGQYV 1 2 ELNALQDELAPFGLVILGFPCNQFGKQEPGENSEILATLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPTSELVGSPDRLFWDPMKIHDVRWNFEKFLVGPNGVPVMRWYHRTPVNTVKLDILNYMKQRVFWGSKK* 0 >GPX3_monDom Monodelphis domestica (opossum) 0 MAGGTLQVSWLLPLVLAAFVQPIWGQEKMK 0 0 TDCYPGITSTIYDYGALTIDGDEYIPFKQYAGKYILFVNVATYuGLTAQYL 1 2 ELNALQEDMKSSGLVILGFPCNQFGKQEPGQNSEILSALK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPPPSELIGTSSRLFWEPMRVHDIRWNFEKFLVGPDGIPIMRWYHRATVSTVKADILEYLQKHSSLDMSA* 0 >GPX3_triVul Trichosurus vulpecula (possum) 0 LSAFVQPIRGQERMK 0 0 TDCYPGITGTIYEYGALTIDGDEYIPFKQYAGKYVLFVNVATYuGLTLQYL 1 2 ELNALQEEFKSLGFVILGFPSNQFGQQEPGQNSEILPALK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPAPSEVVGTPSKLFWEPMKVHDIRWNFEKFLVGPDGVPIMRWYHRATISTVKVDILDYLRKRGSQSRTAEDWQK* 0 >GPX3_macEug Macropus eugenii (wallaby) 0 MAGGTLQVSWLLPLVLSAFVQPIRGQERMK 0 0 TDCYPGITGTIYEYGALTIDGEEYIPFKQYAGKYVLFVNVATYuGLTVQYL 1 2 ELNALQEEFKSFGLVILGFPSNQFGKQEPGQNSEILPALK 2 1 HVRPGGGFVPNFQLFEKGDVNGEKEQKFYTFLK 0 0 NSCPAPSELIGTSKNLFWEPMKVHDIRWNFEKFLVGPDGIPIMRWYHRATISTVKVDILDYLRKRGSQDRTAEDWQK* 0 >GPX3_ornAna Ornithorhynchus anatinus (platypus) 0 MAGPLRPSWILPLVLAGLIQPNQGQDREK 0 2 TDCYSGVSGTVYEYGALTINGEEYIPFKQYMGKPILFVNVATYuGLTLQYL 2 1 ELNALQEELKAYGLVILGFPCNQFGKQEPGDNSEILPGLK 0 0 HVRPGGGFVPNFLLFEKGDVNGDKEQKVFTFLK 0 0 NSCPPPAELLGDPSRLFWSPMKTHDIRWNFEKFLVGPDGVPVMRWYHRATVSTVKADIVRYLRKLNNEQVEEEGRWKK* 0 >GPX3_galGal Gallus gallus (chicken) 0 MGCRAACVLAVLLAGLVPLGQGQEREK 0 0 VKCYDSVRGTIYDYGALTIDGDEYIPFRKYAGKMVLFVNVATYuGLTLQYL 1 2 ELNALQNELGPYGLVVLGFPSNQFGKQEPGQNSEILPALK 2 1 YVRPGGGFVPNFQLFQKGDVNGAKEQKVYSFLK 0 0 NSCPPVAEEFGNPKNLFWEPLRNHDIKWNFEKFLVGTDGVPVMRWYHRANIATVKNDIIAYMRQQRGQ* 0 >GPX3_anoCar Anolis carolinensis (lizard) frag 0 MRGRLKGPWIFPLLLAGLIQPNSAQERQK 0 0 VDCYDSIPGTVYNYGALSIHGDEYIPFQKYAGKLLLFVNVATYuGLTMQYL 1 2 ELNALQTELGRYGLVVLGFPCNQFGKQEPGENSEILPGLR 2 1 YVRPGGGFTPNFQLFQKGDVNGETEQRVYTFLK 0 0 NSCPPVIENFGDPNKLFWSPLKIHDIKWNFEKFLVGTDGKPVMRWYHRTNVATVKSDILRYMRKQLLYG* 0 >GPX3_xenTro Xenopus tropicalis (frog) frag 0 MGVKLRGLLMLPCFLAALIHAQNEMDQKS 0 0 VDCYSSIDGTIYDYGATTLDGTQFIPFKAYQGKYILFVNVATYuGLTMQYQ 1 2 ELNALQEELKNNNFVILGFPSNQFGMQEPGRNDEILPGLR 2 1 YVRPGGKFVPNFQLFEKGDINGRKEQKFYTFLK 0 0 NSCPPVGDNFGSATNRLMWEPIKVNDVKWNFEKFLVGPDGRPVKRWLPRTPVAQVRREIMSYMKLQPGTQRLLMLGLEQK* 0 >GPX3_danRer Danio rerio (zebrafish) frag 0 MGTQSNPWTSVVLLLALMHKIAALSNT 0 0 QACNSAAGDSFHNYGAKTINGTQFIPFSHYAGKHVLVVNVATYuGLTFQYV 1 2 ELNALHEELRHLGFTILGFPCDQFGKQEPGENNEILSALK 2 1 YVRPGNGFVPNFQLFEKGDVNGDGEQALFTFLK 0 0 NACPPVGESFGASNRLFWEPLKVNDIKWNFEKFLLDPDGRPVMRWFPRVNVSEVRADILKYFHQLLATAQ* 0 >GPX3_tetNig Tetraodon nigroviridis (pufferfish) 0 MADFFLQVALFALLGGCSSLETPLKQLCDSTS 0 0 PGTIYDYNAQVLNEDHCVDFSKFRGKPILFVNLATYuRYTHQYK 1 2 DLNALQEELGYTGFLILGFPSNQFGKQEPGAPSEILPGLK 2 1 YVRPGNGFVPNFQLFEKNDVNGENEQGVYTFLK 0 0 NSCPPVGGDLGNSGRLFWEPMKISDIKWNFEKFLVGPDGKPVMRWHPSIQVSEVRAD >GPX3_takRub Takifugu rubripes (fugu) frag 0 0 0 NGTIYDYSAQVLNGPLKVDFSDFRGKFVLFVNVATYuSYTYQYK 1 2 DLNALQEELRDCGFIILGFPSNQFGKQEPGAPAEILPGLK 2 1 YVRPGKGFVPNFQLFSKDDVNGENEQGVYTFLK 0 0 NSCSPVGGDLGDHVGRLFWEPMRISDIKWNFEKFLVGPDGKPIMRWHPSIEVPEVRADIYKHLHENKILD* 0 >GPX3_gasAcu Gasterosteus aculeatus (stickleback) 0 MRPFLTLLLLGLLRPCGAETPLTQRCEF 0 0 TDGTIYNYHAKTLNESYTVNFSDLKGRSVLFINVATYuGYTFQYA 1 2 ELNALHEQMKPLGLTILGFPCNQFGKQEPGQPHEILPGLK 2 1 HVRPGNGFVPNFPLFEKGDVNGKDEQEVFTFLK 0 0 SSCPPVGDLLGDPARLFWGPIKLSDIKWNFEKFLVGPDGKPVMRWHPSVSVSEVKADIHKHLIQLYTQQL* 0 >GPX3_oryLap Oryzias latipes (medaka) frag 0 MRLHFLPQQLLLMMMMMRPMAHGIPLTQKCDSS 0 0 QKCDSSTDGSIYKYHAKTLNGSQTVNFSDYIGKSVLFVNVATYuGYTYQYV 1 2 ELNALHEELKPLGFTIVGFPCNQFGKQEPGQNHEILPGLR 2 1 HVRPGNGFVPNFLLFNKGDVNGKYEQDVFTFLK 0 0 GSCPPVGEDFGNTDRLFWEPLKIGDIKWNFEKFLVGPEGKPVMRWHPSVNVSTVRTDINFLLKLYTQDLFN* 0 >GPX3_calMil Callorhinchus milii (elephantfish) frag 0 0 0 1 2 ELNALHKELQPHGFTILAFPSNQFGKQEPGNNREILLSLK 2 1 YVRPGKGFVPNFLMFQKGDVNGVDEQEHYTYLK 0 0 * 0 >GPX3_leuEri Leucoraja erinacea (skate) 0 MKDTSKEWVLLSLLLAWIHNATSQT 0 0 AYCNSSASGSIYDYGAMAIDGSRYIPLREYMGKTVLILNVASFuGLTSQYV 1 2 ELNALHTDLKSHGFTILAFPCNQFGKQEPGTNQEILPLLK 2 1 YVRPGKGYVPNFQLFQKGDVNGEKEQGLFTYLK 0 0 GSCPPVGDNFGNMDRLFWQTLKISDIKWNFEKFLVGPDGRPVQRWHPLMPVPSVREDVTRYIRSLHRQQTIG* 0 >GPX3_petMar Petromyzon marinus (lamprey) fused exons 1-2 0 MGRRPGGPLPPALNLAALLLLLLLLTGCPRSLAEAQR 00 APCNASATPPGGVFSFCPLSMDGRPVPLERYRGRATLVVNVATFuGLTRTQV 1 2 LLNALMEEFSQHGLVILGFPCNQFGLQEPGTDAAEILNGLR 2 1 YVRPGGGFVPNFPLFKRIDVNGDSEAPLFTFLK 0 0 ASCPSPVERFENTKSLFYEPLKSSDLRWNFDKFLVDAQGRPYMRYDRLAPFELVRSDLVQLLRSQASRTNRVTTR* 0 >GPX3a_braFlo Branchiostoma floridae (amphioxus) frag fused exons 1-2-3 0 MGLSSGLLASLLCLGFLGPGLANKRT 00 EQCSCAAEQGSLHDHHAMLLDGSRNVSLAEYRGTTLLVVTVATFuGFTQQYVGLNALRN 12 KMVEENGHRFEILGFPTNQFGLEEPARNDEILNGIR 2 1 YVRPGNDYVPNFTMFQKGDCNGENEQSLFTYLK 0 0 SCCPPISDVMGISGNKDRLYWKPLKVNDVRWNFEKFLVDPEGRGVKR 2 1 FSSYVTPEDLESVIEEFIQSWNDTHGGDTSGARSARDTGLMSWLN* 0 >GPX3b_braFlo Branchiostoma floridae (amphioxus) frag fused exons 2-3 0 MALSSGLLASLLCLGFLGPGLAQLKRT 00 EECTCTAQDGSFHNHHAMLLDGSRNVSFAEYSGTTLLVVNVATFuGFNEQYPGLNALRD 12 KMVTDNGHRFEILGFPTNNFGLQEPAPNSKILDCVK 2 1 HVNPGNGYVPNFPMFQKADCNGANEQAFYTYMK 0 0 SCCPAISDVFISKIRLYWDPIKNTDIRWNFEKFLVDPAGKAVKR 2 1 FSSFVTPGDLETVIDDFIKNWNNRDDTSGARSSRNSGFLSWL* 0 >GPX3c_braFlo Branchiostoma floridae (amphioxus) 0 MRLVSGLVAVLLCCGVISSEKSIYPKQRADCSCTAEHGSFHDHHAMLLDGSRNVSLAEYRGTTLLVVVVASFuGFTHQYPAMNALKEKMVGQLGFDFDILAFPTNQFGLQEPGTNGEILNVLK 1 1 YVRPGGGYVPNFPLFQKGDCNGENEQSLFTYLK 0 0 SCCPPASDMIVDDKSSLYWKPLRAGDVRWNFEKFLVDPEGKAVVR 2 1 FTPPVEPAEMEPVIEEFLRKWNVKVQREISAVEDLLSDMMIEQ* >GPX3a_cioInt Ciona intestinalis (tunicate) 0 MGHTGVALALVVALFCPALAHRAIGERSKCVSSSRTIYDTPFNFTMLNGTTVPLSKFR 1 2 GEILMIANLASFuGANWQYPLFNALQELEGVTVLGFPCNQFGLQ 0 0 EPGANSEILKILEHVRPGGGFQPNFPMFEKLEVNGENAHPLFKFLKDQCNVVTSQFAPKARLFYEPIQPNDIEWNFHK 0 0 FLVDQEGRARRRYHHNTPPDAAIVRKDIRFLQNN* 0 >GPX3b_cioInt Ciona intestinalis (tunicate) 43% 0 MHQKGAVPAVLLVLGTLAVATSIENSTCFHNT 0 0 FSVYDHSFPTLSGETVNLGEFRGNVSVVINVATYuGATVPQYKAMNALSE 0 0 EYTQSSFVTLAFPCNQFGLQQPEANDEILNGVM 21 YVRPGHGFVPNKKIYFFSKTQVNGGSEDPLFTSIK 0 0 ASCPPTTNNIGITSELYWTPIKANDIYWNWNKFLLDKNGMIR 2 1 YRFGSAVTATQLKPWIDQLLNEK* 0 >GPX3_strPur Strongylocentrotus purpuratus (sea_urchin) ambiguous classifier introns odd no sel 0 MLDAVCYDDAESLSDVAMTKSLSLDDYRGKVVLVVNTASFCTYTYQYPYFNELK 0 0 NEFGDQLAILGFPCNQFWLQEPGVGQEIPNTLRYVRPGGGYEPNFYLNEEKIDVNGPKAHPLFKKLK 0 0 NSCPPVKMEIGDPSNLYWSPMTIGDVTWNFNKFLLDKEGVPFKRYDSVVEPLQLVSDIQLVVDGDYKTVKRPSRPKYATEEL* 0
GPX5: 23 vertebrate sequences
>GPX5_homSap Homo sapiens (human) 0 MTTQLRVVHLLPLLLACFVQTSPKQEKMK 0 0 MDCHKDEKGTIYDYEAIALNKNEYVSFKQYVGKHILFVNVATYCGLTAQYP 1 2 ELNALQEELKPYGLVVLGFPCNQFGKQEPGDNKEILPGLK 2 1 YVRPGGGFVPSFQLFEKGDVNGEKEQKVFSFLK 0 0 HSCPHPSEILGTFKSISWDPVKVHDIRWNFEKFLVGPDGIPVMRWSHRATVSSVKTDILAYLKQFKTK* 0 >GPX5_panTro Pan troglodytes (chimp) 0 MTTQLRVVHLLPLLLACFVQTSPKQEKMK 0 0 MDCHKDEKGTIYDYEAIALNKNEYVSFKQYVGKHILFVNVATYCGLTAQYP 1 2 ELNAPuEELKPYGLVVLGFPCNQFGKQEPGDNKEILPALK 2 1 YVRPGGGFVPSFQLFEKGDVNGEKEQKVFSFLK 0 0 HSCPHPSEILGTFKSISWDPVKVHDIRWNFEKFLVGPDGIPVMRWSHRATVSSVKTDILAYLKQFKTK* 0 >GPX5_macMul Macaca mulatta (rhesus) 0 MTTQLRVVHLLPLLLACFVQTSPKQETMK 0 0 MDCHKDEKGTIYDYEAIALNKNEYVPFKQYVGKHILFVNVATYCGLTAQYP 1 2 ELNALQEELKPYGLVVLGFPCNQFGKQEPGDNKEILPGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKVFSFLK 0 0 HSCPHPSEILGTFKSISWDPVKVHDIRWNFEKFLVGPDGIPVMRWSHRATVSSVKTDILAYLKQFKTK* 0 >GPX5_otoGar Otolemur garnettii (bushbaby) 0 MTAQLRVGYLFPFLLAGFVQTNPRMEKIK 0 0 MDCYKDVKGTIYDYDAFALNGKHYIQFKQYTGKHVLFVNVATYCGLTAQYP 1 2 ELNALQEELKPSGLVVLGFPCNQFGKQEPGDNAEILPGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGERELKVFTFLK 0 0 HACPHPSEILSSFKYISWEPIKVHDVRWNFEKFLVGPNGVPVMRWSHWATVSTVKSDILAYLKQFKTK* 0 >GPX5_musMus Mus musculus (mouse) 0 MVTELRVFYLVPLLLASYVQTTPRPEKMK 0 0 MDCYKDVKGTIYDYEALSLNGKEHIPFKQYAGKHVLFVNVATYCGLTIQYP 1 2 ELNALQEDLKPFGLVILGFPCNQFGKQEPGDNLEILPGLK 2 1 YVRPGKGFLPNFQLFAKGDVNGENEQKIFTFLK 0 0 RSCPHPSETVVMSKHTFWEPIKVHDIRWNFEKFLVGPDGIPVMRWFHQAPVSTVKSDIMAYLSHFKTI* 0 >GPX5_ratNor Rattus norvegicus (rat) 0 MAIQLRVFYLVPLLLASYVQTTPRLEKMK 0 0 MDCYKDVKGTIYNYEALSLNGKERIPFKQYAGKHVLFVNVATYCGLTIQYP 1 2 ELNALQDDLKQFGLVILGFPCNQFGKQEPGDNTEILPGLK 2 1 YVRPGKGFLPNFQLFAKGDVNGEKEQEIFTFLK 0 0 RSCPHPSETVVTSKHTFWEPIKVHDIRWNFEKFLVGPNGVPVMRWFHQAPVSTVKSDILAYLNQFKTI* 0 >GPX5_perMan Peromyscus maniculatus (deer_mouse) 0 MTMQLRVFYLVPLLLASYVQTTPRPEKMK 0 0 MNCYKDVKGTIYDYEALALNGKDRIQFKQYGKHVLFVNVATYCGLTIQYP 1 2 ELNALQEELKPAGLVVLGFPCNQFGKQEPGENIEILPGLK 2 1 YVRPGKGYVPNFQLFAKGDVNGEKEQKIFTFLK 0 0 RCPHPSEIVITHKHISWEPVKVHDIRWNFEKFLVGPDGVPVMRWFHQAPVSTIKADILAYLKQFRTKQG* 0 >GPX5_cavPor Cavia porcellus (guinea_pig) AAKN02051715 0 MTMQLRVFYLVPLLLASYVQTTPRPEKMK 0 0 MDCFKDVDDTIFNYEAVTLGRKELVHFSQFAGKLILVVNVATYCGLTSQYP 1 2 ELNALQEELKPFGLVVLGFPCNQFGKQEPGDNSEILPGLK 2 1 HVRPGGGFVPNFQLFEKGDVNGENEQKIFTFLK 0 0 RSCPHPSELVGSLRRVSWDPIRVHDIRWNFEKFLVGPDGVPVARWFHQAPVSAVREDILAYLKRPPRARGV* 0 >GPX5_oryCun Oryctolagus cuniculus (rabbit) 0 MAAQLRASHLVPFLLAGLVLTDSKPEKMK 0 0 MNCYKDVKGTIYNYEAFTLNGKQRIQFKQYAGKYVLFVNVATYCALTSQYP 1 2 ELNALQEELKPFGLVVLGFPCNQFGKQEPGQNSEILPALK 2 1 YVRPGKGFVPNFQLFEKGDVNGGKEQIFTFL 0 0 HSCPHPSGILGSLRHISWEPIEVHDIRWNFEKFLVGPDGVPVMRWFHLAEVSTVKSDILAYLKQSKPKK 0 >GPX5_ochPri Ochotona princeps (pika) 0 MAAQVQAFCLVPLLLAGFVQTNPNPEKMK 0 0 MDCDKGVEGTIFDYEAFTLDGTEKIRFRQYAGKYVLFINVATYCALTDQYP 1 2 ELNALQDELKPFGLVVLGFPCNQFGKQEPGQNSEILPALK 2 1 VRPGRGFVPNFQLFEKGDVNGRNEQSIFTFLK 0 0 HSCPHPSGTLGLLKHISWEPIKVHDIRWNFEKFLVGPDGVPVMRWFHLAEVSTVKSDILAYLKQSKPTEDAGAEGS* 0 >GPX5_canFam Canis familiaris (dog) 0 MTAWLGASYVLPILLVSFVQTNAKPEKTK 0 0 MDCYKDVKGTIYEYEALTLNGNERIQFKQYAGKHVLFVNVATYCGLTAQYP 1 2 ELNSLQEELKPLGLVVLGFPCNQFGKQEPGENSEILPGLK 2 1 YVRPGRGYVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 LSCPHPSEVLGSFRHISWDPVKVHDIRWNFEKFLVGSDGVPVLRWFHRTPISTVKEDILVYLKQLKMK* 0 >GPX5_felCat Felis catus (cat) 0 MMAQLPASSLFPLLLVGVTCTNPKPEKVK 0 0 MDCYNDVKGTIYEYDALTLNGKEHIQFKQYAGKHVLFVNVATYCGLTAQYP 1 2 ELNALQDELKPWGLVVLGFPCNQFGNQEPGENSEILPGL 2 1 YVRPGRGYVPNFQLFEKGDVNGKTEQKVFSFL 0 0 LSCPQPSEVLGFTGHISWSPVKGHDIRWNFEKFLVGPNGVPVMRWFHQAPISTVRSDILEYLKQPKTK* 0 >GPX5_equCab Equus caballus (horse) 0 MTAQLRASYLFLLLLAGFVQTENNAEKMK 0 0 TDCYKDVKDTIYEYSAVTLNGKEHIQFNQYAGKPVLFVNVATYCGLTAQYP 1 2 ELNTLQEELKPFDLVVLGFPCNQFGKQEPGENSEILPGLK 2 1 YVRPGGGYVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 HSCPHPSDVLGSLKHISWEPIKVHDIRWNFEKFLVGPDGVPVMRWFHKTPVSTVKSDILAYLKQFKIKK* 0 >GPX5_bosTau Bos taurus (cow) 0 MTIQLRASCLFLFFLAGFVQTNSNLEK 0 0 MDCYKDVKGTIYDYDAFTLNGKEHIQFKQYAGKHVLFVNVATYCGLTAQYP 1 2 ELNALQEELKPFGLVVLGFPCNQFGKQEPGENSEILPGLK 2 1 YVRPGGGYVPNFQLFEKGDVNGETEQKVFTFLK 0 0 QSCPHPSEIMGSIKHISWEPIMVRDIRWNFEKFLVGPDGIPVMRWFHRTPVSTVKTDILAYMKQFKTK* 0 >GPX5_susScr Sus scrofa (pig) CU313307 0 MTVQLGAFYLFPLFMAGFVQT 0 0 NSNLEKMDCYKDVTGTIYDYDAFTLNGNEHIQFKQYAGKHVLFVNVATYCGLTAQYP 1 2 ELNTLQEELKPFGLVVLGFPCNQFGKQEPGENSEILLGLK 2 1 YVRPGGGYVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 HSCPHPSELIGSIGYISWEPIRVHDIRWNFEKFLVGPDGVPVMRWVHETPISTVKSDILAYLKQFKTE* 0 >GPX5_eriEur Erinaceus europaeus (hedgehog) 0 MTVYLKASYLFPLLIASFVRASPKPEKMK 0 0 MDCYKDIKGSIYDYDALTLNGKERIQFKQYASKHVLFVNVATYCGLTTQYL 1 2 ELNALQEELKPLGLVVLGFPCNQFGNQEPGENSEILGLK 2 1 YVRPGRGYVPNFQLFEKGDVNRENEQKVFTLI 0 0 HSCPHPSQLLGSLKNISWEPVKVHDIRWNFEKFLVGPDGVPVMRWYHQTAVNTVKSDILTYMKQFKTK* 0 >GPX5_dasNov Dasypus novemcinctus (armadillo) 0 0 0 MYCYKDVKGTIYDYGAATLNGREYIQFRQYAGKHVLFVNVATYCGLTAQYP 2 1 2 0 SQNFEKGEWDSEGEQRIFTLK 0 0 HSCPHPSEILSSSRYISWEPMKVHDIRWNFEKFLVGPDGVPVMRWYHRTPVSTVRADILEYLKQFRTI* 0 >GPX5_loxAfr Loxodonta africana (elephant) gappy tandem 0 2 0 MNCYKDVKGTIYDYDALTLNGKEHIQFKKYAGKHVLFVNVATYCGLTAQYP 1 2 ELNALQEELKPFGLVILGFPCNQFGKQEPGENSEILPGLR 2 1 YVRPGGGYIPNFQLFEKGDVNGEEEQKIFTFLK 0 0 HSCPHPSDILGSSRYIFWEPMKVHDIRWNFEKFLVGSDGIPVMRWSHRAPVSTVKADILEYLKQLKTK* 0 >GPX5_echTel Echinops telfairi (tenrec) 0 MAGQLGASFLLPLFLAGFVQPSPKPEKMK 0 0 MHCDKDVKETIYNYGAFTLLGREYVNFNKFSGKLVLFVNVATYCGLTAQYL 1 2 2 1 YVRPGAGYVPNFQLFEKGDVNGINEQKVFSFLK 0 0 HSCPHPSFILGSPKYIFWEPVKVHDIRWNFEKFLVGPDGVPVMRWYHRAPVSAVRADILHYLKQLKTK*
GPX6: 26 vertebrate sequences
>GPX6_homSap Homo sapiens (human) 0 MFQQFQASCLVLFFLVGFAQQTLKPQNRK 0 0 VDCNKGVTGTIYEYGALTLNGEEYIQFKQFAGKHVLFVNVAAYuGLAAQYP 1 2 ELNALQEELKNFGVIVLAFPCNQFGKQEPGTNSEILLGLK 2 1 YVCPGSGFVPSFQLFEKGDVNGEKEQKVFTFLK 0 0 NSCPPTSDLLGSSSQLFWEPMKVHDIRWNFEKFLVGPDGVPVMHWFHQAPVSTVKSDILEYLKQFNTH* 0 >GPX6_panTro Pan troglodytes (chimp) 0 MFRQFQASCLVLLFLVGFAQQILKPQNRK 0 0 VDCNKGVTGTIYEYGALTLNGEEYIQFKQFAGKHVLFVNVATYuGLAAQYP 1 2 ELNALQEELKNFGVIVLAFPCNQFGKQEPGTNSEILLGLK 2 1 YVCPGSGFVPSFQLFEKGDVNGEKEQKVFTFLK 0 0 NSCPPTSDLLGSSSQLFWEPMKVHDICWNFEKFLVGPDGVPVMRWFHQAPVSTVKSDILEYLKQFNTH* 0 >GPX6_macMul Macaca mulatta (rhesus) 0 MIRQFQACCLVLLFLVGFAQQTLKPQNRK 0 0 VDCNKRVTGTIYEYGALTLNSEEYIQFKQFAGKHVLFVNVATYuGLAAQYP 1 2 ELNALQEELKNFGVVVLAFPCNQFGKQEPGTNSEILLGLK 2 1 YVRPGSGFVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 NSCPPTSDLLGSSSQLFWEPMKVHDIRWNFEKFLVGPDGVPVMRWFHRAPVSTVKSDILEYLKQFNAH* 0 >GPX6_otoGar Otolemur garnettii (bushbaby) 0 MIWQFWASCLFSLFLVGFTQQILKPEQMK 0 0 VDCNKDVAGTIYEYGALTLDGEYIQFKQYAGKHVLFVNVATYuGLAVQYP 1 2 ELNALQEDLRDFGVIILGFPCNQFGKEEPGKSPEILSGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGENEQKVFTFLK 0 0 NSCPPTSDVLGSSKHLFWEPMKVHDIRWNFEKFLVGPNGVPVMRWYHRAPVSKVKSDIQEYLKQDT* 0 >GPX6_tupBel Tupaia belangeri (tree_shrew) frag 0 ACSKGVAGTIYEYGALTLSGEEYIPFRQFAGKHVLFVNVATYuGLAAQYP 1 2 eLNALQEELGSRGLVVLGFPCNQFGKQEPGSNSEILSGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 ELLGTSKQLWEPMKVDIRWNFEKFLVGPDGVPALRWFHKVPMSKVRSDILEYLERSDSQ* 0 >GPX6_musMus Mus musculus (mouse) 0 MAQKLWGSCLFSLFMAALAQETLNPQKSK 0 0 VDCNKGVTGTVYEYGANTIDGGEFVNFQQYAGKHILFVNVASFCGLTATYP 1 2 ELNTLQEELKPFNVTVLGFPCNQFGKQEPGKNSEILLGLK 2 1 YVRPGGGYVPNFQLFEKGDVNGDNEQKVFSFLK 0 0 NSCPPTSELFGSPEHLFWDPMKVHDIRWNFEKFLVGPDGVPVMRWFHHTPVRIVQSDIMEYLNQTSTQ* 0 >GPX6_ratNor Rattus norvegicus (rat) 0 MTQQFWGPCLFSFVMAVLAQETLDPQKSK 0 0 VDCNKGVAGTVYEYGANTLDGGEYVQFQQYAGKHILFVNVASFCGLTATYP 1 2 ELNTLQEELRPFNVSVLGFPCNQFGKQEPGKNSEILLGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGDNEQKVFSFLK 0 0 SSCPPTSELLGSPEHLFWDPMKVHDIRWNFEKFLVGPDGAPVMRWFHQTPVRVVQSDIMEYLNQTRTQ* 0 >GPX6_speTri Spermophilus tridecemlineatus (squirrel) frag 0 0 0 MDCNKGVTGTIYEYGALTLSGEEYIQFKKYAGKHVLFVNVATYuGLTAQYP 1 2 ELNTLQEELKPFNVTVLGFPCNQFGKQEPGKNSEILLGLK 2 1 YVRPGGGYVPNFQLFEKGDVNGDNEQKVFSFLK 0 0 NSCPPTSELLGSSKQLFWEPMKVHDIRWNFEKFLVGPDGVPVMRWFHQIPVSTVKLDILEYLKQFDTQ* 0 >GPX6_dipOrd Dipodomys ordii (kangaroo_rat) frag 0 MVQQFWAFYLFPLFLAGFAQETLKPQEMK 0 0 VDCDKGVTGTIYEYGALTLNGEEYIKFQQYAGKHVLFVNVATFuGLTAQYP 1 2 ELNALQEELKQSNVIVLGFPCNQFGKQEPGKNSEILSGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGENEQKVYTFLK 0 0 NSCPPTSELLGSSKQLFWEPMKIHDIRWNFEKFLVGPDGVPVMRWFHKIPVSTVKSDIIQYLNMSDTQ* 0 >GPX6_cavPor Cavia porcellus (guinea_pig) AAKN02051715 0 MHALLRAAGLGFPLLLLSLAQDTPEAQRMK 0 0 TDCNKTVPGTIYEYGALTLLGEEYIQFKQFAGKHVLFVNVATYuGLTAQYP 1 2 ELNALQDELRSFGVVLLGFPCNQFGRQEPRKNSEILSRLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 NSCPPTSELLGSPRLLFWDPIRVHDIRWNFEKFLVGPDGVPVMRWFHKAPVSVVKSDILEYLKQFEAQ* 0 >GPX6_oryCun Oryctolagus cuniculus (rabbit) frag 0 MIPQFWASCLFLLFLVSFAQETPEPQKMK 0 0 MDCNKEVSGTIYEYGARTLNGEEDIQFQQYAGKYVLFVNVATYCANTAQYP 1 2 2 1 YVRPGGGFVPNFQLFEKGDVNGDKEQKFYTFLK 0 0 NSCPPTSELLGSPRHLFWDPLKIHDIRWNFEKFLVGTDGIPVMRWFHAAPVSTVKADILKYVEQFNTQ* 0 >GPX6_canFam Canis familiaris (dog) 0 MVPQLWASALLPLLLAGLAQPTQNTQKMK 0 0 VDCYNGVMGTIYEYGALTLNNEEYVQFKQYAGKHVLFVNVATYuGLTAQYP 1 2 ELNALQEELKSFGVVVLGFPCNQFGKQEPGKNSEILSGLK 2 1 HVRPGGGFVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 NSCPPTSDLLGSSNQLFWEPMKVHDIRWNFEKFLVGPDGVPVMRWFHQAPVSTVKADILAYLKQLKTE* 0 >GPX6_felCat Felis catus (cat) 0 MAPPLWASCLFPLFLGGLAQPAPKPQTMR 0 0 MDCYTGVTGTIYEYGALTLSDGDYIQFKQYAGKVVLFVNVASYCGLTAQYP 1 2 ELNALQDELKRYGVVVLGFPCNQFGKQEPGKNSEILSGLK 2 1 YVRPGGGFVPSFQLFEKGDVNGENEQKVFTFLK 0 0 0 >GPX6_equCab Equus caballus (horse) 0 MIRQLWASSLFPLFLVGFAQLTPESQKMK 0 0 MDCYKGVTGTIYEYGALTLNGEEYIQFKQYAGKHVLFINVATYuGLTAQYP 1 2 ELNALQEELKPFGVVLLGFPCNQFGKQEPGKNSEILSGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 NSCPPTSDLLGSPKQLFWEPMKVHDIRWNFEKFLVGPDGVPVMRWFHRASVSTVKSDILEYLKQFTPE* 0 >GPX6_bosTau Bos taurus (cow) 0 MILQLWASCLFPLFLVGLAQLTPKQQQMK 1 0 VDCYKGVTGTIYEYGALTLNGEEYIQFKQYVGKHVLFVNVATYuGLTAQYP 1 2 ELNALQEELKPFGVVVLGFPCNQFGKQEPAKNSEILMGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 NACPPTSDLLGSSSQLFWEPMKVHDIRWNFEKFLVGPDGVPVMRWYHRASVSTVKSDMLEYLKQFKSE* 0 >GPX6_turTru Tursiops truncatus (dolphin) frag 0 MDCSKGVTGTICEYGALTLNGEEYIQFKQYAGKHVLFVSVATYuGLAAQYP 1 >GPX6_susScr Sus scrofa (pig) CU313307 0 MTPQFWASCLFSLCLVGFAQLIPKEQKM 0 0 MDCYKGVTGTIYEYGALTLNGEEYIPFKQYAGKHVLFVNVATYuGLTAQYP 1 2 ELNALQEELKPFGVVVLGFPCNQFGKQEPAKNSEILLGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 NSCPPTSDLLGSSNQLFWEPMKVHDIRWNFEKFLVGPDGVPVMRWYHRASVSTVKSDIMEYLKQFKSE* 0 >GPX6_vicVic Vicugna vicugna (vicugna) frag 0 MDCYKGVTGTIYEYGALTLNGEEYIQFKQYSGKPVLFVNVATYuGLAAQYP 1 >GPX6_eriEur Erinaceus europaeus (hedgehog) 0 MIKQFWVSGLFPLFLAGLAQSTLEPQKMK 0 0 VDCYKEVTGTIFEYGALTLNGEEYIKFDQYAGKHVLFINVATYuGLTAQYI 1 2 ELNALQEELKPFNVVLLGFPCNQFGKQEPGKNSEILSGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGENEQKIFTFLK 0 0 HSCPHPSQLLGSLKNLSWEPVKVHDIRWNFEKFLVGPNGVPVMRWFHRAPVSTVKSDILEYLKQFN 0 >GPX6_sorAra Sorex araneus (shrew) frag 0 VDCYEGVTGTIYEYGALTLNSEEYIQFKQYAGKHVLFINVATFuGLTAQYI 1 2 ELNALQEELKPLGVVLLGFPCNQFGKQEPGKNSEILAGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKIFTFLK 0 0 NSCPPTSDVLGSPDHLFWEPMKVHDIRWNFEKFLVGPDGVPVMRWFHQAPVSTVKSDILEYLKR >GPX6_dasNov Dasypus novemcinctus (armadillo) 0 MIRHLHLFPLLLVGFVQPNPNEPTMK 0 0 TDCYKGVTGTIYEYGALTLNGEEYVQFKQYAGKHVLFVNVATYuGLTAQYP 1 2 ELNALQEELRPFGVVVLGFPCNQFGKQEPGKNSEILFGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKVFTFLK 0 0 NSCPPPSELFGSSSQLFWEPLKVHDIRWNFEKFLVGPDGVPVMRWYHRASVSTVKSDMLAYLKQSKTK* 0 >GPX6_choHof Choloepus hoffmanni (sloth) 0 0 0 TDCYKGVTGTIYEYGALTLNGEEYIQFKQYAGKHVLFVNVATYuGLTAQYP 1 2 ELNALQEELKPFGVVVLGFPCNQFGKQEPGKNSEILPGLR 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKIFTFLK 0 0 NSCPPPSELLGSSSQLFWEPMKVHDIRWNFEKFLVGPDGVPVMRWYHRAPVSTVKSDVLDYLKQ* 0 >GPX6_loxAfr Loxodonta africana (elephant) 0 MIRQVWASYLFSLLLVGFVQPSPNQEKMK 0 0 TDCYKGVTGNIYEYGALTLNGEEYIQFKQYVGKHVLFVNVATYuGLTAQYP 1 2 ELNALQEELKPFGLVVLGFPCNQFGKQEPGKNSEILSGLK 2 1 YVRPGGGFVPNFQLFEKGDVNGEKEQKIFTFLK 0 0 NSCPPPSEILGSSSQLFWEPMKVHDIRWNFEKFLVGPDGVPVMRWYHKASVSTVKSDILEYLKQFEIQ* 0 >GPX6_echTel Echinops telfairi (tenrec) frag 0 QTFWASYLFLLFLVGFVHLSLSQKKMK 0 0 tDCYKGVTSTTYESLTRNGEECFQLKWYTGKHVHGVGVATYuGLALQ 1 2 eLNVLQEQMKSFSLVVLAFACSRLEEQEPGRDS 2 1 PQFQLFEKGDVNGEKEQKVYTFLK 0 0 NSCPPPSEILGSSSQLFWEPLKVHDIRWNFEKFLVGPDGVPVMRWYHRAPVSAVRADILEYLKQFEDQ* 0
GPX1: 25 vertebrates sequences
>GPX1_homSap Homo sapiens (human) chr3 2 exons glutathione peroxidase pseudo:chrX 0 MCAARLAAAAAAAQSVYAFSARPLAGGEPVSLGSLRGKVLLIENVASLuGTTVRDYTQMNELQRRLGPRGLVVLGFPCNQFGHQ 0 0 ENAKNEEILNSLKYVRPGGGFEPNFMLFEKCEVNGAGAHPLFAFLREALPAPSDDATALMTDPKLITWSPVCRNDVAWNFEKFLVGPDGVPLRRYSRRFQTIDIEPDIEALLSQGPSCA* 0 >GPX1_panTro Pan troglodytes (chimp) 0 MCAARLAAAAQSVYAFSARPLAGGEPVSLGSLRGKVLLIENVASLuGTTVRDYTQMNELQRRLGPRGLVVLGFPCNQFGHQ 0 0 ENAKNEEILNSLKYVRPGGGFEPNFMLFEKCEVNGAGAHPLFAFLREALPAPSDDATALMTDPKLITWSPVCRNDVAWNFEKFLVGPDGVPLRRYSRRFQTIDIEPDIEALLSQGPSCA* 0 >GPX1_macMul Macaca mulatta (rhesus) 0 MCAARLAAAAYAFSARPLAGGEPVSLGSLRGKVLLIENVASLuGTTVRDYTQMNELQRRLGPRGLVVLGFPCNQFGHQ 0 0 ENAKNEEILNSLKYVRPGGGFEPNFMLFEKCEVNGAGAHPLFAFLREALPAPSDDATALMTDPKLITWSPVCRNDVAWNFEKFLVGPDGVPVRRYSRRFQTIDIEPDIEALLSQGPSSA* 0 >GPX1_tupBel Tupaia belangeri (tree_shrew) 0 MCAARRAAAAQAVYTFSARPLGAEEPMSLGSLRGKVLLIENVASLuGTTVRDYTQMNDLQRRLGIRSLVVLGFPCNQFRHQ 0 0 KSKNEEILKSLKYVRIGDGFEPNFTVFEKCEVNGAKAHPLFAFLREALPAPSDDPCALMTDPKFITWSPVCRNDIAWNFEKFLVGPDGVPVRRYSRRFPTIDIEPDIEALLSQGP* 0 >GPX1_musMus Mus musculus (mouse) 0 MCAARLSAAAVYAFSARPLTGGEPVSLGSLRGKVLLIENVASLuGTTIRDYTEMNDLQKRLGPRGLVVLGFPCNQFGHQ 0 0 ENGKNEEILNSLKYVRPGGGFEPNFTLFEKCEVNGEKAHPLFTFLRNALPTPSDDPTALMTDPKYIIWSPVCRNDIAWNFEKFLVGPDGVPVRRYSRRFRTIDIEPDIETLLSQQSGNS* 0 >GPX1_ratNor Rattus norvegicus (rat) 0 MSAARLSAVAVYAFSARPLAGGEPVSLGSLRGKVLLIENVASLuGTTTRDYTEMNDLQKRLGPRGLVVLGFPCNQFGHQ 0 0 ENGKNEEILNSLKYVRPGGGFEPNFTLFEKCEVNGEKAHPLFTFLRNALPAPSDDPTALMTDPKYIIWSPVCRNDISWNFEKFLVGPDGVPVRRYSRRFRTIDIEPDIEALLSKQPSNP* 0 >GPX1_oryCun Oryctolagus cuniculus (rabbit) 0 MCAARMQSVYSFSAHPLAGGEPVNLGSLRGKVLLIENVASLuGTTVRDYTQMNELQERLGPRGLVVLGFPCNQFGHQ 0 0 ENAKNEEILNSLKYVRPGGGFEPNFMLFQKCEVNGAKAHPLFAFLREALPAPSDDPTALMTDPKFITWSPVCRNDVSWNFEKFLVGPDGVPVRRYSRRFPTIDIEPDIQALLSKGSGGA* 0 >GPX1_canFam Canis familiaris (dog) 0 MCAAALAAAAPSVYAFSARPLAGGEPMSLGSLRGKVLLIENVASLuGTTVRDYTQMNELQRRLGPRGLVVLGFPCNQFGHQ 0 0 ENAKNEEILNSLKYVRPGGGFEPNFTLFEKCEVNGAQAHPLFAFLRESLPAPSDDTTALMTDPKFITWSPVCRNDVAWNFEKFLVGPDGVPVRRYSRRFPTIDIEPDIEALLSQGPSCA* 0 >GPX1_felCat Felis catus (cat) 0 MCAAPLAAAAPSVYSFSARPLAGGEPMSLGSLRGKVLLIENVASLuGTTVRDYTQMNELQRRLGPRGLVVLGFPCNQFGHQ 0 0 ENAKNEEILNSLKYVRPGGGFEPNFTLFEKCEVNGAQAHPLFAFLRQALPAPSDDATALMTDPKFITWSPVCRNDVAWNFEKFLVGPDGVPVRRYSRRFPTIDIEPDIEALLSQGPSCA* 0 >GPX1_equCab Equus caballus (horse) 0 MCAAQLAAARSVYAFSARPLAGGEPLSLGSLRGKVLLIENVASLuGTTVRDYTQMNELQRRLGPRGLVVLGFPCNQFGHQ 0 0 ENAKNEEILNSLKYVRPGGGFEPNFTLFEKCEVNGAQAHPLFAFLREALPAPSDDATALMTDPKFITWSPVCRNDVAWNFEKFLVGPDGVPVRRYSRRFPTIDIEPDIEALLTQGPSCA* 0 >GPX1_bosTau Bos taurus (cow) 0 MCAAQRSAAAPTVYAFSARPLAGGEPFNLSSLRGKVLLIENVASLuGTTVRDYTQMNDLQRRLGPRGLVVLGFPCNQFGHQ 0 0 ENAKNEEILNCLKYVRPGGGFEPNFMLFEKCEVNGEKAHPLFAFLREVLPTPSDDATALMTDPKFITWSPVCRNDVSWNFEKFLVGPDGVPVRRYSRRFLTIDIEPDIETLLSQGASA* 0 >GPX1_eriEur Erinaceus europaeus (hedgehog) 0 MCAAPAAAQSVHAFSARPLVGGEPISLGSLQGKVLLIENVASLuGTTVRDYTQMNELQQRFGPRGLVVLGFPCNQFGHQ 0 0 ENTKNEEILNSLKYVRPGGGFEPNFMLFEKCEVNGAQAHPLFAFLREALPAPSDDATSLMTDPKLITWSPVCRNDVSWNFEKFLVGPDGVPVRRYSRRFPTIDIQPDIEALLAKQPLSS* 0 >GPX1_sorAra Sorex araneus (shrew) 0 MCAAPAAAAMAAQRSVHAFSARPLVGGEPISLGSLQGKVLLIENVASLuGTTVRDYTQMNELQQRFGPRGLVVLGFPCNQFGHQ 0 0 ENTKNEEILNSLKYVRPGGGFEPNFMLFEKCEVNGAQAHPLFAFLREALPAPSDDATSLMTDPKLITWSPVCRNDVSWNFEKFLVGPDGVPVRRYSRRFPTIDIQPDIEALLAKQPLSS* 0 >GPX1_dasNov Dasypus novemcinctus (armadillo) 0 MCAAALSAAQSVHAFSARPLAGGEPVSLGSLRGKVLLIENVASLuGTTVRDYTQMNELQRRLGPRGLVVLGFPCNQFGHQ 0 0 ENAKNEEILNCLKYVRPGGGFEPNFMLFEKCDVNGAQAHPLFAFLREALPAPSDDATALMTDPKFIIWSPVCRNDVAWNFEKFLVGPDGVPVRRYSRRFPTIDIESDIEALLSQGPSCA* 0 >GPX1_loxAfr Loxodonta africana (elephant) 0 MCAAPVAAALSVHAFSARPLAGGEPVNLGSLRGKVLLIENVASLuGTTVRDYTQMNDLQRRLGPRGLVVLGFPCNQFGHQ 0 0 ENGNKEEILNSLKYVRPGGGFEPNFTLFEKCEVNGAQAHPLFAFLREALPAPSDDATALMTDPKFIIWSPVCRNDISWNFEKFLVGPDGVPVRRYSRRFPTIDIEADIEALLSKGPSCA* 0 >GPX1_monDom Monodelphis domestica (opossum) 0 MCAARSVHAFSARLLGSGELLSLASLRGKVLLIENVASLuGTTIRDYTQMNELQKRHGGRGFMVLGFPCNQFGHQ 0 0 ENAKNEEILNSLKFVRPGSGFEPNFMLFEKCEVNGEKAHPLFTFLREALPAPSDDTNSLMTDPKLIIWSPVCRNDISWNFEKFLVGPDGVPVKRYSRRFETINIEKDIEDLLTKMPKEV* 0 >GPX1_ornAna Ornithorhynchus anatinus (platypus) 0 MCASQSVYGFSGRLLRSGEPFSLAALRGKVLLIENVASLuGTTVRDYTQMNELQSLYGPRGLAVLGFPCNQFGHQ 0 0 ENAKNEEILNSLKYVRPGNGFEPNFTMFEKCEVNGEKAHPLFAFLRESLPTPSDDPTSLMNDPKFITWSPVCRNDISWNFEKFLVGPDGVPVKRYSRRFETINIKEDIEVLLGQMSSYFQ* 0 >GPX1_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01BQVNH 454 0 NGEKAHPLFAFLRESLPTPSDDPTSLMNDPKFIIWSPVCRNDISWNFEKFLVGPDGVPLRYSRRFETINIKEDIAMLLDQ >GPX1_galGal Gallus gallus (chicken) 0 MAATGLAGILARPLGAAEPLALSSLRGKVLLVVNVASLuGTTTRDFLQLNELQQRYGPRGLRVLGFPCNQFGHQ 0 0 ENATNEEILRSLEYVRPGNGFKPNFTMFEKCEVNGKGAHPLFAFLREALPFPHDDPSALMTNPQYIIWSPVCRNDVSWNFEKFLVGPDGVPFRRYSRHFETIKLQDDIELLQ* 0 >GPX1_anoCar Anolis carolinensis (lizard) taa-stop 0 MAGLPVRGLAGLLARPLGAKDPVDLGSSRGRVVLVSNVASLuGTTARDFTQFSELQRRFGPRGFVVLAFPCNQFGHQ 0 0 ENATNEEILNSLKYVRPGNGYEPNFTVFEKCEVNGENAHPLFKFLKDELPYPHDDAVSLMANPQNIIWSPVCRNDISWNFEKFLIGPDGKPFKRYSRRFETIKIQDDIEKLQ* 0 >GPX1_xenTro Xenopus tropicalis (frog) 0 MRLAMVSRTVYEFSARLLSAGENTALSQYKGRVLLIENVASLuGTTIRDYTQMSRLQSMYGPRGLQVLAFPCNQFGHQ 0 0 ENSGNQEILNILKHVRPGGGFEPNFPLFEKVDVNGEKEHPLFTFLKGQLPYPSDDSISLMQDPKSIIWSPVRRNDIAWNFEKFLIARNGVPYKRYGRRFETFNIQQDIEKLLDETCE* 0 GPX1_ambTig Ambystoma tigrinum (salamander) 0 MAGKRFYDLSAKLLATGETLSFNAFKGKVVLIENVASLuGTTVRDYTQMNQLQQTLGGRGLVVLGFPCNQFGHQ 0 0 ENTSNEEILNSLKYVRPGKGFEPCFTLFQKCDVNGEKEHPVFSFLKEHLPFPSDDSVSLMTDPKCIIWTPVRRNDISWNFEKFLVGPDGKPYKRYGRRFETINIQPDIEELLKVSHVTSF* 0 >GPX1b_danRer Danio rerio (zebrafish) 0 MAGIKSFYDITAKTLTGEEFKFSSLQGKVVLIENVASLuGTTSRDYTQMNELHERFAEKGLVVLGVPCNQFGYQ 0 0 ENCTNEEILLSLKYVRPGNGYEPKFQLLEKVDVNGKNAHPLFTFLKEKLPFPSDEPMPFMSDPKFIIWSPVCRNDIAWNFEKFLIGSDGVPFKRYSRRFLTSGIDGDIKKLLSIP* 0 >GPX1_tetNig Tetraodon nigroviridis (pufferfish) 0 MAANFYDLTAKLLTGEAFNFSSLQGKVVLIENVASLuGTTTRDYTQMNELHERYASKGLVVLGVPCNQFGHQ 0 0 ENCKNEEILLSLKHVRPGHGFEPKFQLLEKVDVNGKDAHPLFQFLREKLPSPSDDPSALISDPKLIIWSPVCRNDVAWNFEKFLVGPDGVPFKRYSRKFLTSSIESDIKKLLSQA* 0 >GPX1_takRub Takifugu rubripes (fugu) 0 METQFYDLTAKLLTGEAFNFSSLQGKVVLIENVASLuGTTSRDYTQMNELHERYAGQGLVILGVPCNQFGHQ 0 0 ENCKNEEILSSLKYVRPGNGFEPKFQLFEKVDVNGKDAHPLFQFLREKLPFPSDDPTALMSDPKLINWSPVCRNDVSWNFEKFLIGPDGVPFKRYSRKFLTSSIEGDIKKLLSQA* 0 >GPX1_gasAcu Gasterosteus aculeatus (stickleback) 0 MARMAAKFYDLSAKLLTGETFNFSSLQGKVVNVASLuGTTSRDYTQMNELHERYAGKGLVILGVPCNQFGHQ 0 0 ENCKNEEILLSLKYVRPGNGFEPKFQLLEKVDVNGKDAHPLFVFLKGKLPYPSDEPGALMNDPKLILWSPVCRNDVSWNFEKFLIGSDGVPFKRYSRRFLTSDIEGDIKKLLSQA* 0 >GPX1_oryLap Oryzias latipes (medaka) 0 MASKFYSLTAKLLSGEIFNFSVLQGKVVLIENVASLuGTTTRDYTQMNELHERYASKGLVILGVPCNQFGHQ 0 0 ENCKNEEILLSLKHVRPGNGFEPKFQLLEKVDVNGKDAHPLFAFLREQLPTPSDDPAPLMTDPKFIIWSPVCRNDVAWNFEKFLVGPDGTPFKRYSRRFLTSDVEGDIKKLLSQT* 0
GPX2: 37 Vertebrate Sequences
>GPX2_homSap Homo sapiens (human) chr14 2 exons gastrointestinal glutathione peroxidase 2, U in cow pig rat chicken frog trout scorpion 0 MAFIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGYQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKLIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_panTro Pan troglodytes (chimp) 0 MAFIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGYQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKLIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_macMul Macaca mulatta (rhesus) 0 MAFIAKSFYGVSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGYQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPHDDPFSLMTDPKLIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_calJac Callithrix jacchus (marmoset) 0 MAFIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGYQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKFIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_tarSyr Tarsius syrichta (tarsier) 0 MACIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQVSERQCRFPRRLVVLGFPCKQFGHQ 0 0 ...SRTVVDSYVRPGGGYQPTFTLLQKCDVNGQNEHPVFAYLKDKLPYPQDDPHSLMTDPKFIIWSPVRRSDVAWNFEKFLVGPEGEPFRRYSRTFPTISIEPDIKRLLKVAI* 0 >GPX2_otoGar Otolemur garnettii (galago) 0 MAYIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLQYVRPGGGFQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPYDDPLTLMTDPKFIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_micMur Microcebus murinus (mouse_lemur) 0 MAYIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKFVRPGGGYQPTFTLIQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKFIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFHTINIEPDIRRLLKVAI* 0 >GPX2_tupBel Tupaia belangeri (tree_shrew) 0 MAYIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLQYVRPGGGYQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKFIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_musMus Mus musculus (mouse) 0 MAYIAKSFYDLSAIGLDGEKIDFNTFRGRAVLIENVASLuGTTTRDYNQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGYQPTFSLTQKCDVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKLIIWSPVRRSDVSWNFEKFLIGPEGEPFRRYSRSFQTINIEPDIKRLLKVAI* 0 >GPX2_ratNor Rattus norvegicus (rat) 0 MAYIAKSFYDLSAIGLDGEKIDFNTFRGRAVLIENVASLuGTTTRDYTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGFQPTFSLTQKCDVNGQNQHPVFAYLKDKLPYPYDDPFSLMTDPKLIIWSPVRRSDVSWNFEKFLIGPEGEPFRRYSRTFQTINIEPDIKRLLKVAI* 0 >GPX2_cavPor Cavia porcellus (guinea_pig) 0 MAYIAKSFYDLSAISLDGEKIDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ...AQCGGFSDVSWYFEKFLIGPEGEPFRRYSRTFHTINIEPDIKRLLKVAI* 0 >GPX2_speTri Spermophilus tridecemlineatus (squirrel) 0 MAYIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNDLQCRFPRRLVVLGFPCNQFGHQ 0 0 0 >GPX2_ochPri Ochonta princeps (pika) 0 MAYIAKSFYDLTAISLDGEKVDFNTFRGRAVLIENVASLuGTTSRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLQYVRPGGGYQPTFTLVQKCEVNGQNQHPVFAYLKDKLPYPYDDPFSFMTDPRFILWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKLAI* 0 >GPX2_oryCun Oryctolagus cuniculus (rabbit) 0 MAYLAKSFYDLTAVSLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRYPRRLVVLGFPCNQFGHQ 0 0 ENCQDEEILNSLKYVRPGGGYQPTFTLVQKCEVNGQNQHPVFTYLKDKLPYPHDDPFSLMTDPKFIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_canFam Canis familiaris (dog) 0 MAYIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGFQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPYDDPLSLMTDPKFIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_felCat Felis catus (cat) 0 MAYIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGFQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKFIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_equCab Equus caballus (horse) 0 MAYIAKSFYDLSAISLDGEKIDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGFQPTFTLVQKCEVNGQNQHPVFAYLKDKLPYPYDDPLSLMTDPKFIIWSPVCRSDVSWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_myoLuc Myotis lucifigus (microbat) 0 MAFIAKSFYDLSAISLDGEKIDFNTFRGRAVLIENVASLuGTTTRDYTQLNELQCRYPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGDGFQPTFTLAQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKYIIWSPVRRSDVSWNFEKFLIGPEGEPFRRYSRTFQTINIEPDIKRLLKVAI* 0 >GPX2_turTru Tursiops truncatus (dolphin) 0 MAYVAESFYDLSAISLDGEKVDFNTFRGMAVLIENVSSLuHNHRDFTQLNELQCRFPAPVILL... 0 0 ENCQNEEILNSLKYVRPGGGFQPTFTLIQKCDVNGQNEHPVFTYLKDKLPYPYDDPFSLMTDPKFIIWSPVHRSDVSWNFEKFLVGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_bosTau Bos taurus (cow) 0 MAYIAKSFYDLSAISLDGEKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGFQPTFTLVQKCDVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKFIIWSPVRRSDVSWNFEKFLIGPEGEPFRRYSRTFQTINIEPDIKRLLKVAI* 0 >GPX2_vicVic Vicugna vicugna (vicugna) 0 ...LDGEKVDFNTSRGRAVLIENVASLuGTTTRDYTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGFQPTFTLVQKCDVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKFIIWSPVRRSDVSWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_eriEur Erinaceus europaeus (hedgehog) 0 MAYIAKSFYDLSAISLDGEKIDFNTFRGRAVLIENVASLuGTTTRDYTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGFQPTFTLAQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSLLTDPKFIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEADIKRLLKVAI* 0 >GPX2_sorAra Sorex araneus (shrew) 0 MGKKIDFNTFRGRVVLIENVASLuGTTTRDYTQLNELQARFPRRLVVLGFPCNQFGHQ 0 0 0 >GPX2_dasNov Dasypus novemcinctus (armadillo) 0 MGKKIDFNTFRGRVVLIENVASLuGTTTRDYTQLNELQARFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGNDFQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSLMTDPKYIIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_loxAfr Loxodonta africana (elephant) 0 MTHIAKSFYDLSAISLEGDKVDFNTFRGRAVLIENVASLuGTTTRDFTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCKNEEILNSLKYVRPGGGFQPTFTLVQKCEVNGQNEHPVFAYLKDKLPYPYDDPLSLMTDPKFIIWSPVRRSDVSWNFEKFLIGPEGEPFRRYSRTFPTINIEPDIKRLLKVAI* 0 >GPX2_echTel Echinops telafair (hedgehog) 0 MAYIAKSFYDLSATSLDGDKIDFNAFRGRAVLIENVASLuGTTTRDYTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENSQNEEILNSLKYVRPGGGFQPTFTLAQKCEVNGQNEHPVFAYLKDKLPYPYDDPFSFMTDPKFFIWSPVRRSDVAWNFEKFLIGPEGEPFRRYSRTFQTISIEPDIKRLLKVAI* 0 >GPX2_monDom Monodelphis domestica (opossum) 0 MAHIAKSFYDLSATSLEGERIDFNTFRGRAVLIENVASLuGTTTRDYTQLNELQCRFPRRLVILGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGAGFQPNFVLVQKCEVNGQNEHPVFAYLKDKLPYPDDDPFSLMTDPKHIIWSPVRRSDIAWNFEKFLVGPEGEPFRRYSRSFQTINIEPDIKRLLKVAI* 0 >GPX2_ornAna Ornithorhynchus anatinus (platypus) 0 MARIAKSFYDLSATSLNGEKVDFNTFRGRAVLIENVASLuGTTTRDYTQLNELQRLFPRRLVILGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGNGFEPHFVLIQKCEVNGQNEHPVFSYLKAKLPYPEDDPSALMTDPKLIIWSPVRRSDVAWNFEKFLVGPEGEPFRRYSRTFPTIDIEPDIKRLLKVAI* 0 >GPX2_galGal Gallus gallus (chicken) chicken cDNA missing genome 0 MSVPIAKSFYDLSATSLQGEKIDFNTFRGRAVLIENVASLuGTTTRDYTQLNELQCRFPRRLVVLGFPCNQFGHQ 0 0 ENCQNEEILNSLKYVRPGGGFQPTFSLTQKCDVNGQNQHPVFAYLKDKLPYPYDDPFSLMTDPKLIIWSPVRRSDVSWNFEKFLIGPEGEPFRRYSRTFQTINIEPDIKRLLKVAI* 0 >GPX2_xenTro Xenopus tropicalis (frog) cDNA 0 MAYIAKSFYDLYATNIDGEKVDFNVFRGRVVLIENVASLuDTTVRDYTQLNELQTKYPRRLVVLGFPCNQFGYQ 0 0 ENCKNEEILNSLKYVRPGKGFVPGFTLFQKCDVNGKDTHSVFAYLKDKLPVPDNEPAALISDPRYIVWNPVHRSDISWNFEKFLIGPEGEPFKRYNKNFQTISIEPDIQRLLKLTKHLELCVCMYATDNYYMAAHKKI* 0 >GPX2_danRer Danio rerio (zebrafish) 0 MTFIAKTFYDLHATTLEGDTIDFNIYRGRVVLIENVASLuGTTTQDYTQLNELQSRYPHRLVVLGFPCNQFGYQ 0 0 ENCSDGEILNSLKYVRPGEGYKPSFTIFQKCVVNGSDAHPVFSYLKDKLPYPDDDPVTLIQDPKYLVWNPVSRNDISWNFEKFLIGPEGEPFKRYSKKFQTINIEPDIQRLLKLTKN* 0 >GPX2_tetNig Tetraodon nigroviridis (pufferfish) genome frameshifted 0 MTFIAKTFYDLRATTLEGDSLDFNVFRGRVVLIENVASLuGTTVRDFNELNQLQSKYPHRLVVLGFPCNQFGYQ 0 0 ENCSNGEILQSLQNVRPGNGFKPNFTIFEKCDVNGTNTHPVFAYLKDKLPYPDDEPSCLMQDPRFLVWSPVNRTDISWNFEKFLVGPEGEPFKRYSKRYPTIDIEPDIQRLLRLTKT* 0 >GPX2_takRub Takifugu rubripes (fugu) 0 MTFIAKTFYDLRATTLEGDSLDFNVFRGRVVLIENVASLuGTTIRDFSELNQLQSKYPHRLVVLGFPCNQFGYQ 0 0 ENCSNGEILPSLQHVRPGNGFQPNFTLFEKCDVNGTNTHPVFAYLKDKLPYPDDDPSSLMQDPRFLVWSPINRTDISWNFEKFLIGPEGEPFKRYSKKFPTIDIEPDVQRLLRLTKT* 0 >GPX2_gasAcu Gasterosteus aculeatus (stickleback) 0 MTFIAKTFYDLRATTLEGDSVDFNVLRGRVVLIENVASLuGTTPRDYSELNQLQTKYPHRLVVLGFPCNQFGYQ 0 0 ENCKNDEILNSLKHVRPGGGFQPTFTIFEKCDVNGTGTHPVFAYLKDKLPYPDDDPTSLADDPKFLVWSPVSRTDVSWNFEKFLVGPEGEPFKRYSKRFPTIDIEPDIQRLLRLTKN* 0 >GPX2_oryLap Oryzias latipes (medaka) 0 MTYIAKTFYDLKATSLEGDTVDFNVFRGRVVLIENVASLuGTTTRDFNELSQLQSKYPHRLVVLGFPCNQFGYQ 0 0 ENCTNAEILNSLQHVRPGNGFKPNFTIFEKCDVNGVNTHPVFAYLKDKLPYPDDNPSSLMQDPKFLVWSPVNRTDVAWNFEKFLIAPEGEPFKRYSRNFPTIDIEPDIQRLLRLTKA* 0 >GPX2_calMil Callorhinchus milii (elephantfish) tga-cys tag-stop full 0 MANSRKFYGFSTKLLNGQTLNFSKFKGKVVLIENVASLuGTTARDYTQMNELQSRYSREGFAVLGFPCNQFGFQ 0 0 ENCKNDEILKTLKYVRPGGGYEPNFTMFQKSIVNGDGTHPLFAYLKEKLPYPDDDPVSFMKDPQSINWSPVCRSDISWNFEKFLIGPDGEPFKRYSKKFETIQIEPDIQRLLKVAK* 0 >GPX2_leuEri Leucoraja erinacea (skate) 0 MAKAVKSFYDLTAVTLDGDKVDFNVYRGRVVLIENVASLuGTTTRDYTQLNELQTRYSHRFVVLGFPCNQFGFQ 0 0 ENCQNVEILNSLKYVRPGGGYQPNFTMFQKCDVNGAGTHPVFAYLKEKLPVPADDPTSLMKDPKYIIWSPVCRTDISWNFEKFLIGPDGEPFKRYSKSFETIQIEADIQRLLKVTK* 0 >GPX2_torCal Torpedo californica (electric_ray) 0 NLDGDKVDFNVYRGRVVLIENVASLuGTTTRDFTQLNELQTRYPHRFVALGFPCNQFGHQ 0 0 ENCQNDEIRKSLMHIRPGGGFQPNFTMFQKCEVNGSGTHPVFVFLkKKLPFPDDDPICLMKDPKFITWSPVSRSDIAWNFEKFLIGPDGEPFKRYSKTFETIQIESDIQRLLKVTK* 0 >GPX2_petMar Petromyzon marinus (lamprey) cDNA LyEST1379 tga-sel tag-stop full 0 MKSFYELSAKTLGGELVSFSRYRGKVVLVENVASLuGTTTRDFTQLNELQGRYGAQGLAVLGFPCNQFGHQ 0 0 ENSQNEEILNTLKYVRPGSGYEPNFTMFAKCEVNGKDTHPVFEFLKEKLPLPSDDPISFLQDPKHIIWSPVSRSDIAWNFEKFLVGPDGQPFKRYSKKFQTINIEEDLKFLLKQVK* 0 >GPX2a_petMar Petromyzon marinus (lamprey) tandem pair lineage-specific duplication 0 MAVAASQVAGMKSFYELSAKTLGGELVSFSRYRGKVVLIENVASLuGTTTRDFTQLNELQGRYGAQGLAVLGFPCNQFGHQ 0 0 ENSQNEEILNTLKYVRPGSGYEPNFTMFAKCEVNGKDTHPVFEFLKEKLPLPSDDPISFLQDPKHIIWSPVSRSDIAWNFEKFLVGPDGQPFKRYSKKFQTINIEEDLKFLLKQVK* 0 >GPX2b_petMar Petromyzon marinus (lamprey) 0 MAAAAAGALRSFYELSAKTLGGELVNFSRYRGKVVLIENVASLuGTTTRDFTQLNELQGRYGAQGLAVLGFPCNQFGHQ 0 0 ENTQNEEILNILKYVRPGSGYEPNFTMFAKCEVNGKDTHPVFEFIKEKLPLPSDDPISFLQDPKSIIWSPVSRSDIAWNFEKFLIGPDGQPFKRYSRKFHTIGIEDDIKLLLKQAS* 0 >GPX2_eptBur Eptatretus burgeri (hagfish) BJ648558 tga-sel tag-stop frag 0 SARGIAKSFYELSARNLAGuELVQFSKFRDKVVLIENVASLuGTTSRDFTQMNQLHQRLGVHGLVVLGFPCNQFGHQ 0 0 ENATNEELLQSLKYVRPGRGFEPNFPIFDKCEVNGANAHPLFTFLKEHLPLPSDNPTCFMSDCKSIIWSPVQRSDIAWNFEKFLIAPNGEPFRRYSKLYQTIDLEPDICKLLGL* 0 >GPX2A_braFlo Branchiostoma floridae (amphioxus) full 0 MAAATAVKSFFELSAKALSGEMVSFSRYQGKVVLVENVASLuGTTVRDFTQLNELAAMFGDKLAILGFPCNQFGHQ 0 0 ENATNEEILNSLKYVRPGNGYEPKFDMFSKVQVNGSDAHPVFAYLREKLPIPADSENAFLIMNDPKCVIWSPVTRTDIAWNFEKFLIGPDGQPIKRFSRYFQTIDIKNDIEALLK* 0 >GPX2B_braFlo Branchiostoma floridae (amphioxus) frag 0 HVRNFFDLSGQALSGAGAGDIIHFSRYTGKVVLVTNVASACYLTTREFTQ 0 0 LNNLMHLYGLHGLVILAFCCNQFGHSEPFENDEIVKCLRYVRPGPPFQ 0 0 PSFQLFVKCDVNGSRTHRVFDFLKDRLPYPSDDNTMLVAESSEITWNPVK 0 0 RNDITYNFEKFLIGRDGQPYRRYSYKTPPSRLHQDIKRLLGIE* 0 >GPX2_strPur Strongylocentrotus purpuratus (sea_urchin) ILDISCFVSLIYCLILDFYFCIS 0 MSTGSKSTIYGFTVASLDGKERQLSDYQGKVCMIINVASLuGTTTRDYHQLNELTSKYGSRGFEVLAFPCNQFGHQ 0 0 ENCKNEEILDLLKYVRPGQGYSPQ 21 FPIFSKVTVNGKDTHPLYTFLKTTLPFPSDLAAGAGSVLMNDPKGIIWNPVRRYDISWNFEKFLIGQDGVPFGRYSPKLPPADLAGDIEKLL* 0
GPX4: 26 vertebrate sequences
>GPX4_homSap Homo sapiens (human) at chr19:10549651057778 7 exons glutathione peroxidase 4 0 MSLGRLCRLLKPALLCGALAAPGLAGTM 0 0 CASRDDWRCARSMHEFSAKDIDGHMVNLDKYR 2 1 GFVCIVTNVASQuGKTEVNYTQLVDLHARYAECGLRILAFPCNQFGKQ 0 0 EPGSNEEIKEFAAGYNVKFDMFSKICVNGDDAHPLWKWMKIQPKGKGILGN 2 1 AIKWNFTK 0 0 FLIDKNGCVVKRYGPMEEPL 0 0 VIEKDLPHYF* 0 >GPX4_panTro Pan troglodytes (chimp) 0 MSLGRLCRLLKPALLCGALAAPGLAGTM 0 0 CASRDDWRCARSMHEFSAKDIDGHMVNLDKYR 2 1 GFVCIVTNVASQuGKTEVNYTQLVDLHARYAECGLRILAFPCNQFGKQ 0 0 EPGSNEEIKEFAAGYNVKFDMFSKICVNGDDAHPLWKWMKIQPKGKGILGN 2 1 AIKWNFTK 0 0 FLIDKNGCVVKRYGPMEEPL 0 0 VIEKDLPHYF* 0 >GPX4_macMul Macaca mulatta (rhesus) 0 MNLGRLCRLLKPALLCGALAAPGLAGTM 0 0 CASRDDWRCARSMHEFSAKDIDGHMVNLDKYR 2 1 GFVCIVTNVASQuGKTEVNYTQLVDLHARYAECGLRILAFPCNQFGKQ 0 0 EPGSNEEIKEFAAGYNVKFDMFSKICVNGDDAHPLWKWMKIQPKGKGILGN 2 1 AIKWNFTK 0 0 FLIDKNGCVVKRYGPMEEPL 0 0 VIEKDLPHYF* 0 >GPX4_calJac Callithrix jacchus (marmoset) 0 MSLGRLCRLLKPALLCGALAAPGLAGTM 0 0 CASQDDWRSARSMHEFSAKDIDGHTVNLDKYR 2 1 GFVCIVTNVASQuGKTQVNYTQLVDLHARYAECGLRILAFPCNQFGKQ 0 0 EPGSNEEIKEFAAGYNVKFDMFSKICVNGDDAHPLWKWMKIQPKGKGTLGN 2 1 AIKWNFTK 0 0 FLVDKNGCVVKRYGPMEEPQ 0 0 VIEKDLPCYF* 0 >GPX4_musMus Mus musculus (mouse) 0 MSWGRLSRLLKPALLCGALAAPGLAGTM 0 0 CASRDDWRCARSMHEFSAKDIDGHMVCLDKYR 2 1 GFVCIVTNVASQuGKTDVNYTQLVDLHARYAECGLRILAFPCNQFGRQ 0 0 EPGSNQEIKEFAAGYNVKFDMYSKICVNGDDAHPLWKWMKVQPKGRGMLGN 2 1 AIKWNFTK 0 0 FLIDKNGCVVKRYGPMEEPQ 0 0 VIEKDLPCYL* 0 >GPX4_ratNor Rattus norvegicus (rat) 0 MSWGRLSRLLKPALLCGALAVPGLAGTM 0 0 CASRDDWRCARSMHEFAAKDIDGHMVCLDKYR 2 1 GCVCIVTNVASQuGKTDVNYTQLVDLHARYAECGLRILAFPCNQFGRQ 0 0 EPGSNQEIKEFAAGYNVRFDMYSKICVNGDDAHPLWKWMKVQPKGRGMLGN 2 1 AIKWNFTK 0 0 FLIDKNGCVVKRYGPMEEPQ 0 0 VIEKDLPCYL* 0 >GPX4_cavPor Cavia porcellus (guinea_pig) 0 MFLNRLSRLLKPALVCGALATPGLIGTM 0 0 CASRDDWRCARSMHEFATKDIDGHMVCLDKYR 2 1 GCVCIVTNVASQuGKTDVNYTQLVDLHARYAECGLRILAFPCNQFGRQ 0 0 EPGSNQEIKEFAAGYNVRFDMYSKICVNGDDAHPLWKWMKVQPKGRGMLGN 2 1 AIKWNFTK 0 0 FLIDKNGCVVKRYGPMEEPQ 0 0 VIEKDLPCYL* 0 >GPX4_felCat Felis catus (cat) frag 0 0 0 CASRDDWRCAQSMHEFSAKDIDGHMVNLDKYR 2 1 GFVCIVTNVASQuGKTEVNYTQLVDLHARYAECGLRILAFPCNQFGRQ 0 0 EPGSNAEIKEFAAGYNVKFDMFSKICVNGDDAHPLWKWMKVQPKGRGILGN 2 1 AIKWNFTK 0 0 FLIDKNGCVVKRYCPMEDPL 0 0 VIEKDLPCYL* 0 >GPX4_bosTau Bos taurus (cow) 0 MSFSRLYRLLKPALLCGALAAPGLASTM 0 0 CASRDDWRCARSMHEFSAKDIDGRMVNLDKYR 2 1 GHVCIVTNVASQuGKTDVNYTQLVDLHARYAECGLRILAFPCNQFGRQ 0 0 EPGSNAEIKEFAAGYNVKFDLFSKICVNGDDAHPLWKWMKVQPKGRGMLGN 2 1 AIKWNFTK 0 0 FLIDKNGCVVKRYGPMEEPL 0 0 VIEKDLPCYL* 0 >GPX4_susScr Sus scrofa (pig) frag 0 ALLCGTLAVPGLAGTM 0 0 CASRDDWRCARSMHEFSAKDIDGHMVNLDKYR 2 1 GYVCIVTNVASQuGKTEVNYAQLVDLHARYAECGLRILAFPCNQFGRQ 0 0 EPGSDAEIKEFAAGYNVKFDMFSKICVNGDDAHPLWKWMKVQPKGRGMLGN 2 1 AIKWNFTK 0 1 FLIDKNGCVVKRYGPMEEPQ 0 0 VIEKDLPCYL* 0 >GPX4_echTel Echinops telfairi (tenrec) frag 0 CASRDDuRCARSMHEFSAKDIDERMVNLDKYR 2 1 GFVCIVTNVASQuGKTDVNYTQLVDLHARYAECGLRILAFPCNQFGRQ 0 0 EPGSNAEIKEFAAGYNVKFDMYSKVCVNGDDAHPLWKWMKVQPK 2 1 FVDKNGCVVKRYAPMEEP 0 0 VIEKDLPCYL* 0 >GPX4_monDom Monodelphis domestica (opossum) 0 MSFSRLYRLVKPALLCAALAAPGLSSSM 0 0 CASRNDWRCARSMHDFSAKDIDGRLVSLDKYK 2 1 GYVCIVTNVASQuGKTDVNYTQLVDLHARYAENGLRILAFPCNQFGRQ 0 0 EPGSNAEIREFAAGYNVKFDMYSKICVNGDDAHPLWKWMKIQPRGKGILGN 2 1 AIKWNFTK 0 0 FLIDKDGCVVKRYGPEEPQ 0 0 VIEKDLPCYL* 0 >GPX4_ornAna Ornithorhynchus anatinus (platypus) 0 MSLFRLARLAKPLLLGGAVAVPALRRTM 0 0 CASPDDWRCANSIYEFSAEDIDGKLVSLEKYR 2 1 GKVCIITNVASKuGKTEVNYTQLVDLHAQYAGQGLRILGFPCNQFGRQ 0 0 EPGTNSEIKEFAAGYNVKFDMFSKVCVNGDEAHPLWKWLKDQPKGKGTLGK 2 1 AIKWNFTK 0 0 FLIDKEGQVVKRYGPMDEPR 0 0 VIEKDLPCYL* 0 >GPX4_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01DM9S9 454 0 LFRLARFAKPLLLGGAVAVPALRRTM 0 0 CASPDDWRCANSIYDFSAEDIDGNSVSLEKYR 2 1 GKVCIITNVASKuGKTEVNYTQLVDLHAQYVEQGLRILGFPCNQFGKQ 0 0 EPGTNSEIKEFAAGYNVKFDMFSKVCVNGDEAHPLWKWLKDQPKGKGTLGN 2 1 AIKWNFTK 0 0 FLIDREGQVVKRYGPMDEPR 0 0 VIEKDLPCY* 0 >GPX4_galGal Gallus gallus (chicken) frag 0 AAVRGWRLVRGAARRM 0 0 AQADEWRSATSIYDFHARDIDGRDVSLEQYR 2 1 GFVCIITNVASKuGKTAVNYTQLVDLHARYAEKGLRILAFPCNQFGKQ 0 0 EPGDDAQIKAFAEGYGVKFDMFSKIEVNGDGAHPLWKWLKEQPKGRGTLGN 2 1 AIKWNFTK 0 0 FLINREGQVVKRYSPMEDPY 0 0 VIEKDLPAYL* 0 >GPX4_taeGut Taeniopygia guttata (finch) 0 MGWASAVRGALCCARAAAAAGAVRGPGLGPGPGPGPVRSM 0 0 CAQADDWRSAKAIYDFHALDIDGNDVSLEKYR 2 1 GDVCIITNVASKuGKTAVNYTQLVDLHARYAERGLRILGFPCNQFGKQ 0 0 EPGDNAQIKAFAENYGVKFDMYSKIDVNGDDAHPLWKWMKEQPKGRGTLGN 2 1 AIKWNFTK 0 0 FLINREGQVVKRYSPMEDPY 0 0 VIEKDLPAYL* 0 >GPX4_anoCar Anolis carolinensis (lizard) frag 0 EPGTEADIKAFAAGYGVKFDMFSKIEVNGEGAHPLWKWMKSQPKGRGTLGN 2 1 AIKWNFSK 0 0 FLINKEGQVVKRYSPMDDPV 0 0 VIEKDLPAYL* 0 >GPX4_xenTro Xenopus tropicalis (frog) 0 MLNCLCRSIKSTVLLSSVTGGLRSLTM 0 0 CAQVADWKAAKSIYEFSAVDIDGNEVSLEKYR 2 1 GYVCIIVNVASKuGKTPVNYTQLEELHAKYAEKGLRILGFPCNQFGKQ 0 0 EPGDESQIKVFAASYKVEFDMFSKIDVNGDGAHPLWKWMKDQPKGHGTLGN 2 1 AIKWNFTK 0 0 FLINREGAVVKRFSPMEDPV 0 0 VIEKDLPSYL* 0 >GPX4b_danRer Danio rerio (zebrafish) 0 MWLFQRALLVGAVGSKSFARAM 0 0 CAQANDWQSAKSIYEFSAIDIDGNDVSLEKYR 2 1 GYVCIITNVASKuGKTPVNYTQLAAMHVTYAEKGLRILGFPCNQFGKQ 0 0 EPGSEAEIKEFAKGYNAEFDLFSKIDVNGDAAHPLWKWMKEQPKGRGTLGN 2 1 NIKWNFTK 0 0 FLIDREGQVVKRYGPMDDPSV 0 0 VEKDLPKYL* 0 >GPX4_tetNig Tetraodon nigroviridis (pufferfish) 0 MQLLGSAVLFSVLLQVL 0 0 SASTEDWQRATSIYNFSAVDIDGNVVSLEKYR 2 1 GNVVIITNVASKuGKTPVNYSQFAEMHVKYAERGLRILAFPSNQFGNQ 0 0 EPGTEAQIKQFAQSYNAQFDMFSKIDVNGPDAHPLWKWLKEQPNGGGFMGN 2 1 GIKWNFTK 0 0 FLIDREGQVVKRYRPLDDPS 0 0 VENDLPQYLANNS* 0 >GPX4_takRub Takifugu rubripes (fugu) 0 MQLLGSAVLFSVLLQAMVGPNTRATL 0 0 AVPTEDWQTATSIYNFSAVDIDGNVVSLEKYR 2 1 GDVVIITNVASKuGKTPVNYSQFAEMHAKYAERGLRILAFPSNQFGNQ 0 0 EPGTEAQIKQFAQSYNAQFDLFSKIEVNGPDAHPLWKWLKEQPNGSGFMGN 2 1 SIKWNFTK 0 0 FLIDREGQVVKRYGPLDDPS 0 0 VVEKDLPTYL* 0 >GPX4_gasAcu Gasterosteus aculeatus (stickleback) 0 MRLLGSTVLFSVLLQAM 0 0 SAPTEDWQTATSIYDFSATDIDGNLVSLEKYK 2 1 GDVVIITNVASKuGKTPLNYSQFAAMHATYSERGLHILAFPTNQFGNG 0 0 ERDNETQIKQLAQSYNARFDIFSKIDVNGPTAHPLWKWLKDQPNGKGFMGN 2 1 SIKWNFTK 0 0 FLINREGQVMKRYGPLDDPS 0 0 VVEKDLPKYL* 0 >GPX4_oryLap Oryzias latipes (medaka) 0 MLLTGCTVLFSVLLQVM 0 0 STPEDWQRATSIYDFNATDIDGNLVSLEKYR 2 1 GNVVIITNVASKuGKTVVNYSQFAKMHAAYAERGLRILAFPSNQFGNQ 0 0 EPGNETQIKSFAESFDAHFDLFSKIDVNGPSAHPLWKWLKEQPHGRGLFGN 2 1 SIKWNFT 0 0 FLINKEGQVVKRYGPMDD 0 0 VVEKDLPKYL* 0 >GPX4_calMil Callorhinchus milii (elephantfish) frag 0 0 0 AQVADWKAAKSIYEFSAVDIDGNEVSLEKYR 2 1 GLVCIITNLASKuGRTAVNYTQLAKMHAKLAEQGLRILGFPCNQFGKQ 0 0 EPGSNAEIKEFAKGFDVNFDMFSKVEVNGENAHPLWKWLKDQPKGKGTLG 2 >GPX4_squAca Squalus acanthias (spiny dogfish) N-frag 0 VPSPDSGIFIQLAFRVGLAAVNEQARAAATM 0 0 SAQPADWQKATSIFEFDAKDIDGNEVSLEKYK 2 1 GFVSIITNVASKuGKTSVNYAQLAHMYTQYADAGLRILGFPCNQFGKQ 0 0 EPGNEEVIKEFAKGYNVKFDMYSKIDVNGDNAHPLWKWMKEQPKGKGLLGN 2 1 AIKWNFTK 0 0 FLINKEGQVVKRYSPMDDP 0 0 VVIEKDLPKYL* 0 >GPX4_petMar Petromyzon marinus (lamprey) 0 MAESADTWKDAKSMYEFSAKDIDGNEVSLEKYK 2 1 DHVCIVVNVASKuGKTAVNYNQLAALHKQYAESHGLCILAFPCNQFGKQ 0 0 EPGTDAEIKEFVQQYNVKFDMFSKIDVNSDSAHPLWKWMKSQPNGRGTLYN 2 1 AIKWNFTK 0 0 FLINKQGQVVKRYSPVDDPV 0 0 GIEKDFLPIL* 0 >GPX4_eptBur Eptatretus burgeri (hagfish) BJ646422 tga-sel frag 0 GTM 0 0 ASNGEWQTAKNMFEFSAMDIDGNNVSLEKYR 2 1 GHVSIVVNVASKuGKTTVNYTQLSAMHAKYADSHGLRILAFPCNQFGKQ 0 0 EPGTDAEIKAFAAGYDVHFDLFSKIMVNGDDAHPLWKWMKSQRYGHGTLGN 2 1 AIKWNFTK 0 0 FLIDKEGQVVKRYGPIDDPV 0 0 VI * 0 >GPX4a_braFlo Branchiostoma floridae (amphioxus) 0 MQSAVHVCTQ 00 ATGGEEWKNAMSIYEFSAKDIDGNEVSLEKYR 2 1 DHVCLIVNVACKuGGTNVNYTQLQELHDKYAESKGLCILAFPCNQFGGQ 0 0 EPWPEPEIKKWVTDKFGVKFDMFSKINVNGKDAHPLWKYLKSKQGGTLID 2 1 AIKWNFSK 0 0 FLINKEGQAVKRYGPNVKPL 0 0 EIEKDFEPYW* 0 >GPX4b_braFlo Branchiostoma floridae (amphioxus) frag 0 SVDPNKWRKTRFIYEFEAKDIDGNMISFEKYRY 2 1 GQPLLIVNVASRCGGTDRNYKQLMDLYRKYGEKGLRILAFPCNQFHNQ 0 0 EPYIERDIKEFVTTRYGVSFDMFSKIHVLGPETHPIYNWLVNTTRGTLGE 2 >GPX4a_cioSav Ciona savignyi (tunicate) 56% one exon 0 MAGTAVAGFAKHIHTSQVLASSNTNWKDAASIYEFNALDITGNN VSLDKYKGNVCIIVNVATQuGLTKSNYTQLESLYEKYSKDGLRILAFPCNQFGNQEPKSN AEILKFAKDTFNVQFDMFAKIDVNGDNAIPLYKYLKSGKNTGGFLTDAIKWNFTKFLVDK EGKPVERYAPKDKPFDMESHIKSLL* 0 >GPX4b_cioInt Ciona savignyi (tunicate) 58% one exon 0 MPKCKHSLFILLLSCILTYFLLPGKETFSMAGKDVKDIYGFTVNDIDDQ EVSLSKYKGHVCIIVNVASEuGFTKVNYEQLQQLYGKYSQQGLKILAFPCNQFGKQEPKP NADIKKFATENYGVTFDLFSKINVNGDNAIPLYKFLKTHKNTTGTLVNAIKWNFTKFLVT KQGIPYKRFAPNAKPLDMVKDIEELLNQ* 0 >GPX4_strPur Strongylocentrotus purpuratus (sea_urchin) N-frag 0 WKTAENIYAFTATDIDGNEVSLEKYK 2 1 GHVCLIVNVASSuGLTDKNYKQLVEMYQQFADSKGLRILAFPCNQFGGQ 0 0 EPKPNEEIKKFAQDKYGAKFDLFSKIDVNGDNTIPLYKYLKIKQKGFLGN 2 1 RIKWNFSKFLVNKEGIPVKRYGPPTEPKDIVKDLSKYF* 0
GPX7: 19 vertebrate sequences
>GPX7_homSap Homo sapiens (human) chr1:5243796152444640 3 exons 7 no genomic U 0 MVAATVAAAWLLLWAAACAQQEQDFYDFKAVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTDQHYRALQQLQRDLGPHHFNVLAFPCNQFGQQEPDSNKEIESFARRTYSVSFPMFSKIAVTGTGAHPAFKYLA 1 2 QTSGKEPTWNFWKYLVAPDGKVVGAWDPTVSVEEVRPQITALVRKLILLKREDL* 0 >GPX7_panTro Pan troglodytes (chimp) 0 MVAATVAAAWLLLWAAACAQQEQDFYDFKAVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTDQHYRALQQLQRDLGPHHFNVLAFPCNQFGQQEPDSNKEIESFARRTYSVSFPMFSKIAVTGTGAHPAFKYLA 1 2 RTSGKEPTWNFWKYLVAPDGKVVGAWDPTVSVEEVRPQITALVRKLILLKREDL* 0 >GPX7_macMul Macaca mulatta (rhesus) 0 MVAAAAWLLLWAVACAQREQDFYDFKAVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTDQHYRALQQLQRDLGPHHFNVLAFPCNQFGQQEPDSNKEIESFARRTYSVSFPMFSKIAVTGTGAHPAFKYLA 1 2 RTSGKEPTWNFWKYLVAPDGKVVGAWDPTVSVEEVRPQITALVRKLILRKREDL* 0 >GPX7_musMus Mus musculus (mouse) 0 MVAAVATAWLLLWAAACAQSEQDFYDFKAVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTDQNYRALQQLQRDLGPHHFNVLAFPCNQFGQQEPDTNREIENFARRTYSVSFPMFSKIAVTGTGAHPAFKYLT 1 2 QTSGKEPTWNFWKYLVDPDGKVVGAWDPTVPVAEIKPRITEQVMKLILRKREDL* 0 >GPX7_ratNor Rattus norvegicus (rat) 0 MVAAVATAWLLLWAAACTQSEQDFYDFKAVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTDQNYRALQQLQRDLGPYHFNVLAFPCNQFGQQEPDSNREIENFARRTYSVSFPMFSKIAVTGTGAHPAFKYLT 1 2 QTSGKEPTWNFWKYLVAPDGKVVGAWDPTVPVEEIKPRITEQVMKLILQKREDL* 0 >GPX7_canFam Canis familiaris (dog) 0 MVALVAAWLLLWAAACAPREQDFYAFQAVNIRGKLVSLEKYRGS 0 0 VSLVVNVASACGFTDGHYRSLQQLQRDLGPRHFTVLAFPCNQFGQQEPDGDREIESFARRTYGVSFPMFSKVAVTGTGAHPAFKYLT 1 2 QTSGKEPTWNFWKYLVAPDGKVVGAWDPTVSVEDIRPQITALVRKLILKKREDL* 0 >GPX7_bosTau Bos taurus (cow) 0 MVAARAAAWLLLAAAACAPREQDFYDFKAVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTDQHYRALQQLQRDLGPHHFNVLAFPCNQFGQQEPDSNKEIESFARRTYSVSFPMFSKIAVTGTGAHPAFKYLT 1 2 GTSGKEPTWNFWKYLVAPDGKVIGAWDPTVSVEEIRPQITALVRKLILKKREDL* 0 >GPX7_susScr Sus scrofa (pig) 0 MVAALVAAWLLLAAAARAQREQDFYDFKAVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTDQHYRALQQLQRDLGPHHFNVLAFPCNQFGQQEPDSNKEIESFARRTYSVSFPMFSKIAVTGTGAHPAFKYLT 1 2 QTSGKEPTWNFWKYLVAPDGKVVGAWDPTVSVEEIRPQITALVRKLILKKREDL* 0 >GPX7_monDom Monodelphis domestica (opossum) mrna error MLGRQTSCRPLPLVPLLLFLASPATPGPKESDFYNFKVVNIRGKWV macEug 0 MLGRQAPCCPLPLGPLLFLLASPATPGPKESDFYNFKVVNIRGNWVSLEKYRGS 0 0 VSLVVNVASECGYTDQHYRALQQLQRELGPYHFNVLAFPCNQFGHQEPDTNKEIENFARKTYGVSFPMFSKIAVRGTGANAAFKYLT 1 2 ESSGEEPTWNFWKYLVAPNGKVVGTWDSTVPVEEIRPKIHELVGKLILGKKDEL* 0 >GPX7_ornAna Ornithorhynchus anatinus (platypus) LOC100090031 0 MVAATVAAAWLLLWAAACAQQEQDFYDFKAVNIRGKLVSLEKYKGR 0 0 VSLVVNVASECGFTDQHYRALQQLQKDLGPSHFNVLAFPCNQFGQQEPDGNKEIESFVRKTYGVSFPMFSKIAVSGAGANAAFKFLT 1 2 ESSGEEPTWNFWKYLVSPDGKVVNSWDSTVSVEEVRPQITALVRKLILLKREDL* 0 >GPX7_tacAcu Tachyglossus aculeatus (echidna) EUPZL4S02GXFL4 0 QLQKDLGPSHFNVLAFPCNQFGQQEPDSNKEIESFVRKTYGVSFPMFSKIAVSG >GPX7_galGal Gallus gallus (chicken) 0 MLLAITALLLLAFSATQQKETDFYTFKVVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTDSHYKALQQLQKDLGPYHFNVLAFPCNQFGQQEPDTNKEIESFARKTYGASFPMFSKVAVSGAGAIPAFKYLI 1 2 DSTGEEPTWNFWKYLVDPNGKVVKAWDSTVSVEEIRPHVTELVRKIILKKKDEL* 0 >GPX7_taeGut Taeniopygia guttata (finch) 0 MLLAIAALLLLAFSATRQKEPDFYTFKVVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGYTDSHYKALQQLQRDLGPYHFNVLAFPCNQFGQQEPDTNKEIESFARKTYGASFPMFSKITVSGAGAIPAFKYLI 1 2 DSTGEEPTWNFWKYLVDPNGKVVKAWDSTVSVEEIRPHVTELVRKIILKKKDEL* 0 >GPX7_anoCar Anolis carolinensis (lizard) 0 MLLAVGALLFLAFSASQTKEDNFYAFKVVNIRGKLVSLEKYRGA 0 0 VSLVVNVASECGYTDNHYKDLQKLQRELGPYHFNVLAFPCNQFGQQEPNTNKEIESFARKTYGVSFPMFSKIAVTGNGANPAFRYLV 1 2 ESTNEEPTWNFWKYLVDPNGKAVKAWDSTVTIAEIKPHITELVRNIILKKRDEL* 0 >GPX7_xenTro Xenopus laevis (frog) 0 MYLAALVLLLLLPPSLQKSRDFYTFKVVNIRGKLVSLEKYRGS 0 0 VTLVVNVASECGFTDSHYKALQQLQRDLGSHHFNVLAFPCNQFGQQEPNSDREIENFVRKNYSASFPMFSKTAVTGSGANTAFKYLI 1 2 ESSGKEPDWNFWKYLVGPEGKVVDAWGPTVSVAEVRPHITSLVRNIILKKKDEL* 0 >GPX7_danRer Danio rerio (zebrafish) 0 MGMFLRAFTLITLVCLLEAKQKDFYTFKVVNSRGRLVSLEKYRGS 0 0 VSLAVNVASECGYTDEHYKDLQQLQKDFGPFHFNVLAFPCNQFGQQEPGSDKEIDSFVRRVYGVSFPIFSKIAVVGIGANNAYKYLV 1 2 EASRKEPTWNFWKYLIDTDGKVVDAWGPEVSVKEIRPRITEMVRKLIIKRKEEL* 0 >GPX7_gasAcu Gasterosteus aculeatus (stickleback) 0 MMLPAALAVLLTIFSLIETKEKDFYTFKVVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTEEHYKDLQQLQRDFGPYHFNVLAFPCNQFGQQEPGSDKEIDSFVRRVYGVSFPLFSKIAVVGTGANNVYKYLV 1 2 ESSENEPDWNFWKYLVDVNGKVLNAWGPKVSVKEIRPKIAEMVRQIIIKKKEEL* >GPX7_gadMor Gadus morhua (cod) 0 MLLAITALLLLAFSATQQKETDFYTFKVVNIRGKLVSLEKYRGS 0 0 VSLVVNVASECGFTDSHYKALQQLQKDLGPYHFNVLAFPCNQFGQQEPDTNKEIESFARKTYGASFPMFSKVAVSGAGAIPAFKYLD 1 2 ESTGEEPTWNFWKYLVDPNGKVVKAWDSTVSVEEIRPHVTELVRKIILKKKDEL* 0 >GPX7_braFlo Branchiostoma floridae (amphioxus) 0 MAAGQANFLLALLLSLASSAISQDDFYSFTAKDIKGKTVLLDKYRGK 0 0 VSLVVNVASECGYTDGHYRELVRLQDHLAPTKHFNVLAFPCNQFGGQEPMGNSAIAQFAKSMYKANFPMFSKIDVVGREAHPAYKYLA 1 2 ESTQAPPTWNFWKYLVDPNGKVITAWPPHNAVVDIWSKVESAVNRARWDKGQRSTEL* 0 >GPX7_cioInt Ciona intestinalis (tunicate) 49% 0 MAREVLRVLVLLAPLFGVFSENKHNGFYDYNVKTFDGETVSLKKYIGK 0 0 VSLVVNVASECGYTDEHYKELTALQNELVQKEQPFTVLAFPCNQFGEQEPHDNHDIQEFASSEYKASFPIFAKIDVRDRDAHPAYEFLR 1 2 RSTGQEPQWNFWKYLLDGSGNVINEWGPSISVSQVKDEILKAIQKLKNTHAEL* 0 >GPX7_strPur Strongylocentrotus purpuratus (sea_urchin) frag no sel 0 0 0 SSLVVNVASDCGYTDRTYRDLVDLSKDPQFEDRLNILAFPCNQFGHQEPQSNEEIKHYVKALYDVEFPLFAKVEVKGYDADPAWQYLT 1 2 SNAHQEPTWNFWKFLVDSEGKVKDAWGPDVPIHSIYGELLNEALMADPYHYHHAHFDDEF* 0
GPX8: 30 vertebrate sequences
>GPX8_homSap Homo sapiens (human) chr5 LOC493869 tga-stop inside intron 1 of CDC20B 0 MEPLAAYPLKCSGPRAKVFAVLLSIVLCTVTLFLLQLKFLKPKINSFYAFEVKDAKGRTVSLEKYKGK 0 0 VSLVVNVASDCQLTDRNYLGLKELHKEFGPSHFSVLAFPCNQFGESEPRPSKEVESFARKNYGVTFPIFHKIKILGSEGEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEPIEVIRPDIAALVRQVIIKKKEDL* 0 >GPX8_panTro Pan troglodytes (chimp) 0 MEPLAAYPLKCSGPRAKVFAVLLSMVLCTVTLFLLQLKFLKPKINSFYAFEVKDAKGRTVSLEKYKGK 0 0 VSLVVNVASDCQLTDRNYLGLKELHKEFGPSHFSVLAFPCNQFGESEPRPSKEVESFARKNYGVTFPIFHKIKILGSEGEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEPIEVIRPDIAALVRQVIIKKKEDL* 0 >GPX8_macMul Macaca mulatta (rhesus) 0 MEPLAAYPLKCSGPRAKVFAVLLSMVLCTVTLFLLQLKFLKPKINSFYAFEVKDAKGRTVSLEKYKGK 0 0 VSLVVNVASDCQLTDRNYLALKELHKEFGPSHFSVLAFPCNQFGESEPRPSKEVESFARKNYGVTFPIFHKIKILGSEGEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEPIDVIRPDIAALVRKLIIKKKEDL* 0 >GPX8_otoGar Otolemur garnettii (bushbaby) 0 MEPLTVYPSKCSGPKAKVLAVLLSMVLCTGTLFLLQLKFLKPKINSFYTFEVKDAKGRPVSLEKFKGK 0 0 VSLVVNVASDCQLTDRNYLALKELHREFGPSHFSVLAFPCNQFGESEPRPSKEVESLARKNYGVTFPIFHKIKILGSDAEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEPIEAIRPDIAALVRQMIIKKKEDL* 0 >GPX8_tupBel Tupaia belangeri (tree_shrew) 0 MEPLTTYSLKCSRPKAKIFAVLLSMGLCTVMLFLLQLKFLKLKINSFYTFEVKDAKGNIVSLEKFKGK 0 0 VSLVVNMTSDCQLTDKNYLALKELHKEFGPFHFLASVFPCNQFGELEPYPSKEVEAFARKNYGITSPIFHKIKILGSEVEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEPIEVIRPDIAALVRQVIIKKKEDL* 0 >GPX8_musMus Mus musculus (mouse) 0 MEPFAAYPLKCSGPKAKIFAVLLSMVLCTVMLFLLQLKFLKPRTNSFYSFEVKDAKGRTVSLEKFKGK 0 0 ASLVVNVASDCRFTDKSYQTLRELHKEFGPYHFNVLAFPCNQFGESEPKSSKEVESFARQNYGVTFPIFHKIKILGPEAEPAFRFIV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEPLEAIRPHVSQMIGQIILKKKEDL* 0 >GPX8_ratNor Rattus norvegicus (rat) 0 MEPLGAYPLKCSGPKAKIFAVLLSMVLCTVMLFLLQLKFLKPRINSFYSFEVKDAKGRMVSLEKFKGK 0 0 ASLVVNVASDCRFTDKSYETLRELHKEFGPYHFNVLAFPCNQFGESEPKSSKEVESFARKNYGVTFPIFHKIKILGPEAEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEPLEAIRPHVSQMIGQIILKKKEDL* 0 >GPX8_cavPor Cavia porcellus (guinea_pig) 0 MEPLSVHPLKCSGPKAKVFAVLLSMVLCTMMLFLLQLKFLRPKINSFYAFEVKDAKGRVVSLEKFKGK 0 0 VTLVVNVASDCQLTEKNYLAMKELHKQFGPFHFSVLAFPCNQFGESEPRSSQEVESFVRQNYGVTFPIFHKIKILGSEAEPAFKFLI 1 2 DSSKREPRWNFWKYLVNPEGQVVKYWRPEEPVEAIKPDIVELIRQIIVKKKEDL* 0 >GPX8_oryCun Oryctolagus cuniculus (rabbit) 0 MEPLSAYPVKCSGPKAKVFAVLLSMLLCTVTLFLLQLKFLKPKTNSFYAFEVKDAKGRTVSLEKFKGK 0 0 VTLVVNVASDCQLTDRNYLALKELHKEFGPYHFSVLAFPCNQFGDSEPRPSKEVESFARSTYGVTFPIFHKIKILGPEAEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPDGQVVKFWRPEEPIDVMKPDIAALIRQIIIKKKEDL* 0 >GPX8_canFam Canis familiaris (dog) 0 MEPLTAYPLRCSGSKAKVFAVLLSMVLCTVMLFLLQLKFLKPKINSFYTFEVKDAKGRTVSLEKFKGK 0 0 VTLVVNVASDCQLTDRNYLALQELHKEFGPFHFSVLAFPCNQFGESEPRPSKEVVSFARNNYGATFPIFHKIKILGSEADPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKSWRPEEPIEVIRPEIAALVRQMILRKKDDL* 0 >GPX8_equCab Equus caballus (horse) 0 MEPLTAYPLRCSGPKAKVFAVLLSMVLCTVMLFLLQLKFLKPKINSFYAFEVKDARGRMVSLEKFKGK 0 0 VALVVNVASDCQLTDRNYLGLQELHKEFGPFHFSVLAFPCNQFGESEPRPSNEVVSFARNNYGATFPIFHKIKILGSEADPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEPIDVIRPEIAALIRQMILKKKEDL* 0 >GPX8_bosTau Bos taurus (cow) 0 MEPLTAYPLRCSGPKAKAFAVLLSMVLCTVMLFLLQLKFLKPKINSFYTFEVKDANGRVVSLEKFKGK 0 0 VALVVNVASDCQLTDRNYLALQELHKEFGPFHFSVLAFPCNQFGESEPRPSKEVVSFARNNFGVTFPIFHKIKILGSEAEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKTWRPEEPIEVIRPEIAALIRQMIIKKKEDL* 0 >GPX8_oviAri Ovis aries (sheep) 0 MEPLTAYPLRCSGPKAKAFAVLLSMILCTVMLFLLQLKFLKPKINSFYTFEVKDANGRAVSLEKFKGK 0 0 VALVVNVASDCQLTDRNYLALQELHKEFGPFHFSVLAFPCNQFGESEPRPSKEVVSFARNNFGVTFPIFHKIKILGSEAEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKSWRPEEPIEVIRPEIAALIRQMIIKKKEDL* 0 >GPX8_susScr Sus scrofa (pig) 0 MEPLTAYPLRCSGPKAKVFAVLLSMVLCTVMLFLLQLKFLKPKINSFYNFEVKDAKGRTVSLEKFKGK 0 0 VALVVNVASDCQLTDRNYLALQELHKEFGPFHFSVLAFPCNQFGESEPRPSKEVLSFARNNYGVTFPIFHKIKILGSEAEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKYWRPEEPIEVIRPEIAALIRQMIIKKKEDL* 0 >GPX8_eriEur Erinaceus europaeus (hedgehog) 0 MEPLTTYPLRCSGPKAKVFAVLLSMVLCTVMLLLLQLKFLKPKINSFYTFEVKDAKGQPVSLEKFKGR 0 0 VSLVVNVASDCQLTDRNYLALQGLHKEFGPSHFSVLAFPCNQFGDSEPRPSKEVESFARNNYGVTFPIFHKIKILGPEAEPAFRFLV 1 2 DSTKKEPRWNFWKYLVNPEGQVVKSWRPEEPIEVIRPEIAALVRQMIIKKKEDL* 0 >GPX8_dasNov Dasypus novemcinctus (armadillo) 0 MEPLTAYSLKSSGSKAKLFAVLLSLVLCTVMLFLLQLKFLRPKINSFYTFEVKDAKGRPVSLEKFKGK 0 0 VSLVVNVASDCQLTDRNYLALQKLHKDFGPSHFTVLAFPCNQFGDSEPRPSKEVESFARNNYGVTFPIFHKIKILGSEAEPAFRFLI 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEHIEAIRPDIAALIRQMIIKKKEDL* 0 >GPX8_loxAfr Loxodonta africana (elephant) 0 MEPLTAYPLKCSGPKAKVFAVFLSMVLCTVLLFLLQLKFLKPRINSFYTFEVKDAKGRAVSLEKFKGK 0 0 VSLVVNVASDCKLTDRNYLALQELHREFGPFHFSVLAFPCNQFGDSEPRPSKEVESFARSNYGVTFPIFHKIKILGSEAEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKFWRPEEPIEAIRPDIAALVRQMIIKKKEDL* 0 >GPX8_monDom Monodelphis domestica (opossum) 0 MEPFAAPTPLKCSIPKARVIAVLLSMVLCSVTLFLLQLKYLKPRIKDFYSFEVKDSRGRTVSLEKYRGK 0 0 ASLIVNVATNCQHTDRNYLALQELHREFGSFHFNVLAFPCNQFGESEPGLSHEIEAFVKANYGVTFPIFHKIKILGSEGEPAFKFLVD 1 2 SSKKEPRWNFWKYLVNPEGKVVKYWRPEESLEAIRPDVAALVRQIIIKKKEDL* 0 >GPX8_triVul Trichosurus vulpecula (possum) 0 MEPLTAYPLRCSGPKAKAFAVLLSMILCTVMLFLLQLKFLKPKINSFYTFEVKDANGRAVSLEKFKGK 0 0 VALVVNVASDCQLTDRNYLALQELHKEFGPFHFSVLAFPCNQFGESEPRPSKEVVSFARNNFGVTFPIFHKIKILGSEAEPAFRFLV 1 2 DSSKKEPRWNFWKYLVNPEGQVVKSWRPEEPIEVIRPEIAALIRQMIIKKKEDL* 0 >GPX8_ornAna Ornithorhynchus anatinus (platypus) 0 MEPLLSSPSKCSVAKARVLAVLLSLVLCTAILCLLQLRSIKPKIKTDFYSFQVKDSKGRTVALEKFRGK 0 0 VSLIVNVASNCQYTERNYLALQELHREFGPSHFSVLAFPCSQFGDSEPGPSQEIEAFAKGNYGATFPIFHKIKILGSEAEPAFRYLI 1 2 DSSKKEPRWNFWKYLVNPEGKVVKFWRPEEPFEIIKPDIAALVRQIIMKKKEDL* 0 >GPX8_galGal Gallus gallus (chicken) 0 MEPLTAAYPLKYSVPKARVFVVFLSMVFCTAVLCLLQLRFFKPKTKDFYSFEIKDSRGRIVSLEKYRGK 0 0 ATLVVNVASYCQHTDKNYIALQDLHREFGPSHFTVLAFPCNQFGESEPGSSQEIEAFAKGNYGVTFPVFHKIKILGSEAEPAFKFLI 1 2 DSSKKEPRWNFWKYLVSPEGKVVKFWRPEEPIESIKPEIASLIRQIIMKKREDL* 0 >GPX8_melGal Meleagris gallopavo (turkey) frag 0 MEPLAANPLKCSIPKARVIAVLLSMVLCSVTLFLLQLRYLKPKIKDFYSFEVKDSRGRTVSLEKYRGK 0 0 VSLIVNVASNCQHTDRNYLALQQLHREFGSFHFNVLAFPCNQFGESEPRPSREIEAFVKSNYGVTFPVFHKIKILGSEGEPAFRFLI 1 2 DSSKKEPRWNFWKYLVNPEGKVVKFWRPDESLEAIR 0 >GPX8_xenTro Xenopus tropicalis (frog) 0 MEPLSPYPLKYSSPKAKVFLVFLSMVLCTGLVCVLQLKFLRAKGGDFYSYEVTDAKGRTVALSKYRGK 0 0 VTLVVNVASECGFTDSHYKALQQLQRDLGSYHFNVLAFPCNQFGQQEPNSDREIENFVRKNYSASFPMFSKTAVTGTGVNSAFKYLI 1 2 DSTKTKPRWNFWKYLVNPEGQVVKYWRPDETAEIIRPEVASLVRQIIMKKKEDL* 0 >GPX8_danRer Danio rerio (zebrafish) 0 MEALGGYPSKSSASRAGLFKVLLSVALCMGSLYLLQNKLSKSRKTKDFYSYEVKDARGRTVSLEKYRGK 0 0 ATLVVNVASGSELTEQSYRALQELHRELGTSHFNVLAFPCSQYGDTESGTSREIEAFAKSNYGVTFPIFNKIKIMGSEAEPAFRFLT 1 2 DSVQKIPKWNFWKFLVSPEGQVVRFWKPEEPVSDIRKEATTLVRNIILKKRQEL* 0 >GPX8_tetNig Tetraodon nigroviridis (pufferfish) 0 MEALGGYPTRSSNPKAKKLTVLLSMTVGVGCLLLLQTQLLKPRPSDFYSFEVKDAKGRTVSLEKYRGK 0 0 ASLVVNVASRSEQTESNYRSLQELHRELGPSHFNVLAFPCAQFGETETGSSRDSEAFAKATYGVTFPFFSRIKIMGSEAEPAFRFLT 1 2 DSVQKVPRWNFWKFLVNPEGKVVRFWRTDEPMESIRREVTALVREIILKKRVEL* 0 >GPX8_takRub Takifugu rubripes (fugu) 0 MEALGGYPTRSSNPKAKKLTVLLSMTVGVGCLFLLQTQLLKPRPSDFYSFEVKDAKGRTVSLEKYRGK 0 0 ASLVVNVASYSEQTESNYRYLQELHRELGTSHFNVLAFPCGQFGDTEPGTSRDSEAFAKATYGVTFPFFNRIKIMGSEAEPAFRFLT 1 2 DSVHKVPKWNFWKFLVNPEGKVVRFWRTDEPLESIRQEVTMLVREIIVKKRVEL* 0 >GPX8_gasAcu Gasterosteus aculeatus (stickleback) 0 MEALGGYPAKSSSPKARTLRVVVSMTVGVGCLLLLQTQLNPRKPADFYAFAVRDAKGRPVSLEKYRGK 0 0 ASLVVNVASRSEQTEANYVSLQELHRQLGPSHFNVLAFPCGQFGDTETGTSRDIEAFARSTYGVTFPFFSRIKIMGSEADPAFKFLT 1 2 DSVQKTPKWNFWKFLVNPEGRVVRFWRTDEPVDSVRQEVTALVREIILKKRVEL* 0 >GPX8_oncMyk Oncorhynchus mykiss (trout) frag 0 VKDAKGRSVSLERYRGK 0 0 ASLVVNVASHSEHTEMNYRSLQELHRELGTSHFNVLAFPCGQFGETEMGASRDIEAYAKNTYGVTFTIFSKVKIMGSEAEPAFKFIT 1 2 DSVKKVPKWNFWKFLVNPEGQVVRFWKAEEPVDSVRQEAITMVRQIILKKRTEL* 0 >GPX8_oryLap Oryzias latipes (medaka) 0 MEELGGYPTKSSLRKAKMLRVLLSMTVGVGCLFLLQIRLRIKNSSDFYSFEVKDAKGRTVSLEKYRGK 0 0 ASLVVNVASHCEHTEANFRFLQDLHRELGTAHFNVLAFPCGQFGTTELGLSRDIEAFAKNTYGLTFPFFGKIRTFGSKAEPAFKFLT 1 2 DSVQKVPKWNFWKFLVDPEGKVVRFWKTEEPMESIRQEAISLVRQIILKKRAEL* 0 >GPX8_calMil Callorhinchus milii (elephantfish) frag 0 NSKGKTVSLEKYRGK 0 0 ASLVVNVASQCEHTELSYRSLQQLQKELGPMHFNVLAFPCNQFGDSEPGSNYDIESFAKANYGVTFPIFSKIKVLGSEAHPAFRFLV 1 2 DSTNKEPRWNFWKYLVNPEGQVIKVWKTEEPLAAIKQEVSALVAKIILKKREEL* 0 >GPX8_squAca Squalus acanthias (spiny_dogfish) 0 MESVPAFRLKSWTPKARIFAVFTCMLVCLAALCVLQFKLFKHKNKDFYSFQVLNSKGKPVSLEKYRGK 0 0 GSLVVNVASNCEHTEKNYRGLQELQRELGPMHFNVLAFPCNQFGKSEPGSNSQIDNFAKTNHGVTFPIFGKIRVLGNESEPAFRYLI 1 2 TSKMEPRWNFWKYLVNPKGQVIKVWRTEEPMASIKQEVSELVANIILKKRDEF* 0
Basal selenoproteins: 28 platypus 18 echidna 19 elephantshark 18 lamprey 8 hagfish 14 amphioxus 6 tunicate 4 urchin
Outgroups are exceedingly important in determining ancestral characters along the evolutionary stem leading to placental mammals. The most important species are hagfish, lamprey, coelocanth/lungfish, and platypus/echidna. However only a fraction of the likely selenoprotein repertoire is available because of incomplete data.
These sequencs are current as of 6 May 2008. They derive from cdna at GenBank (both hagfish and lamprey selenoproteins were surprisingly well represented), at NCBI WGS contigs for cartilaginous fish and at the UCSC lamprey genome browser. Hagfish has no genome project underway so its transcripts have been intronated by homology. Platypus has both a genome assembly and a 454 transcrip project; echidna has only the latter. One interesting finding -- in addition to startling differences in degree of conservation of different selenoprotein -- is that SEPHS1 from both hagfish and lamprey have (ancestral) selenocysteine unlike mammals which have cysteine.
Summary of availablity by species: >DIO1_ornAna Ornithorhynchus anatinus (platypus) >DIO2_ornAna Ornithorhynchus anatinus (platypus) >DIO3_ornAna Ornithorhynchus anatinus (platypus) >GPX1_ornAna Ornithorhynchus anatinus (platypus) >GPX2_ornAna Ornithorhynchus anatinus (platypus) >GPX3_ornAna Ornithorhynchus anatinus (platypus) >GPX4_ornAna Ornithorhynchus anatinus (platypus) >GPX7_ornAna Ornithorhynchus anatinus (platypus) >GPX8_ornAna Ornithorhynchus anatinus (platypus) >MSRB1_ornAna Ornithorhynchus anatinus (platypus) >MSRB2_ornAna Ornithorhynchus anatinus (platypus) >MSRB3_ornAna Ornithorhynchus anatinus (platypus) >SELH_ornAna Ornithorhynchus anatinus (platypus) >SELI_ornAna Ornithorhynchus anatinus (platypus) >SELK_ornAna Ornithorhynchus anatinus (platypus) >SELM1_ornAna Ornithorhynchus anatinus (platypus) >SELM2_ornAna Ornithorhynchus anatinus (platypus) >SELO_ornAna Ornithorhynchus anatinus (platypus) >SELS_ornAna Ornithorhynchus anatinus (platypus) >SELT_ornAna Ornithorhynchus anatinus (platypus) >SELU1_ornAna Ornithorhynchus anatinus (platypus) >SEP15_ornAna Ornithorhynchus anatinus (platypus) >SEPHS1_ornAna Ornithorhynchus anatinus (platypus) >SEPHS2_ornAna Ornithorhynchus anatinus (platypus) >SEPP1_ornAna Ornithorhynchus anatinus (platypus) >SEPP2_ornAna Ornithorhynchus anatinus (platypus) >SEPW1_ornAna Ornithorhynchus anatinus (platypus) >SEPW_ornAna Ornithorhynchus anatinus (platypus) >DIO2_tacAcu Tachyglossus aculeatus (echidna) >GPX1_tacAcu Tachyglossus aculeatus (echidna) >GPX4_tacAcu Tachyglossus aculeatus (echidna) >GPX5_tacAcu Tachyglossus aculeatus (echidna) >GPX7_tacAcu Tachyglossus aculeatus (echidna) >MSRB1_tacAcu Tachyglossus aculeatus (echidna) >SELH_tacAcu Tachyglossus aculeatus (echidna) >SELK_tacAcu Tachyglossus aculeatus (echidna) >SELM__tacAcu Tachyglossus aculeatus (echidna) >SELO_tacAcu Tachyglossus aculeatus (echidna) >SELS_tacAcu Tachyglossus aculeatus (echidna) >SELT_tacAcu Tachyglossus aculeatus (echidna) >SELT_tacAcu Tachyglossus aculeatus (echidna) >SELU1_tacAcu Tachyglossus aculeatus (echidna) >SEP15_tacAcu Tachyglossus aculeatus (echidna) >SEPHS1_tacAcu Tachyglossus aculeatus (echidna) >SEPHS2_tacAcu Tachyglossus aculeatus (echidna) >SEPP1_tacAcu Tachyglossus aculeatus (echidna) >DIO1_calMil Callorhinchus milii (elephantfish) >DIO2_calMil Callorhinchus milii (elephantfish) >DIO3_calMil Callorhinchus milii (elephantfish) >GPX2_calMil Callorhinchus milii (elephantfish) >GPX2_calMil Callorhinchus milii (elephantfish) >GPX3_calMil Callorhinchus milii (elephantfish) >GPX4_calMil Callorhinchus milii (elephantfish) >GPX8_calMil Callorhinchus milii (elephantfish) >SELO_calMil Callorhinchus milii (elephantfish) >SELO_calMil Callorhinchus milii (elephantfish) >SELS_calMil Callorhinchus milii (elephantfish) >SELU1_calMil Callorhinchus milii (elephantfish) >SELU1_calMil Callorhinchus milii (elephantfish) >SEPHS1_calMil Callorhinchus milii (elephantfish) >SEPHS1_calMil Callorhinchus milii (elephantfish) >SEPP2_calMil Callorhinchus milii (elephantfish) >SEPP2_calMil Callorhinchus milii (elephantfish) >SEPW2_calMil Callorhinchus milii (elephantfish) >SEPW_calMil Callorhinchus milii (elephantfish) >DIO2a_petMar Petromyzon marinus (lamprey) >DIO2b_petMar Petromyzon marinus (lamprey) >GPX2a_petMar Petromyzon marinus (lamprey) >GPX2b_petMar Petromyzon marinus (lamprey) >GPX3_petMar Petromyzon marinus (lamprey) >GPX4_petMar Petromyzon marinus (lamprey) >MSRB3_petMar Petromyzon marinus (lamprey) >SELI_petMar Petromyzon marinus (lamprey) >SELM1_petMar Petromyzon marinus (lamprey) >SELO_petMar Petromyzon marinus (lamprey) >SELS_petMar Petromyzon marinus (lamprey) >SELT_petMar Petromyzon marinus (lamprey) >SELU3_petMar Petromyzon marinus (lamprey) >SELW_petMar Petromyzon marinus (lamprey) >SEP15_petMar Petromyzon marinus (lamprey) >SEPHS1_petMar Petromyzon marinus (lamprey) >SEPN1_petMar Petromyzon marinus (lamprey) >SEPW_petMar Petromyzon marinus (lamprey) >GPX2_eptBur Eptatretus burgeri (hagfish) >GPX4_eptBur Eptatretus burgeri (hagfish) >SELM1_eptBur Eptatretus burgeri (hagfish) >SELT_eptBur Eptatretus burgeri (hagfish) >SELW_eptBur Eptatretus burgeri (hagfish) >SEP15_eptBur Eptatretus burgeri (hagfish) >SEPHS1_eptBur Eptatretus burgeri (hagfish) >SEPW_eptBur Eptatretus burgeri (hagfish) >DIO1_braFlo Branchiostoma floridae (amphioxus) >DIO3_braFlo Branchiostoma floridae (amphioxus) >GPX2A_braFlo Branchiostoma floridae (amphioxus) >GPX2B_braFlo Branchiostoma floridae (amphioxus) >GPX3a_braFlo Branchiostoma floridae (amphioxus) >GPX3b_braFlo Branchiostoma floridae (amphioxus) >GPX3c_braFlo Branchiostoma floridae (amphioxus) >GPX4a_braFlo Branchiostoma floridae (amphioxus) >GPX4b_braFlo Branchiostoma floridae (amphioxus) >GPX7_braFlo Branchiostoma floridae (amphioxus) >SELI_braFlo Branchiostoma floridae (amphioxus) >SELK_braFlo Branchiostoma floridae (amphioxus) >SELU1_braFlo Branchiostoma floridae (amphioxus) >SEPHS1_braFlo Branchiostoma floridae (amphioxus) >GPX3a_cioInt Ciona intestinalis (tunicate) >GPX3b_cioInt Ciona intestinalis (tunicate) >GPX4a_cioSav Ciona savignyi (tunicate) >GPX4b_cioInt Ciona savignyi (tunicate) >GPX7_cioInt Ciona intestinalis (tunicate) >SELI_cioInt Ciona intestinalis (tunicate) >GPX2_strPur Strongylocentrotus purpuratus (sea_urchin) >GPX3_strPur Strongylocentrotus purpuratus (sea_urchin) >GPX4_strPur Strongylocentrotus purpuratus (sea_urchin) >GPX7_strPur Strongylocentrotus purpuratus (sea_urchin) >SELM2_strPur Strongylocentrotus purpuratus (sea_urchin) >SELW_strPur Strongylocentrotus purpuratus (sea_urchin) >SELO_spu Strongylocentrotus purpuratus (sea_urchin) >SELN1_strPur Strongylocentrotus purpuratus (sea_urchin) >SELU1_strPur Strongylocentrotus purpuratus (sea_urchin) >SELU2_strPur Strongylocentrotus purpuratus (sea_urchin) 2nd copy
Temporary fragments of sea urchin selenoproteins needing more work... >SELU1_strPur Strongylocentrotus purpuratus (sea_urchin) tga confirmed 0 MALLWGAAAAAVGGLLMANMDFWLPRGGAATTEAYLAGAVLQLLGETKLQAQLSLVDEAVRTFKAKDLWKDTGAVVLAVRRPGUSLCREEAKE LSSLKPELDALGIPLYAVLLEPGGYKEFLPFFSGEVFLDTETRFYGEEKRNISLVGLLRI STLLQVRENKKQGIQGNYIGHGLTLXGIFVIGPGDRGYSWNIAXRTLAITHQMR >SELU2_strPur Strongylocentrotus purpuratus (sea_urchin) 2nd copy CD317360 84% id also Sec PRVRRDGGDLFLWEKQACVIHFMRRFGuSACRLGASELDSLRPQLDEADVRLVGIGLEDL GAQEFLDGGFWKGDLFIDQQQKSYQGLGFKKYSLMSIMKAVFSAKTRAALSRASDKKIKA NLKGDKLQIGGTLIISKGGEKVLVDFKQEAPGDHIPLQTVLDAFXIKGETPKTEGAEGGT VVCXDDVCQMK* 0 >SELO_strPur Strongylocentrotus purpuratus (sea_urchin) tga confirmed CD336539 fragments 35-end NSIIGNIASRSLLAVQRGHHRFSDSRSFSKSPHKMARIDKLNFDNRVLRTLPIDPETENYARQVPGACFSRVKPTPVANPKTVAYSVPAMKLLDLSEEELKREDFPAYFSGSKPLPGSDPAAHCYCGHQFGNFAGQLGDGAAMLVCILFESYFRNL...MNATNPKFILRNYIAENAIKEAENGDFSEVHRVLKLLESPFSDDVSLPFIEHTSELAGDEDAGASKPVEPTISPSNTSTVNSCTGVAYNSRPPEWAMELRVSUSS- >SELN1_strPur Strongylocentrotus purpuratus (sea_urchin) HAEFQLNEPPSHPFWFTPAQFIGHLIIRKDASHVKYF NMHVPSNRSLNVDMEWMNGPNEVENMEVDIGFMPKMELHAPNPSSQVTIYSESGEVLYEPSNQQEMSSGTIHWDEEISMEEALVLLEKEMYPFKKIEYMPLNAAFKRTHA
These fragmentary sequences suggest echidna retains the indicated gene but do not establish whether selenocysteine is present.
>MSRB1_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01EKBYH 454 FCSFFGGEVFQNHFETGIYVCARCGYELFSSRSKFEHSSPWPAFTETIHPDSVAKREEPGRPGAFKVSCGKCGNGL >SELH_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01B810Z 454 RALSLAAPHLPILLNPRQPRRNSFEVTLFGPDGTRTELWSGIKKGPPRRLKFPEPEMLADLLRSSLA >SELK_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01DCGU7 454 VMVYISNGHVLNGQNRSPWSLSYIKDFFWGILDFIIMFFKSMIHPNVKRGCRNSSSDSKYDDGRGPPGYPRRGMGRINHSNGPNPPPMAGG >SELM__tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01AWAS8 454 LNFRYEELERIPLSHMTRAEINQLVQDLGFYRKADRDAPVPPEFQQAPAK >SELT_tacAcu Tachyglossus aculeatus (echidna) EUEMSW402BA71P 454 VFFLSNMIENQCMSTGAFEITLNDVPVWSKLESGHLPSMQQLVQILDNEMKLNVHMDSIPHHRS >SELT_tacAcu Tachyglossus aculeatus (echidna) EUGXWLM01DXGYB 454 YRHIASFLSVFKLVLIGLIIVGKDPFAFFGMQAPSIWQWGQENKVYACMMVFLLSNMIENQCMSTVAFEITLNDVPVWSKLESGHLPSMQQLVQILDNEMKLLNVHMDSIPHHRS >SEPHS2 Tachyglossus aculeatus (echidna) EUPZL4S02I1W5B QRIRRGKLVVSREDVELAYQEAMFSMAMLNRTAAGLMHTFNAHAATDI >SEPP1_tacAcu Tachyglossus aculeatus (echidna) EUEMSW408ERBZB 454 GELERHAPPGVPVYAQDGPDPDVWSILGGGKDDFLVYDRCGRLTFHIRLPFSFLHFPYVEAAVLFSHRHDFCGNCSYY