PRDM11: giant missing exon: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 27: | Line 27: | ||
PRDM11 must also share a certain heritage with ZNF862. This protein is structured as a partial internal repeat of (KRAB ZNF_TTF)x2 + hATC dimerization domains. ZNF852 ends in a very large penultimate phase 2 exon weakly alignable with PRDM11 along most of its length (28% identity over positions 19-690). Neither protein contains C2H2-type zinc fingers. PRDM7/9 has a single KRAB domain but it is not readily alignable via Blastp with either KRAB domain of ZNF852. | PRDM11 must also share a certain heritage with ZNF862. This protein is structured as a partial internal repeat of (KRAB ZNF_TTF)x2 + hATC dimerization domains. ZNF852 ends in a very large penultimate phase 2 exon weakly alignable with PRDM11 along most of its length (28% identity over positions 19-690). Neither protein contains C2H2-type zinc fingers. PRDM7/9 has a single KRAB domain but it is not readily alignable via Blastp with either KRAB domain of ZNF852. | ||
This implies a complex history of gene duplication coupled with domain shuffling -- PRDM7/9 shares a closely related PR(SET) domain with PRDM11 (but nothing else), which shares the large two-domain terminal exon (but nothing else) with ZNF852, which shares a KRAB domain intronated like that of PRDM7/9 (but nothing else). | This implies a complex history of gene duplication coupled with domain shuffling -- PRDM7/9 shares a closely related PR(SET) domain with PRDM11 (but nothing else), which shares the large two-domain terminal exon (but nothing else) with ZMYM1 and ZNF852, the latter of which shares a KRAB domain intronated like that of PRDM7/9 (but nothing else). | ||
<br clear=all> | <br clear=all> | ||
=== Correcting the gene model === | === Correcting the gene model === | ||
The curated NCBI reference gene model for human PRDM11 (NM_020229) is unsatisfactory given the human genome project has been out for 10 years. The sequence begins with a dubious first coding exon that has no phylogenetic support for translation even in placental mammals. This exon is more likely non-coding 5'UTR that happens (in human and a few primates) to contain an in-frame ATG codon for methionine (no statistical surprise). Initial methionines are very difficult to recognize as the Kozak sequence is too weak to provide a definitive signature. The best resolution would come from mass spectroscopy of in vivo protein | The curated NCBI reference gene model for human PRDM11 (NM_020229) is unsatisfactory given the human genome project has been out for 10 years. The sequence begins with a dubious first coding exon that has no phylogenetic support for translation even in placental mammals. This exon is more likely non-coding 5'UTR that happens (in human and a few primates) to contain an in-frame ATG codon for methionine (no statistical surprise). Initial methionines are very difficult to recognize as the Kozak sequence is too weak to provide a definitive signature. The best resolution would come from mass spectroscopy of in vivo protein though post-translational trimming can muddy the waters there.. | ||
[[Image:PRDM11SpliceLast.gif|left]] | [[Image:PRDM11SpliceLast.gif|left]] | ||
More seriously, the reference sequence terminates by reading through a splice junction to the first encountered stop codon, thereby omitting the gigantic terminal coding exon -- ironically one already identified as a standalone gene EAW68047 in the [http://www.ncbi.nlm.nih.gov/pubmed/ | More seriously, the reference sequence terminates by reading through a splice junction to the first encountered stop codon, thereby omitting the gigantic terminal coding exon -- ironically one already identified as a standalone gene EAW68047 in the [http://www.ncbi.nlm.nih.gov/pubmed/11181995 Venter group paper]. This exon is joined to the rest of the gene by the human transcript DR731303 which links it in the correct reading phase to the properly shortened preceding exon and by similar transcripts in dog, chicken, finch and frog (DN430942, BU271565, DC286485, DN081198/CK800288). | ||
Although some transcripts skip it, this exon has no transcripts of its own to support standalone gene status. Its phase 2 splice junction has been conserved for many billions of years of branch length in bony vertebrates. It contains two well-established Pfam domains proving that it is not conserved-non-coding dna as concluded by some bioinformatic tools. A string of 2166 bp implausibly has an open reading frame of this length without a stop codon unless it encodes a protein. | Although some transcripts skip it, this exon has no transcripts of its own to support standalone gene status. Its phase 2 splice junction has been conserved for many billions of years of branch length in bony vertebrates. It contains two well-established Pfam domains proving that it is not conserved-non-coding dna as concluded by some bioinformatic tools. A string of 2166 bp implausibly has an open reading frame of this length without a stop codon unless it encodes a protein. | ||
Various gene prediction tools find this extending exon (Genscan, Geneid, N-Scan, SGP, UniGene, Exoniphy) while others fail to predict it (Ensembl, Encode, CCDS, MGC, Vega, and AceView). Oddly the predicted NCBI gene models in birds and lizards all correctly contain the exon (XM_421099 XM_003206406 XM_002199814 XM_003214639) but this information never | Various gene prediction tools find this extending exon (Genscan, Geneid, N-Scan, SGP, UniGene, Exoniphy) while others fail to predict it (Ensembl, Encode, CCDS, MGC, Vega, and AceView). Oddly the predicted NCBI gene models in birds and lizards all correctly contain the exon (XM_421099 XM_003206406 XM_002199814 XM_003214639) but this information was never related to human genome annotation. | ||
No further attention will be paid to the erroneous gene model on this site. It is impossible to understand protein function when 60% of the protein and two informative domains have been dropped. | No further attention will be paid to the erroneous gene model on this site. It is impossible to understand protein function when 60% of the protein and two informative domains have been dropped. | ||
| Line 450: | Line 450: | ||
CRNMTLLFNTAYHLAMEGRPYYDFRPLAELLRKCELRVVDQYMNEGDCQILIHHIARALREDLIERIRQSPFLSIILDGQSEDLLADTVAVYVQYTSNDGPPATEFLSLQELALPTTESYLQGIDRAFSALGIRLQDERPS | CRNMTLLFNTAYHLAMEGRPYYDFRPLAELLRKCELRVVDQYMNEGDCQILIHHIARALREDLIERIRQSPFLSIILDGQSEDLLADTVAVYVQYTSNDGPPATEFLSLQELALPTTESYLQGIDRAFSALGIRLQDERPS | ||
VGLGIDGVNITAGLRANLYMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRLTAATLCEETEFLGDIRAVKW | VGLGIDGVNITAGLRANLYMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRLTAATLCEETEFLGDIRAVKW | ||
=== Varying levels of conservation within PRDM11 === | === Varying levels of conservation within PRDM11 === | ||
| Line 628: | Line 588: | ||
Difference Alignment of Final Exon Region of PRDM11 Used in Establishing Amniote Phylogenetic Tree | Difference Alignment of Final Exon Region of PRDM11 Used in Establishing Amniote Phylogenetic Tree | ||
<font color=green>homSap WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEMCRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALmagentaLVERIRQSPCLSVILDGQSDDLLA Homo sapiens (human)</font> | <font color=green>homSap WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEMCRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALmagentaLVERIRQSPCLSVILDGQSDDLLA Homo sapiens (human)</font> | ||
<font color=green>musMus ................................................................................................................................................I........... Mus musculus (mouse)</font> | <font color=green>musMus ................................................................................................................................................I............... Mus musculus (mouse)</font> | ||
<font color=green>canFam ................................................................................................................................................I........... Canis familiaris (dog)</font> | <font color=green>canFam ................................................................................................................................................I............... Canis familiaris (dog)</font> | ||
<font color=green>loxAfr ................................................................................................................................................I........... Loxodonta africana (elephant)</font> | <font color=green>loxAfr ................................................................................................................................................I............... Loxodonta africana (elephant)</font> | ||
<font color=green>monDom ...........................................................................................F.................................................F..I........I.. Monodelphis domestica (opossum)</font> | <font color=green>monDom ...........................................................................................F.................................................F..I........I...... Monodelphis domestica (opossum)</font> | ||
<font color=brown>ornAna .............................................................................................................................................F..I........... Ornithorhynchus anatinus (platypus)</font> | <font color=brown>ornAna .............................................................................................................................................F..I............... Ornithorhynchus anatinus (platypus)</font> | ||
<font color=brown>galGal ...........................................................................................Y.................................................F..I........V.. Gallus gallus (chicken)</font> | <font color=brown>galGal ...........................................................................................Y.................................................F..I........V...... Gallus gallus (chicken)</font> | ||
<font color=brown>melGal ...........................................................................................Y.................................................F..I........V.. Meleagris gallopavo (turkey)</font> | <font color=brown>melGal ...........................................................................................Y.................................................F..I........V...... Meleagris gallopavo (turkey)</font> | ||
<font color=brown>anaPla ...........................................................................................Y.................................................F..I........V.. Anas platyrhynchos (duck)</font> | <font color=brown>anaPla ...........................................................................................Y.................................................F..I........V...... Anas platyrhynchos (duck)</font> | ||
<font color=brown>taeGut ...........................................................................................Y.................................................F..I........... taeGut Taeniopygia guttata (finch)</font> | <font color=brown>taeGut ...........................................................................................Y.................................................F..I............... taeGut Taeniopygia guttata (finch)</font> | ||
<font color=brown>strCam ...........................................................................................Y.................................................F..I........... strCam Struthio camelus (ostrich)</font> | <font color=brown>strCam ...........................................................................................Y.................................................F..I............... strCam Struthio camelus (ostrich)</font> | ||
<font color=#CC66CC>allMis ...........................................................................................Y.................................................F..I........... | <font color=#CC66CC>allMis ...........................................................................................Y.................................................F..I............... allMis Alligator mississippiensis (alligator)</font> | ||
<font color=#CC66CC>allSin ...........................................................................................Y.................................................F..I........... allSin Alligator sinensis (alligator)</font> | <font color=#CC66CC>allSin ...........................................................................................Y.................................................F..I............... allSin Alligator sinensis (alligator)</font> | ||
<font color=#CC66CC>croSia ...........................................................................................Y.................................................F..I........... croSia Crocodylus siamensis (crocodile)</font> | <font color=#CC66CC>croSia ...........................................................................................Y.................................................F..I............... croSia Crocodylus siamensis (crocodile)</font> | ||
<font color=blue>carIns ...........................................................................................F.................................................F..I........... Carettochelys insculpta (turtle)</font> | <font color=blue>carIns ...........................................................................................F.................................................F..I............... Carettochelys insculpta (turtle)</font> | ||
<font color=blue>podUni ...........................................................................................F.................................................F..I........... Podocnemis unifilis (turtle)</font> | <font color=blue>podUni ...........................................................................................F.................................................F..I............... Podocnemis unifilis (turtle)</font> | ||
<font color=blue>traScr ...........................................................................................F.................................................F..I........... Trachemys scripta (turtle)</font> | <font color=blue>traScr ...........................................................................................F.................................................F..I............... Trachemys scripta (turtle)</font> | ||
<font color=blue>pelSin ...Q.......................................................................................F...................................S.............F..I........W.. Pelodiscus sinensis (turtle)</font> | <font color=blue>pelSin ...Q.......................................................................................F...................................S.............F..I........W...... Pelodiscus sinensis (turtle)</font> | ||
<font color=#00CC66>anoCar ..........................................................R..........................M.....C............................................V....F..I........... Anolis carolinensis (lizard)</font> | <font color=#00CC66>anoCar ..........................................................R..........................M.....C............................................V....F..I............... Anolis carolinensis (lizard)</font> | ||
<font color=#00CC66>hemBow ..........................................................R..........................M.....C...S.............................................F..I........... Hemidactylus bowringii (gecko)</font> | <font color=#00CC66>hemBow ..........................................................R..........................M.....C...S.............................................F..I............... Hemidactylus bowringii (gecko)</font> | ||
<font color=#00CC66>sciRee ..........................................................R..........................M.....CE...........................................V....F..I........... Scincella reevesii (skink)</font> | <font color=#00CC66>sciRee ..........................................................R..........................M.....CE...........................................V....F..I............... Scincella reevesii (skink)</font> | ||
<font color=#00CC66>dibBou ..........................................................R..........................M.....C.................................................F..I........... Dibamus bourreti (skink)</font> | <font color=#00CC66>dibBou ..........................................................R..........................M.....C.................................................F..I............... Dibamus bourreti (skink)</font> | ||
<font color=#00CC66>najAtr ..........................................................R..........................I.....C.........................................I..V....F..I........... Naja atra (cobra)</font> | <font color=#00CC66>najAtr ..........................................................R..........................I.....C.........................................I..V....F..I............... Naja atra (cobra)</font> | ||
<font color=#6699FF>batYen ......S.................................................................S....L.......V.....Y..............R..................................F..I......EY... Batrachuperus yenyuanensis (salamander)</font> | <font color=#6699FF>batYen ......S.................................................................S....L.......V.....Y..............R..................................F..I......EY....... Batrachuperus yenyuanensis (salamander)</font> | ||
<font color=#6699FF>ichBan ......S....................................................................................F..............R..................................F..I......E..I. Ichthyophis bannanicus (caecilian)</font> | <font color=#6699FF>ichBan ......S....................................................................................F..............R..................................F..I......E..I..... Ichthyophis bannanicus (caecilian)</font> | ||
<font color=#6699FF>xenTro ......S..............................................................................M.....Y..............R..................................F..I......E.... Xenopus tropicalis (frog)</font> | <font color=#6699FF>xenTro ......S..............................................................................M.....Y..............R..................................F..I......E........ Xenopus tropicalis (frog)</font> | ||
<font color=#6699FF>ranNig ......S..............................................................................M.....Y..............R..........................I.......F..I......E.... Rana nigromaculata (dark-spotted frog)</font> | <font color=#6699FF>ranNig ......S..............................................................................M.....Y..............R..........................I.......F..I......E........ Rana nigromaculata (dark-spotted frog)</font> | ||
<font color=green>homSap DTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAFSALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTAATLCEETEFLGDIRAVRW Homo sapiens (human)</font> | <font color=green>homSap DTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAFSALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTAATLCEETEFLGDIRAVRW Homo sapiens (human)</font> | ||
| Line 683: | Line 643: | ||
<font color=#6699FF>ranNig ...........N..............ALPT......GI..............R.S....I..V....G...NL<font color=magenta>Y</font>.............................................................L..................K. Rana nigromaculata (dark-spotted frog)</font> | <font color=#6699FF>ranNig ...........N..............ALPT......GI..............R.S....I..V....G...NL<font color=magenta>Y</font>.............................................................L..................K. Rana nigromaculata (dark-spotted frog)</font> | ||
=== | === PRDM11 relationship to ZNF862 and ZMYM1 === | ||
ZNF862 is | ZNF862 is otherwise the most closely related human protein to PRDM11, aligning along almost all of the terminal exon of PRDM11 with low percent identity 195/697 (28%)and gaps totaling 34/697 (5%). The two Pfam domains are not noticably better conserved than bulk exon. ZNF862 too has a very long terminal exon containing the uncommon transcription factor zinc finger type (ZNF_TFF) and a dimerization domain (hATC) often paired with it. | ||
The structure of ZNF862 suggests a partial internal duplication at some point in its history. Its doubled KRAB domain is also found undoubled in PRDM7/9 though percent identity is negligible. Still, the commonality in domains suggests related functionality. ZNF862 reportedly has a pseudogene (NG_024481) as well. | |||
>ZNF862_homSap Homo sapiens (human) | Another human gene, ZMYM1, also has a long terminal exon containing the transcription factor zinc finger type and dimerization domain. This exon is phase 0 unlike the phase 2 terminal exon of PRDM11 and ZNF862. ZMYM1 contains four zinc fingers of MYM-type in its earlier exons somewhat reminiscent of C2H2 domains of PRDM9. The alignment below shows how poorly these domains are conserved between the two proteins. | ||
[[Image:Prdm11znfttfhatdim.png]] | |||
It follows that PRDM11 shares a distant domain shuffling history with these two proteins as well (which are all quite diverged today). ZNF862 is well-conserved back to monotreme but hard to locate earlier; ZMYM1 only has clear full-length counterparts within placental mammals. Rapid divergence could make it difficult to find earlier orthologs if they existed. At face value, this suggests a major expansion of this overall class of domain-shuffled proteins in early mammals, with only a few members -- like PRDM11 -- having a history back into early vertebrates. | |||
>ZNF862_homSap Homo sapiens (human) aka KIAA0543 <font color=blue>KRAB</font> <font color=#990099>ZNF_TFF</font> <font color=brown>hATC dimerization dimerization</font> | |||
0 MEPRESGK 0 | 0 MEPRESGK 0 | ||
0 AP<font color= | 0 AP<font color=blue>VTFDDITVYLLQEEWVLLSQQQKELCGSNKLVAPL 1 | ||
2 GPTVANPELFRKFGRGPEPWL</font>GSVQGQRSLLEHHP 1 | |||
2 GKKQMGYMGEMEVQGPTRESGQSLPPQKKAYLSHLSTGSGHIEGDWAGRNRKLL<font color= | 2 GKKQMGYMGEMEVQGPTRESGQSLPPQKKAYLSHLSTGSGHIEGDWAGRNRKLL<font color=#990099>KPRSIQKSWFVQFPWLIMNEEQTALFCSACREYPSIRDKRSRLIEGYTGPFKVETLKYHAKSKAHMFCVNALAARDPIWAARFR</font> | ||
SIRDPPGDVLASPEPLFTADCPIFYPPGPLGGFDSMAELLPSSRAELEDPGGDGAIPAMYLDCISDLRQKEITDGIHSSSDINILYNDAVESCIQ 0 | |||
0 | 0 DPSAEGLSEEVPVVFEELP<font color=blue>VVFEDVAVYFTREEWGMLDKRQKELYRDVMRMNYELLASL 1 | ||
2 GPAAAKPDLISKLERRAAPWI</font>KDPNGPKWGKGRPP 1 | |||
2 GNKKMVAVREADTQASAADSALLPGSPVEARASCCSSSICEEGDGPRRIKRTY<font color= | 2 GNKKMVAVREADTQASAADSALLPGSPVEARASCCSSSICEEGDGPRRIKRTY<font color=#990099>RPRSIQRSWFGQFPWLVIDPKETKLFCSACIERPNLHDKSSRLVRGYTGPFKVETLKYHEVSKAHRLCVNTVEIKEDTPHTALV</font> | ||
PEISSDLMANMEHFFNAAYSIAYHSRPLNDFEKILQLLQSTGTVILGKYRNRTACTQFIKYISETLKREILEDVRNSPCVSVLLDSSTDASEQACVGIYIRYFKQMEVKESYITLAPLYSETADGYFETIVSALDELDI | |||
PFRKPGWVVGLGTDGSAMLSCRGGLVEKFQEVIPQLLPVHCVAHRLHLAVVDACGSIDLVKKCDRHIRTVFKFYQSSNKRLNELQEGAAPLEQEIIRLKDLNAVRWVASRRRTLHALLVSWPALARHLQRVAEAGGQIG | |||
HRAKGMLKLMRGFHFVKFCHFLLDFLSIYRPLSEVCQKEIVLITEVNATLGRAYVALESLRHQAGPKEEEFNASFKDGRLHGICLDKLEVAEQRFQADRERTVLTGIEYLQQRFDADRPPQLKNMEVFDTMAWPSGIEL | |||
ASFGNDDILNLARYFECSLPTG<font color=brown>YSEEALLEEWLGLKTIAQHLPFSMLCKNALAQHCRFPLLSKLMAVVVCVPISTSCCERGFKAMNRIRTDERTKLSNEVLNMLMMTAVN</font>GVAVTEYDPQPAIQHWYLTSSGRRFSHVYTCAQVPARSPA 1 | |||
2 SARLRKEEMGALYVEEPRTQKPPILPSREAAEVLKDCIMEPPERLLYPHTSQEAPGMS* 0 | 2 SARLRKEEMGALYVEEPRTQKPPILPSREAAEVLKDCIMEPPERLLYPHTSQEAPGMS* 0 | ||
>ZMYM1_homSap Homo sapiens (human) Q5SVZ6 <font color=green>Zinc finger MYM-type</font> <font color=#990099>ZnF_TTF</font> <font color=brown>hATC_dimerization</font> | |||
0 MKEPLLGGECDKAVASQLGLLDEIKTEPDNAQ 0 | |||
0 EYCHRQQSRTQENELKINAVFSES 1 | |||
2 ASQLTAGIQLSLASSGVNKMLPSVSTTAIQVSCAGCKKILQKGQTAYQRKGSA<font color=green>QLF<font color=red>C</font>SIP<font color=red>C</font>ITEYISSASSPVPSKRT<font color=red>C</font>SN<font color=red>C</font>SK 2 | |||
1 DILNPKDV</font>ISVQLEDTTSC<font color=green>KTF<font color=red>C</font>SLS<font color=red>C</font>LSSYEEKRKPFVTICTNSILTK<font color=red>C</font>SM<font color=red>C</font>QKTAI 0 | |||
0 IQYEV</font>KYQNVK<font color=green>HNL<font color=red>C</font>SNA<font color=red>C</font>LSKFHSANNFIMNC<font color=red>C</font>EN<font color=red>C</font>GTYCYTSSSL</font>SHILQMEGQSHYFNSSKSITAYKQ 0 | |||
0 KPAKPLISVPCKPLKPSDEMIETTSDLGKT<font color=green>ELF<font color=red>C</font>SIN<font color=red>C</font>FSAYSKAKMESSS 1 | |||
2 VSVVSVVHDTS</font>TELLSPKKDTTPVISNIVSLADTDVALPIMNTDVLQ 1 | |||
2 DTVSSVTATADVIVD 0 | |||
0 LSKSSPSEPSNAVASSSTEQPSVSPSSSVFSQHAIGSSTEVQKDNMKSMKISDELCHPKCTSKVQKVKG<font color=#990099>KSRSIKKSCCADFECLENSKKDVAFCYSC | |||
QLFCQKYFSCGRESFATHGTSNWKKTLEKFRKHEKSEMHLKSLEFWREYQFCDGAVSD</font>DLSIHSKQIEGNKKYLKLIIENILFLGKQCLPLRGNDQSVSSVNKGNFLELLEMRAKDKGEE | |||
TFRLMNSQVDFYNSTQIQSDIIEIIKTEMLQDIVNEINDSSAFSIICDETINSAMKEQLSICVRYPQKSSKAILIKERFLGFVDTEEMTGTHLHRTIKTYLQQIGVDMDKIHGQAYDSTT | |||
NLKIKFNKIAAEFKKEEPRALYIHCYAHFLDLSIIRFCKEVKELRSALKTLSSLFNTICMSGEMLANFRNIYRLSQNKTCKKHISQSCWTVHDRTLLSVIDSLPEIIETLEVIASHSSNT | |||
SFADELSHLLTLVSKFEFVFCLKFLYRVLSVTGILSKELQNKTIDIFSLSSKIEAILECLSSERNDVYFKTIWDGTEEICQKITCKGFKVEKPSLQKRRKIQKSVDLGNSDNMFFPTSTE | |||
EQYKINIYYQGLDTILQNLKLCFSEFDYCKIKQISELLFKWNEPLNETTAKHVQEFYKLDEDIIPELR<font color=brown>FYRHYAKLNFVIDDSCINFVSLGCLFIQHGLHSNIPCLSKLLYIALSWPITS | |||
ASTENSFSTLPRLKTYLCNTMGQEKLTGPALMAVEQEL</font>VNKLMEPERLNEIVEKFISQMKEI* 0 | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 02:17, 14 February 2012
See also: PRDM9: meiosis and recombination
Introduction: PRDM11
No vertebrate genome outside of mammals encodes a protein closely related to either PRDM7 or PRDM9. Since the latter are responsible for initiation of meiosis -- which arose very early in single-cell eukaryotes -- this raises questions about the meiotic process in the ancestral amniote, how that precedes without PRDM7/9 in contemporary birds and reptiles, and how PRDM7/9 arose and -- in mammals -- displaced the older mechanism.
A 2003 study reported that PRDM7 expression was strongly elevated in chicken auditory epithelia suggesting an auxiliary role outside meiosis. However bird genomes (and for that matter, all earlier vertebrates) don't contain either PRDM7 or PRDM9. Re-inspection of the short human probes (which align to the center of the PR(SET) domain) suggest that chicken PRDM11 expression was inadvertently studied as the best fit among PR(SET) domains in chicken. Auditory epithelia of course are not meiotic tissue. Other than this article and a high-thruput structural determination, PRDM11 has never been the subject of a published study.
These PRDM11 domains must share a close history of descent in the distant past with those of PRDM7/9 as the closest match among PRDM* genes still present in extant reptiles to the newly arisen chimeric mammalian PRDM7/9. Although PRDM11 lacks the KRAB domain and terminal zinc finger array and indeed has quite a different distal domain structure (a ZNF_TTF type zinc finger followed by a ubiquitous hATC dimerization Pfam motif), its comparative genomics and current functions are of considerable interest.
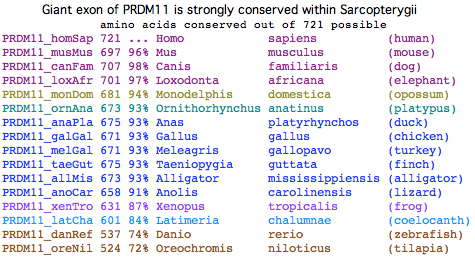
PRDM11 is a much older gene than PRDM7/9, one that arose in early bony vertebrates and persisted in all descendent lineages including human. Its domain structure indicate a role in regulation of transcription; its methylation capability could mark up a histone for meiosis but it seems to lack any ability to recognize specific sequences in dna (in the manner of the zinc fingers of PRDM7/9).
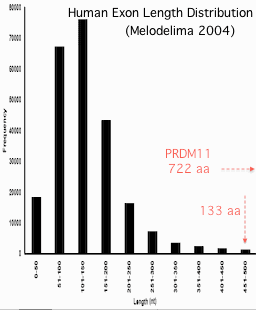
The final exon of PRDM11 contains a very large open reading frame of 722 amino acids (2166 bp). This is an extreme outlier, given the size distribution of exons (averaging ~55 amino acids) in the human proteome. In fact, this exon (and a homolog) may be the second largest of all known exons, with only the gigantic first exon of the microtubule-associated protein MAP1A (2678 amino acids) being larger.
It is not known how such large exons arise; in both cases here internal tandem duplication can be ruled out (though MAP1A has numerous small regions of low complexity). Another option is that these genes once had normal intron densities but these were lost by recombination with retroprocessed mature mRNA or came to be read through as coding (lost splicing capacity).
This latter origin is not plausible in the case of PRMD11 because the last exon exhibits very strong conservation along its entire length (87% identical between human and frog).
This extraordinary invariance over 721 residues is a bit mysterious since only a quarter of the exon falls into the two identifiable domains (182 residues or 25%). The other conserved residues lack a deep phylogenetic history so are unlikely to constitute a cryptic domain. Perhaps they are critical to interactions with other binding partners.
PRDM11 must also share a certain heritage with ZNF862. This protein is structured as a partial internal repeat of (KRAB ZNF_TTF)x2 + hATC dimerization domains. ZNF852 ends in a very large penultimate phase 2 exon weakly alignable with PRDM11 along most of its length (28% identity over positions 19-690). Neither protein contains C2H2-type zinc fingers. PRDM7/9 has a single KRAB domain but it is not readily alignable via Blastp with either KRAB domain of ZNF852.
This implies a complex history of gene duplication coupled with domain shuffling -- PRDM7/9 shares a closely related PR(SET) domain with PRDM11 (but nothing else), which shares the large two-domain terminal exon (but nothing else) with ZMYM1 and ZNF852, the latter of which shares a KRAB domain intronated like that of PRDM7/9 (but nothing else).
Correcting the gene model
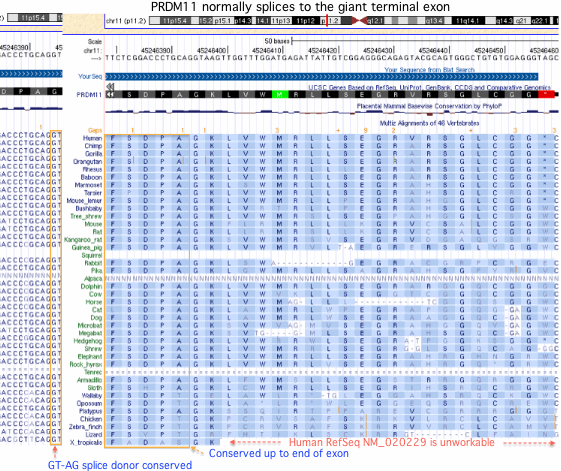
The curated NCBI reference gene model for human PRDM11 (NM_020229) is unsatisfactory given the human genome project has been out for 10 years. The sequence begins with a dubious first coding exon that has no phylogenetic support for translation even in placental mammals. This exon is more likely non-coding 5'UTR that happens (in human and a few primates) to contain an in-frame ATG codon for methionine (no statistical surprise). Initial methionines are very difficult to recognize as the Kozak sequence is too weak to provide a definitive signature. The best resolution would come from mass spectroscopy of in vivo protein though post-translational trimming can muddy the waters there..
More seriously, the reference sequence terminates by reading through a splice junction to the first encountered stop codon, thereby omitting the gigantic terminal coding exon -- ironically one already identified as a standalone gene EAW68047 in the Venter group paper. This exon is joined to the rest of the gene by the human transcript DR731303 which links it in the correct reading phase to the properly shortened preceding exon and by similar transcripts in dog, chicken, finch and frog (DN430942, BU271565, DC286485, DN081198/CK800288).
Although some transcripts skip it, this exon has no transcripts of its own to support standalone gene status. Its phase 2 splice junction has been conserved for many billions of years of branch length in bony vertebrates. It contains two well-established Pfam domains proving that it is not conserved-non-coding dna as concluded by some bioinformatic tools. A string of 2166 bp implausibly has an open reading frame of this length without a stop codon unless it encodes a protein.
Various gene prediction tools find this extending exon (Genscan, Geneid, N-Scan, SGP, UniGene, Exoniphy) while others fail to predict it (Ensembl, Encode, CCDS, MGC, Vega, and AceView). Oddly the predicted NCBI gene models in birds and lizards all correctly contain the exon (XM_421099 XM_003206406 XM_002199814 XM_003214639) but this information was never related to human genome annotation.
No further attention will be paid to the erroneous gene model on this site. It is impossible to understand protein function when 60% of the protein and two informative domains have been dropped.
Crystal structure of PRDM11
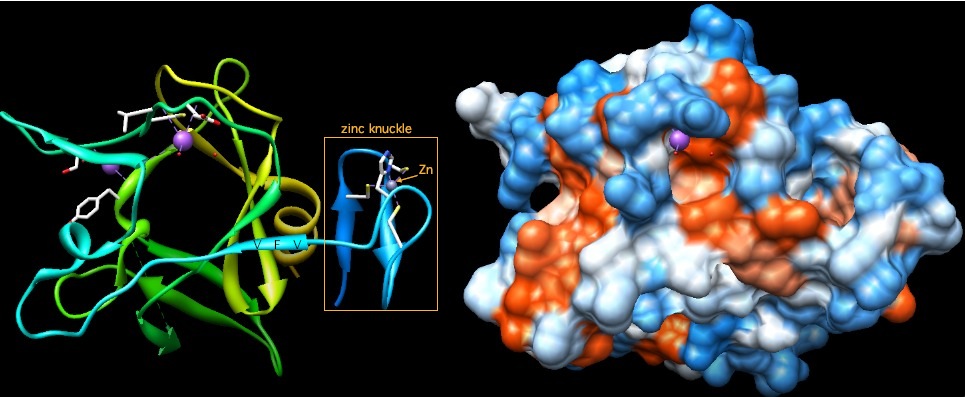
Here we are very fortunate to have a pre-publication entry at PDB (3RAY) that covers the zinc knuckle and PR(SET) domain of human PRDM11. The four zinc binding residues (3 cysteins and 1 histidine are clearly identified. Since this is the closest match PRDM7/9 have at PDB -- and PRDM11 is the closest match for the PR(SET) domain -- the setting for modelling PRDM7/9 is quite favorable. Note the two genes have identical positioning and phasing of 4 consecutive introns but earlier and later exons have no detectable homology.
>PRDM11_homSap 3RAY zinc knuckle PR(SET) domains
GDSSAMEVEPKKLKGKRDLIVPKSFQQVDFWFCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDT
SGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWLIVDKNNRYKSIDGSDETKANWMRYVVISREEREQNLLAFQH
SERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLARGEKRLQREKSEQVLDNPEDLRGPIHLSVLRQGK
introns 12 b1b aaaaaa b2b 00 21 BB4BB bb3bbb
PRDM11: QQVDFWFCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEV--IPKGHIFGPYEGQISTQDKSAGF-FSWLIVDKNNRYKSIDGSDETKANWMRYVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSM
Q D+ +CE CQ +F+D C HGPP FV D+ V G P+R+AL++P G+ + ++ + NE +P G FGPYEG+I+ +++A +SWLI N Y+ +DG D++ ANWMRYV +R++ EQNL+AFQ+ +I++R CR IRPG L VWY ++Y + L
PRDM9: QDDDYLYCEMCQNFFIDSCAAHGPPTFVKDSAVDKGHPNRSALSLPPGLRIGPSGIPQAGLGVWNEASDLPLGLHFGPYEGRITEDEEAANNGYSWLITKGRNCYEYVDGKDKSWANWMRYVNCARDDEEQNLVAFQYHRQIFYRTCRVIRPGCELLVWYGDEYGQELGIK
introns 12 00 21
The two Chimera displays of this region of PRDM11 show the hydrophobicity surface and secondary structure ribbons. The two bright purple atoms are sodium ions -- their significance is unknown. The zinc knuckle on the right shows how the zinc atom is chelated, creating a rigid knob from a short stretch of peptide.
This short type of zinc finger is not thought to bind a dna trinucleotide in the manner of the C2H2 zinc fingers in the terminal array of PRDM7/9; however it may serve as a protein recognition site for binding partners of PRDM11.
The percentage identity between PRDM11 and PRDM7/9 means 3RAY provides a satisfactory template for the latter. Note the 24 residues between the two domains are as well conserved as the domains themselves. This is explained in part by three inter-domain residues VFV extending a 3-strand beta sheet within the PR(SET) domain; a later alpha helix formed by DRAALT and following IP also appears nestled against the PR(SET) domain. The motif TIP is especially deeply conserved in all 16 human PR(SET) proteins.
Crystallographic structures for PRDM4 (2L9Z) and PRDM10 (3IXH) confirm the conservation of structural relationships observed between the two domains in PRDM11. This suggests that the two domains are not structurally independent but are instead fold co-adaptively. Indeed they are found in the same configuration in PRDM4, PRDM6, PRDM10 and PRDM15. However the nine other PR(SET) domains diverging earlier in the domain tree lack the zinc knuckle. The zinc knuckle domain is not known to occur independently.
Other domains in the respective proteins differ, implying that they have more relaxed structural interactions with the knuckle-PR(SET) pair befitting the mix-and-match assembly of chimeric proteins. However intronation greatly restricts chimeric gene formation because conjoined domains must must sustain a consistent reading frame.
Curated PRDM11 reference sequences
The reference sequence for the human gene PRDM11 appears to be wrong in two respects: translating a segment of 5'UTR and running out into a stop codon rather than splicing to the large terminal exon. This gene model is not pursued further here for lack of comparative genomics and because the omitted exon contains two domains presumably vital to the role of PRDM11 in regulation of gene transcription. That is, a methylase domain alone without the transcription factor zinc finger and dimerization domain is a dubious functional proposition.
>PRDM11_homSap Homo sapiens (human) original refSeq 511 aa model not supported PhosS 3RAY coverage knuckle SET 0 MLKMAEPIASLMIVECRACLRCSPLFLYQREK 0 0 DRMTENMKECLAQTNAAVGDMVTVVKTEVCSPLRDQEYGQPC 2 1 SRRPDSSAMEVEPKKLKGKRDLIVPKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGSDETKANWMR 2 1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR 1 2 GEKRLQREKSEQVLDNPEDLRGPIHLSVLRQGKSPYKRGFDEGDVHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLVIRKVPKYQDDAYSQCATTMTHGVQNIGQTQGEGDWKVPQGVSKEPGQLEDEEEEPSSFKADSPAEASLASDPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPPVLPPQVLELPEFSDPAGKLVWMRLLSEGRVRSGLCGG* 0
The sequences below use a gene model of human PRDM11 consistent with transcript data and vertebrate comparative genomics. The coding regions consists of seven exons and 1233 total amino acids, including the second largest exon in the entire proteome (722 amino acids). An early serine is post-translationally modified to phosphoserine (according to its UniProt entry Q9NQV5). There are four domains conserved in all species: an early zinc knuckle, a PR(SET) methylation domain, a ZNF_TTF transcription factor zinc finger and a distal hATC dimerization domain.
All species containing the gene have these four domains and the same introns. While much older than PRDM7/9, PRDM11 cannot be traced back earlier than rayfinned fish and may have arisen in stem bony vertebrates though chondrichthyian data is admittedly meagre. Fish sequences are showing major signs of divergence in early exons and are also highly variable in the sixth exon. The PR(SET) domain of PRDM11 is the most closely related such domain found in non-mammalian amniotes (which lack and never had PRDM7/9).
>PRDM11_homSap Homo sapiens (human) corrected 511+722 aa PhosS 3RAY coverage knuckle SET ZnF_TTF hATC_dimerization no early zinc finger or terminal array syn PFM8 related ZNF 862 0 MTENMKECLAQTNAAVGDMVTVVKTEVCSPLRDQEYGQPC 2 1 SRRPDSSAMEVEPKKLKGKRDLIVPKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGSDETKANWMR 2 1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR 1 2 GEKRLQREKSEQVLDNPEDLRGPIHLSVLRQGKSPYKRGFDEGDVHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLVIRKVPKYQDDAYSQCATTMTHGVQNIGQTQGEGDWKVPQGVSKEPGQLEDEEEEPSSFKADSPAEASLASDPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPPVLPPQVLELPEFSDPA 1 2 ASESMVSGPAIMEDDDQEVDSADESVSNDMMTATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSVILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAF SALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTAATLCEETEFLGDIRAVRWIIGEQN VLNALIKDYLEVVAHLKEVSSQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYIFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAMKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRIFVKACQVFDLAAWPRSSEELMSYGKEDMVQIFDHLEAIPTFSRDVCREGLDPRGSLLMEWRELKADYYTKNGFKDLISHICKYKQRFPLLNK IIQVLKVLPTSTACCEKGRNALQRVRKNHRSRLTLEQLSDLLTIAVNGPPITNFDAKRALDSWFEEKSGNSYALSAEVLSRMSALEQKPALQTMDHGTEFYPDI* 0 >PRDM11_musMus Mus musculus (mouse) blat PhosS 3RAY coverage knuckle SET ZnF_TTF hATC_dimerization 0 MTENMKECLAHTKAAVGDMVTVVKTEVCSPLRDQEYGQPC 2 1 SRRLEPSSMEVEPKKLKGKRDLIVTKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDAGGESDVRCINEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGSDETKANWMR 2 1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGERLRVWYSEDYMKRLHSMSQETIHRNLAR 1 2 GEKRLQREKAEQALENPEDLRGPTQFPVLKQGRSPYKRSFDEGDIHPQAKKKKIDLIFKDVLEASLESGNVEARQLALSTSLVIRKVPKYQDDDYGRAALTQGICRTPGEGDWKVPQRVAKELGPLEDEEEEPTSFKADSPAEASLASDPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPSLLPPQVLELPEFSDPA 1 2 ASESMVSGPAIMEDDDQEVDSADESVSNDVMTATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAF AALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTASTLCEETEFLGDIRAVRWIIGEQN VLNALIKDYLEVVAHLKDVSSQTQRADASAIALALLQFLMDYQSMKLIYFLLDVIAVLSRLAYIFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAVKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRVFVKACQVFDLAAWPRNSEELLSFGKEDMVQIFDHLEAIPAFSRDVCREGTDPRGSLLMEWRDLKADYYTKNGFKDLLSHICKYKQRFPLLNK IIQVLKVLPTSTACCEKGRSALQRVRKNHRSRLTLEQLSDLLTIAVNGPPIANFDAKRALDSWFEEKSGNSYTLSAEVLSRMSALEQKPMLHVVDHGSEFYPDM* 0 >PRDM11_canFam Canis familiaris (dog) blat PhosS 3RAY coverage knuckle SET ZnF_TTF hATC_dimerization 0 MTENMKECLAQTKAAVGDMVTVVKTEVYSPLRDQEYGQPC 2 1 SRRPDPSTMEVEPKKLKGKRELIMPKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKEASGENDVRCINEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGSDETKANWMR 2 1 YVVISREEREQNLLAFQHSERIYFRACRDIRPGERLRVWYSEDYMKRLHSMSQETIHHNLAR 1 2 GEKRLQREKSEQALDNPEDLRGPIQLPVLRQGKSPYKRGFDEGDAHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLVIRKVPKYQDDAYSRCAMTMSHGVQNVSRTQGEGDWKIPQGASKEPGPLEDEEEEPSSFKADSPAEASLASDPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPPVLPAQVLELPEFSDPA 1 2 ASESMVSGPAIMEDDDQEVDSADESVSNDMLTAADEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAF SALGIRLQDEKPTVGLGIDGANVTASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTASTLCEETEFLGDIRAVRWIIGEQN VLNALIKDYLEVVAHLKDVSSQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYVFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAMKNLRVAE AKFQSVREKICQKTQVILAQRFDSRSRIFVKACQVFDLAAWPRSSEELMSYGKEDMVQIFDHLEAIPTFSRDVCREGLDPRGSLLMEWRELKADYYTKNGFKDLIGHICKYKQRFPLLNK IIQVLKVLPTSTACCEKGRNALQRVRKNHRSRLTLEQLSDLLTIAVNGPPIANFDAKRALDSWFEEKSGNSYALSAEVLSRMSALEQKPVLQTVDHGSEFYPDI* 0 >PRDM11_loxAfr Loxodonta africana (elephant) blat PhosS 3RAY coverage knuckle SET ZnF_TTF hATC_dimerization 0 MTENMKECLAKTKAAVGDMVPVVKTEVCSPLHDQEYGQPC 2 1 SRRPDPSAMDVEPKKLKGKRDLIMPKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKEASGENDVRCISEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSVDGSDETKANWMR 2 1 YVVISREEREQNLLAFQHSERIYFRVCRDIRPGERLRVWYSQDYMKRLHSMSQETIHRNLAR 1 2 GEKRLQREKSEQALDNPEDLRGSIQLPVLRQGKSPYKRGFDEGDLHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLVIRKVPKYQDDTYSRCATTMSHGVQNVSRTQGEGDWKIPQGASKEPGPTEDEEEEPSSFKADSPAEASLASDPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPPVLPAQVLELPEFSDPA 1 2 ASESMVSGPAIMEDDDQEVDSADESVSNDIMTATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSGTESYLQALDRAF STLGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTASTLCEETEFLGDIRAVKWIIGEQN VLNALIKDYLEVVAHLKDISSQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYVFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAMKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRIFVKACQVFDLAAWPRSSEELVSYGKEDMVQIFDHLEAIPTFSRDVCREGLDPRGSLLMEWRELKADYYTKNGFKDLISHVGKYKQRFPLLNK IIQILKVLPTSTVCCEKGRNALQRVRKNHRSRLTLEQLSDLLTIAVNGPPIANFDAKRALDSWFEEKSGHSYTLSAEVLSRMSALEQKPVLQAIDHGTEFYPDI* 0 >PRDM11_monDom Monodelphis domestica (opossum) blat PhosS knuckle SET ZnF_TTF hATC_dimerization 0 MTENLKACLAHTQASMGEMVTVKTEVCSPRRDQEYGQPW 2 1 SGRPDPPSMEVEPKKPKGKRELIMTKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPMGIPDRAALTIPPGMEVVKEASGQSDVRCMNEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNHYKSIDGTDETKANWMR 2 1 YVVISREEREQNLMAFQHSEKIYFRVCRDIRPGERLRVWYSEDYMKRLHSMSQETIQRNLTR 1 2 GDKKLQREKPEKALDHQEDLRGPLQLTVLRHGKSAYKRGFDEVDAHPPPKKKKIDLIFKDVLEASLETSKIEEHPLAPGTPLVLRKAPKFHTEDVYDQCGMAISHGPQDLSRNQGEKEWKAPQGASYGPSKDTSLLEDEEEEPSSFKADSPAEASLASDLHELPTTSFCPNCIRLKKKIRELQAELDMLKSGKLPEPPLLPPQVPELPEFSDPT 1 2 ASESVVSVPTMLEDDDQEVDSADESVSNEMITATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYFDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDILADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRLTASTLCEETEFLGDIRAVRWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYIFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGVAVKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRTFVKACQVFDLAAWPRSTEELMSYGKEDMIQIFDHLETIPSFSREICREGMDPRGSLLMEWRELKADYYTKNGFKDLISHICKYKQRFPLLNKI IQILKVLPTSTACCEKGRVALQRVRKNNRSRLTLEQLSDLLTIAVNGPPIANFDAKRALDSWFEEKSGNSYTLSAEVLSRMSALDQKPMLPTMDHGSEFYSDL* 0 >PRDM11_ornAna Ornithorhynchus anatinus (platypus) blat PhosS knuckle SET ZnF_TTF hATC_dimerization 0 MTENLKDCLAQTQASMGEMVTV KTEVCSPHRDQEYGQPW 2 1 SGRPDPSSMEVEPKKLKGKRDLIMSKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPPGIEVVKEASGENDVRCMNEVIPKGHIFGPYEGQISTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGTDETKANWMR 2 1 YIVISREEREQNLLAFQHSERIYFRVCRDIRPGERLRVWYSEDYMKRLHSMSQETIHRNLTR 1 2 GEKKLLREKTDKAPESQEDLRGPLQLTVLKQGKSPYKRSCDEGDAHPQTKKKKIDLIFKDVLEASLESAKIDEHQLATSTPLAFKKMPKFQAEDVFERCGAILPHGTQSFGRTHSEGDWKLGHGTPYGPSKEKGLLEEDQGEPSPIKVDSPTEASLTGDSQELPTTSFCPNCIRLKKKIRELQAELDMLKSGQLPEPPLVPPQVPELPEFSDPA 1 2 ASESLVSIPTILEDDDPEVDSADESVSNDMIAATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF AALGVRLQDEKPTVGLGVDGANVTASLRAGMFMTVRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRLTAATLCEETEFLGDIRAVRWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYVFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGVAVKNLRVAE AKFQSIREKICQKAQVILAQRFDSRSRTFVKACQVFDLAAWPRSSEELVSYGREDMVQILEHLEAIPSFSREVCREGADPRGALLTEWRELKADYYTKNGFKDLIGHVGKYKQRFPLLNK VIQILKVLPTSTACCEKGRSALQRVRKNHRSRLTLEQLSDLLTIAVNGPPIAHFDAKRALDSWFEEKSGNSYALSAEVLSRMSSLDQKPMLQSVDHGSEFYPDM* 0 >PRDM11_allMis Alligator mississippiensis (alligator) scaffold:58581 PhosS knuckle SET ZnF_TTF hATC_dimerization 0 MSENLKDCLIQTQTSLGEMVTIKTEACSPHRDQEYGQPC 2 1 SGRPDPQSMEIEPKKLKGKRDLIMTKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRASLTIPPGMEVVKEPNGENDVRCMNEVIPKGHIFGPYEGQISSQDKSAGFFSWL 0 0 IVDKNNRYKSIDGTDETKANWMR 2 1 YVIISREEREQNLMAFQHSERIYFRTCRDIRPGERLRVWYSEDYMKRLHSMSQETINRNLTR 1 2 GDKKSQREKSEKNMENQEDMRGPLQLTTLKQGKSPYKRSCEEAESHPQTKKKKIDLIFKDVLEASLESAKLEEHQLTTSTPLSIRKASKYQTEDVFERCGTTIQHSSPNLSRNRSEGEWKVPHSSSFSTAKEMGLLEDEEEEPLSLKADSPTEPSLASTQGNSHEIPTTSFCPNCIRLKKKIRELQAELDMLRSGKLPEQPALAPQVPELQEFSDPT 1 2 ASESIISVPTIMEDDDQEVDSADESVSNDMIAATDEPSKMSSVTGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPKLMCELRVTAATLCEETEFLGDIRAVKWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYVFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAVKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRTFVKACQVFDLAAWPRSTEELMSYGKEDMVQIFEHLETVPSFSREVCREGMDIRGSLLMEWRELKVDYYTKNGFKDLLGHICKYKQRFPLLNK IVQILKVLPTSSACCEKGRNALQRVRKNNRSRLTLEQLSDLLTIAVNGPAIANFDCKRALDSWFEEKSGNSYALSAEMLSRMSSLDQKPMLQSMDHGSEFYPDI* 0 >PRDM11_galGal Gallus gallus (chicken) XM_421099 PhosS knuckle SET ZnF_TTF hATC_dimerization 0 MSENLKDCLNQTQASLGEMVTIKTEACSPHRDQEYGQPC 2 1 SGRLDPQSMDVEPKKLKGKRDLIMTKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPPGIEVVKEPSGENDVRCMNEVIPKGHIFGPYEGQISSQDKSAGFFSWL 0 0 IVDKNNRYKSIDGTDETKANWMR 2 1 YVIISREEREQNLMAFQHSERIYFRACRDIRPGEKLRVWYSEDYMKRLHSMSQETINRNLTT 1 2 GDKKLQKEKSEKNADNQEDTRAPLHFTTLKQGKSPYKRSYDEGESHPQTKKKKIDLIFKDVLEASLESAKFEEKQLATSTPLSTRATSKYQAEEIFERCSSAMQHGSLNLSRNRSEEEWKAPHGSSFSSAKEVGVLEDEEEEPLSLKADSPTELSLASAEGNSHEIPTTSFCPNCIRLKKKIRELQAELDMLRSGKLPEPPVLPPQVPELQEFSDPT 1 2 ASESIISVPTIMEDDDQEVDSADESVSNEMIAATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDVLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTASTLCEETEFLGDIRAVKWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYVFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAVKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRTFVKACQVFDLAAWPRSTDELMSYGKEDMVQIFEHLETVPSFSREVCREGMDTQGSLLMEWRELKVDYYTKNGFKDLLSHICKYKQRFPLLNK IVQILKVLPTSSACCEKGRNALQRVRKNNRSRLTLEQLSDLLTIAVNGPPIANFDCKRALDSWFEEKSGNSYALSAEMLSRMSSLDQKPMLQSVDHGSEFYPDI* 0 >PRDM11_melGal Meleagris gallopavo (turkey) blat/XM_003206406 PhosS knuckle SET ZnF_TTF hATC_dimerization 0 MSENLKDCLNQTQASLGEMVTIKTEACSPHQDQEYGQPC 2 1 SGRPDPQSMDVEPKKLKGKRDLIMTKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPPGIEVVKEPSGENDVRCMNEVIPKGHIFGPYEGQISSQDKSAGFFSWL 0 0 IVDKNNRYKSIDGTDETKANWMR 2 1 YVIISREEREQNLMAFQHSERIYFRACRDIRPGEKLRVWYSEDYMKRLHSMSQETINRNLTT 1 2 GDKKLQKEKSEKNTDNQEDTRGPLQFTMLKQGKSPYKRSYDEGESHPQTKKKKIDLIFKDVLEASLESAKFEEKQLATSTPLSTRATSKYQAEEIFERCSGAMQHLSRNRSEEEWKAPHGSSLSSAKEVGVLEDEEEEPLSLKADSPTELSLASAEGNSHEIPTTSFCPNCIRLKKKIRELQAELDMLRSGKLPEPSVLPPQVPELQEFSDPT 1 2 ASESIISVPTIMEDDDQEVDSADESVSNEMIAATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDVLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTASTLCEETEFLGDIRAVKWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYVFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAVKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRTFVKACQVFDLAAWPRSTDELMSYGKEDMVQIFEHLETVPSFSREVCREGMDTQGSLLMEWRELKVDYYTKNGFKDLLSHICKYKQRFPLLNK IVQILKVLPTSSACCEKGRNALQRVRKNNRSrlTLEQLSDLLTIAVNGPPIANFDCKRALDSWFEEKSGNSYALSAEMLSRMSSLDQKPMLQSVDHGSEFYPDi* 0 >PRDM11_anaPla Anas platyrhynchos (duck) blast/HQ902403 PhosS knuckle SET ZnF_TTF hATC_dimerization 0 MSENLKDCLNQTQASLGEMVTIKTEACSPHRDQEYGQPc 2 1 SGRPDPQSMDVEPKKLKGKRDLIVTKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPPGMEVVKEPSGENDVRCMNEVIPKGHIFGPYEGQISSQDKSAGFFSWL 0 0 IVDKNNRYKSIDGTDETKANWMR 2 1 YVIISREEREQNLMAFQHSERIYFRACRDIRPGEKLRVWYSEDYMKRLHSMSQETINRNLTR 1 2 GDKRLQREKSEKNVENQEDMRGPLQLTTLKQGKSPYKRSCDEGESHPQTKKKKIDLIFKDVLEASLESAKFEENQLATSTPLSIRTASKYQAEDIFERCGTAMQHGSLNLSRNRSEEEWKIPHGSSFSSAKEVGILDDDEEEPLSLKADSPTELSLASAQGNSHEIPTTSFCPNCIRLKKKIRELQAELDMLRSGKLPEPPELPPQVPELQEFSDPT 1 2 ASESIISVPTIMEDDDQEVDSADESVSNDMIAATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDVLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYVFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAVKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRTFVKACQVFDLAAWPRSTDELMSYGKEDMVQIFEHLETVPSFSREVCREGMDTRGSLLMEWRELKVDYYTKNGFKDLLSHICKYKQRFPLLNK IVQILKVLPTSSACCEKGRNALQRVRKNNRSRLTLEQLSDLLTIAVNGPPIANFDCKRALDSWFEEKSGNSYALSAEMLSRMSSLDQKPMLQSMDHGSEFYPDI* 0 >PRDM11_taeGut Taeniopygia guttata (finch) XM_002199814 PhosS knuckle SET ZnF_TTF hATC_dimerization 0 MSENLRDCLIQTQASLREMVTIKTEACSPHRDQEYGQPC 2 1 SGRPDPQSMEMEPKKLKGKRDVIMTKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPPGMEVVKEPSGENDVRCMNEVIPKGHIFGPYEGQISSQDRSAGFFSWL 0 0 IVDKNNRYKSIDGTDETKANWMR 2 1 YVIISREEREQNLMAFQHSERIYFRACRDIHPGEKLRVWYSEDYMKRLHSMSQETMNRSFTS 1 2 GDKMLQNENSEKNVENQEDARGALQFTTLKQGKSPYKRSCDEGESHPQTKKKKIDLIFKDVLEASLESAKFEENQLATSTPLSLRRASKYQAEDIFEQCGNAMQRSSLSLSRNQSESEWRVPHSSSFISAKEMSILEDEEEEPLSLKADSPTELSLASAQGNSHEIPSTSFCPNCIRLKKKIRELQAELDMLRSGKLPEAPVLPPQVPELQEFSDPT 1 2 ASESIISVPTIMEDDDQEVDSADESVSNDMIAATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYVFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGIAVKNLRVAE AKFQSIREKICQKTQVILAQRFDSRSRTFVKACQVFDLAAWPRSTDELMSYGKEDMVQIFEHLETVPSFSREVCREGMDTRGSLLMEWRELKVDYYTKNGFKDLLSHICKYKQRFPLLNK IVQILKVLPTSSACCEKGRSALQRVRKNNRSRLTLEQLSDLLTIAVNGPPIANFDCKRALDSWFEEKSGNSYALSAEMLSRMSSLDQKPMLQSMDHGSEFYPDI* 0 >PRDM11_anoCar Anolis carolinensis (lizard) blat/XM_003214639 PhosS knuckle SET ZnF_TTF hATC_dimerization 0 MSEKLNDCLGEMVTIKTEPCSPCREEEYGQLW 2 1 SSRKVDSQSVDVEPKKLKGKQDLIMSKSFQQVDFW 1 2 FCESCQEYFVDECPNHGPPMFLSDAPVPIGIPDRAALTVPPGMEVVKEANGERDVRCVGEIIPKGRIYGPYEGKLSSQDKSAGFFSWL 0 0 IVDKNNRYKSIDGTDETTSNWMR 2 1 YVAISREEREQNLMAFQHSERIYFRTCRDIRPGERLRVWYSEDYMKRLHSMSQETINRNLTR 1 2 GDKKSLRERSERNTENQMEMLYPLELTISKQGKSPYKRCSEEGVSQPQAKKKKIDLIFKDVLEASLESTKMEEHKVTRNSAPSTRKSSRFREQDASESCGTGMQHNSPTHSGSRN EDEWKVPHGPSFSVSKETGLLEDEGEEPLSFKLNSPTDLTLAPIDDEALGLPTTSLCPNCIRLKKKIRELQAELNMLRSGKLVEPPLLPPQVPEYQAFSYPT 1 2 ASETIMSVPTIMEDDDQEVDSADESVSNDMITATDEPSKMSAVTRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYRLRMHPEKTEEM CRNMTLLFNTAYHLAMEGRPYCDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERVRQSPFLSIILDGQSDDLLADTVAVYVQYISSDGPPATEFLSLQELGFSATDSYIQALDRAF SSLGIRLQDERPSVGLGIDGANITASLRANMYMTIRKTLPWLLCLPLMIHKPHLEVLDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAFIFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEENFRESFNGVAVKNLRVAE AKFQSIREKICQKTQVILAQRFEPRTRAFVKACQVFDLAMWPRSAEELMSYGREDMVQIFDHLEAVPTFSTDIIREGMDTRGSLLMEWRELKVDYYTKNGFKDLISHICKYRQRFPLLNK IIQILKVLPTSTACCEKGRNALQRVRKNNRSRLTLEQLSDLLTIAVNGPPIANFEAKRALDSWFEEKSSNSYALSAEMLSRMSSLDHKPMLQSMDHGSEFYPDI* 0 >PRDM11_xenTro Xenopus tropicalis (frog) blat/CF781198 PhosS 3RAY coverage knuckle SET ZnF_TTF hATC_dimerization 1 MSEISKECRVAFSPSLGDIVRVKREVGSPVEEQGYGHFR 2 1 SSVGPNSRCLDMEPKRLKEKRESTMSKSLQQVDFW 1 2 FCESCQEYFVDECPSHGPPILVPDTLVPIGMPERAALSVPCGIEVVKDSSGESEVRCVNEVIPKGHMFGPYEGQICSQDKSSGFFSWL 0 0 IVDNNNRYKSIDGTDEAYANWMR 2 1 YVVISREEREQNLMAFQHSEKIYFRTCRDIQPGEKLRVWYSEDYMKRLHSMSQETINRNLTQ 1 2 GDKRLLRENNERLLENQEDVKGTFPLATLKQGKSLYKRSCEEVDLHPQTKKKKIDLIFKDVLEASLETARIDEYHLVTSSPLSGQKKNPKYLYENHGDRCRMNRQCSSPQNQIRNMRDWKAKHVSASGLNRQASFPEDEVEDHSSVKAESPTESSAIGNVDEIPTTSFCPNCIRLKKKIRELQAELEMLRSEKMAETSQMTNQINEIPEFADAS 1 2 APEGVAIATTMIDDDEQEVDSADESVSNDMMAATDEPSKMSAGSGRRIRRFKQEWLKKFWFLRYSSTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLAMEGRPYYDFRPLAELLRKCELRVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSEDLLADTVAVYVQYTSSDGPPATEFLSLQELGLPTTESYLQGIDRAF SALGIRLQDERPTVGLGVDGANITAGLRANLYMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRLTAATLCEETEFLGDIRAVKWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADTSAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYVFQGEYLLVSQVDEKVEEAIQEISRLTDSPGEYLQEFEENFRESFNGIALKNLRVAE AKFQSIREKICQKTQVTLAQRFDSRSRMFVKACQVFDLSTWPRTTEELINYGEEDMLQIYELLETIPNFLHDLGREVADTRGNLLMEWRELKADYCTKNGFKDLIGHICKYKQRFLFLNK IVQILKVLPTSTACCEKGRNALQRLRKNNRSRLTLDQMSDLLAIAVNGAPIANFDAKRALDSWFEEKSGNSYSLSAEMLSKMSSLDQKPLLQPMEHGSEYYQDI* 0 >PRDM11_latCha Latimeria chalumnae (coelocanth) AFYH01005054 knuckle SET ZnF_TTF hATC_dimerization 1 2 1 STGAAETPRIEGEPKRTKGKLETIMAKTLQQVDFW 1 2 FCEECQEHFVVECPTHGPPVFTMDTPVPVGMPERAALTAPPGIHIVKGSNGEIDVECVDEVVQKGRIFGPYEGQITTQDKSAGFFSWL 0 0 IVDKNNRYKSIDGTDETKANWMR 2 1 YVVISRDEREQNLLAFQHSEKIYFRASRNLHPGERLRVWYSDEYMKRLHSMSQETIDRNLTA 1 2 GNLKLQRENSEEGWDAQENLRGMLLKQGKSSYKRGNDEAESHQQPKRKKIDLIFKDVLEASLESSKLEGNSLATSSPLPLKKPIKFQLEDVLQKPEYYYKHVSQLLGRGEVEWKSHQKSGCSLPDNEDSDRIKEESPDEVPGNSTEEDPEDVPTTSFCPNCIRLKKKIRELQEELDRLRSGQPPASPQLPQVQELLEPPG 2 VQEAHPSVSLMEDDDQEVDSADESVSNDMIAASDETSKITVGASRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTVLFNTAYHLALEGRPYLDFRPLSDLLRKCELKVVDQYMNEGDCQILIHHIARALQEDLIERIRQSPFLSVILDGQTDDILADTVAVYIQYTTSDGPPATEFLSLQELGCVTTDSYVQAVDRAF AVFGLRLQDQRNVVGLGVDGTCLTAGLRANLFMTIRKTLPWLLCLPFMVHKPHLEVLDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKWIIGEQN VLNALIKDYLEVVAHLKDVSGQTQRADAAAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAFVFQGEYLLVCQVDDKIEEAIQEISRLSDSPGEYLMEFEENFRESFNGIALKNLRVAE AKFQSIREKICQKTQVILAQRFDNRSRPFIKACQMLDIATWPRSTDDLKNFGEEEIMVIYEQLELVPTFAREVCREGTDNRGSLVMEWRELKADFYSKNGFKDLIGHICKYRQRFPILNR VLQILKVLPSSAACCEKGRSALQRIRKNNRSRLTLDQLNDLLTIAINGPSIANFDAKRALDSWFEEKSGNSYALSAEVLNRMSADQKPMLQGMDFVSDFYPDI* 0 >PRDM11_danRef Danio rerio (zebrafish) blat/EB776339/EB946706/BX088562/XM_688756 knuckle SET ZnF_TTF hATC_dimerization 0 MADSSTNPDHSSMEAEGECSTS 2 1 ASNEKSAEEPNKRLKVEHERYSSFW 1 2 FCEECKKYYLEDCPTHGPPVFVPDTPVVSGVPNRAALTAPSGIEVRRNGDKVDVYCMDEKIPKGALFGPYKGQIMASDKPSGPYSWM 0 0 IVDKDSKYKFIDGSDEATANWMR 2 1 FIHITSDESEQNLSAFQHGDQIYFRVCHRLKVGEKLGVWYSSEYMKRLQSVSRDSIDHNLDT 1 2 GVKSEDQEEPKGPVLRSAMHGRRTLSKHGSDEAENQPQAKKKKIDLIFKDVLEASLEANQSQNNPLNSTLSFPRARTNVCQVFCHPDAESKESVFTSGLVSMDHHEIGFDGKCIKMENTEEDEALTSEGPSTSFCPNCVRLKRRIRELEAELHRLRGQGHAEVKPVPASEMLAGEDHR 1 2 DTMTPIPAALEEDDQDVDSADESISADLLVAADESSKLSVGSGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMMTLLNAAYHLALEGRPFSDLRPLAELLKKCDLKVVDQYMNENDCQILIQHISRAFKEDLAEKIRLSPFLSIIMDGQNDDLLADMVAVYVQFTTTDGLPATEFLSLQRLCGGNVEGYLQAVDRAF GVLGLRLQDMLVVGLGIDGSNISSSLRANLYVAIRKTIPWVLCLPVMIHRPHLEVLDAISGKELSCLEDLENNLKQLLSFYRYSPRLMAELRSSAPTLSEETEFLGDIRAVRWIIGEPN VLNALIKDYLEVVAHLKTISNQTQRGDAAAIALSLLQFLLDYQSVKLIYFLLDVIAVFSRLAFIFQGDYLLVSQVDAKIEDAIHEIGQLVDSPGEYLQEFEDNFRESFNGVDLKNLRVAE SKFQSIREKICQQSQCILAQRFEPRSRTVVQACQVLDLASWPINRDDLGAYGEEEILVIFDHLETIPSSGRERSIERTDARGSLVVEWRDLKADYCSVNGFKEVVSHIFRYKQRYPLLN HILQIVRVLPTSTTCCDKGRGSLQKVRRNSRSRLTLDQINDLLTLAVNGPPIGSFDGKRALDSWFEEKSGNSISLSTEVLSRMSTTEQKSVLHNMDMNAEYYPDV* 0 >PRDM11_oreNil Oreochromis niloticus (tilapia) XM_003458287 knuckle SET ZnF_TTF hATC_dimerization first exons uncertain 0 MASENCRQIASCLQIEAEKAWRRSEPGRALC 1 2 fCEECQDCFHKECPSHGPPLFIQDTHAAPGTANRAALTVPSGLEVFSEEDEVDVRCVDAIYPKGALFGPYEGELVSKDRSSGFFSWI 0 0 IVDVNNTYQSIDGSDETKANWMR 2 1 YVRTSSEESDRNLTAFQHGKNIYFRVCRALVAGEKLRVWYSDDYIRRLHCVSQESIDRNLDT 1 2 GPGKDFKSRCLQSALQGKLSKQLSEESDGQPPAKRKKIDLIFKDVLEASLEESGKFRSRSGQPSEYKVPALVSRFDSSETGFGIPNLKVEEKEEEENQNTEKPSTSFCPNCVKLKRRILELEEELSRLRGEQRDAAASATSEQTQPQRDQAPPHPEQGPIEDFQ 1 2 GMEPLTPTQVVLDEDDQDVDSADESIAADLVISPEDSSKLSSGGGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CKNMMLLFNAAYHLALEGRPFSDLRSLAELLKKCELKVVDQYMNEGDCQILIHHIARAVKEDLAEKMRLSPFLSVIMDAQNDDLFSDMVAVYVQFVTNEGSPNTEFLSLQRLTVANVDGYLQVMDRAF GVLGLRFQDLLVVGLGVDGTNISSGMRANLYIAVQKTFPWILCLPIMIHRPHLEVLDAISGKELSCLEDLENNLKQLLSFYRYSPRMMAELRSTAPTLSEETEFLGDIRAIRWIIGEPN VLNALIKDYLEVVAHLKEISSQTQRADAAAIALTLLQFLMDYQSVKLIYFLLDIIAILSRLAFTFQGEYLLVSQVEAKIEEAIQEIGQLVDCPGEYLQEFEENFRESFNGVALKNLRVAE SKFQSIREKICNRSQSILSQRLDLQSRSFAKACKVLDLSTWPSNHEDLQAYGDEEIKIIFNHLESIPTAAQEGSQTEARGSLVVEWKDLKADYYSMNGFKEVIGHICRYKQRFPLLN RIVQVIRVLPSSTACCDKGRGSLQKMCKNNRSRLTLEQMNDLLTVAINGPPIANFDGKRALDSWFEEKSGSSYSLSAEVLNRMSAADQKCVLHSVDVNAEFYPDV* 0
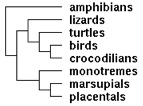
PRDM11 fragmentary sequences from the unrecognized terminal exon were utilized in a Dec 2011 study by XX Shen et al of informative loci to determine the topology of the amniote tree, resolving the taxonomic position of turtles: outgroup to crocodillians + birds. (Note a microRNA-based analysis on the same date proposed outgroup to lizards.) Because that study sequenced species rarely represented at GenBank such as skink, salamander, caecilian and amphibia, translated sequences are reproduced here and supplemented with the appropriate region of the full length sequences PRDM11 above (which were not all used in the study). The fragment studied begins 16 residues within the ZNF_TTF zinc finger and terminates well before the end of the exon. The difference alignment shows a number of group synapomorphies consistent with turtle placement (though one gene alone does not suffice to reliably determine the species tree).
>PRDM11_homSap Homo sapiens (human) revised 511+722 aa WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSVILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAF SALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTAATLCEETEFLGDIRAVRW >PRDM11_musMus Mus musculus (mouse) blat WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAF AALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTASTLCEETEFLGDIRAVRW >PRDM11_canFam Canis familiaris (dog) blat WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAF SALGIRLQDEKPTVGLGIDGANVTASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTASTLCEETEFLGDIRAVRW >PRDM11_loxAfr Loxodonta africana (elephant) blat WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSGTESYLQALDRAF STLGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTASTLCEETEFLGDIRAVKW >PRDM11_monDom Monodelphis domestica (opossum) blat WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYFDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDILADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRLTASTLCEETEFLGDIRAVR >PRDM11_ornAna Ornithorhynchus anatinus (platypus) blat WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF AALGVRLQDEKPTVGLGVDGANVTASLRAGMFMTVRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRLTAATLCEETEFLGDIRAVRW >PRDM11_galGal Gallus gallus XM_421099 WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDVLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTASTLCEETEFLGDIRAVKW >PRDM11_melGal Meleagris gallopavo (turkey) blat/XM_003206406 WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDVLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTASTLCEETEFLGDIRAVKW >PRDM11_anaPla Anas platyrhynchos (duck) blast/HQ902403 WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDVLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_taeGut Taeniopygia guttata (finch) XM_002199814 WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_strCam Struthio camelus (ostrich) HQ902400 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_allMis Alligator mississippiensis (alligator) scaffold:58581 WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAF SSLGIRLQDEKPTIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPKLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_allSin Alligator sinensis (alligator) HQ902411 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAFSSLGIRLQDEKPT IGLGVDGANITASLRADLFMTIRKALPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPKLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_croSia Crocodylus siamensis (crocodile) HQ902406 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYYDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFSTTDSYLQALDRAFSSLGIRLQDEKP TIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPKLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_carIns Carettochelys insculpta (turtle) HQ902407 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYFDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFCTTDSYLQALDRAFSSLGIRLQDEKPT IGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_podUni Podocnemis unifilis (turtle) HQ902402 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYFDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFCTTDSYLQALDRAFSSLGIRLQDEKPT IGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_traScr Trachemys scripta (turtle) HQ902399 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYFDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQELGFCTTDSYLQALDRAFSSLGIRLQDEKP TIGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_pelSin Pelodiscus sinensis (turtle) HQ902412 frag ZnF_TTF WFLQYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYFDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARSLREDLVERIRQSPFLSIILDGQSDDWLADTVAVYVQYTSSDGPPATEFLSLQELGFCTTDSYLQALDRAFSSLGIRLQDEKPT IGLGVDGANITASLRANLFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_anoCar Anolis carolinensis (lizard) blat/XM_003214639 WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYRLRMHPEKTEEM CRNMTLLFNTAYHLAMEGRPYCDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERVRQSPFLSIILDGQSDDLLADTVAVYVQYISSDGPPATEFLSLQELGFSATDSYIQALDRAF SSLGIRLQDERPSVGLGIDGANITASLRANMYMTIRKTLPWLLCLPLMIHKPHLEVLDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_hemBow Hemidactylus bowringii (gecko) HQ902409 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYRLRMHPEKTEEM CRNMTLLFNTAYHLAMEGRPYCDFRSLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYISSDGPPATEFLSLQELGFSTADSYIQALDRAFSSLGIRLQDEKPS VGLGMDGANITASLRANMYMTIRKTLPWLLCLPLMVHKPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_sciRee Scincella reevesii (skink) HQ902404 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYRLRMHPEKTEEM CRNMTLLFNTAYHLAMEGRPYCEFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERVRQSPFLSIILDGQSDDLLADTVAVYVQYISSDGPPATEFLSLQELGFSTTDSYIQALDRAFSSLGIRLQDEKPS VGLGIDGANITASLRANMYMTIRKTLPWLLCLPLMVHKPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_dibBou Dibamus bourreti (skink) HQ902405 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYRLRMHPEKTEEM CRNMTLLFNTAYHLAMEGRPYCDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSDDLLADTVAVYVQYISSDGPPATEFLSLQELGFSATDSYIQALDRAFSSLGIRLQDEKPT VGLGVDGANITASLRASMYMTIRKTLPWLLCLPLMVHKPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAVKW >PRDM11_najAtr Naja atra (cobra) HQ902408 frag ZnF_TTF WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYRLRMHPEKTEEM CRNMTLLFNTAYHLAIEGRPYCDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLIERVRQSPFLSIILDGQSDDLLADTVAVYVQYVSCDGPPATEFLSLQELGFSTTDSYVQALDRAFSSLGMRLQDEKPS VGLGIDGANITASLRANIYMTIRKTLPWLLCLPLMVHKPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRAIKW >PRDM11_batYen Batrachuperus yenyuanensis (salamander) HQ902410 frag ZnF_TTF WFLRYSSTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRSMTLLLNTAYHLAVEGRPYYDFRPLAELLRKCELRVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSEYLLADTVAVYVQYTSNDGPPATEFLSLQELGVPTTESYLQAIDRAFSALGIRLQDEKPT VGLGVDGFNITAGLRANMYMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRVTAATLCEETEFLGDIRGVRW >PRDM11_ichBan Ichthyophis bannanicus (caecilian amphibian) HQ902398 frag ZnF_TTF WFLRYSSTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLALEGRPYFDFRPLAELLRKCELRVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSEDLIADTVAVYVQYTSCDGPPATEFLSLQEIGLSTAESYLQGIDRAFSALGIRLQDEKPT VGLGIDGANITAGLRANMYMTIRKTLPWLLCLPFMVYRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRTTASTLCEETEFLGDIRAVRW >PRDM11_xenTro Xenopus tropicalis (frog) blat/CF781198 WFLRYSSTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLAMEGRPYYDFRPLAELLRKCELRVVDQYMNEGDCQILIHHIARALREDLVERIRQSPFLSIILDGQSEDLLADTVAVYVQYTSSDGPPATEFLSLQELGLPTTESYLQGIDRAF SALGIRLQDERPTVGLGVDGANITAGLRANLYMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRLTAATLCEETEFLGDIRAVKW >PRDM11_ranNig Rana nigromaculata (dark-spotted frog) HQ902401 frag ZnF_TTF WFLRYSSTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEM CRNMTLLFNTAYHLAMEGRPYYDFRPLAELLRKCELRVVDQYMNEGDCQILIHHIARALREDLIERIRQSPFLSIILDGQSEDLLADTVAVYVQYTSNDGPPATEFLSLQELALPTTESYLQGIDRAFSALGIRLQDERPS VGLGIDGVNITAGLRANLYMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRLTAATLCEETEFLGDIRAVKW
Varying levels of conservation within PRDM11
The alignment below shows a difference alignment of PRDM11 sequences representative of tetrapod phylogeny. Many additional placental mammal sequences are available but are not shown because PRDM11 does not vary much in that clade over placental time scales. Actinopterygii sequences -- which begin to significantly diverge -- are shown only where they illuminate conservation of functional domains.
Note that conservation is very uneven along the protein. In particular, exon 6 has two small patches exhibiting conservation but on the whole is changing very rapidly and erratically in terms of amino acid properties and is developing indels. It is skipped over in some transcripts, possibly to no ill effect. This pattern suggests only the two patches are currently contributing to function in human. There may exist species where this exon is no longer used (exonic pseudogenization). This is possible because exon 6 and exon 7 both have phase 2 splice acceptors. The first two exons also exhibit high variability likely due to minimal selective pressure on the amino acid sequence.
In contrast, the zinc knuckle and PR(SET) domain are very conserved as is the large terminal exon 7. The extraordinary conservation of the TTF-type zinc finger suggests a recognition target conserved throughout bony vertebrates and that (unkown) protein itself will be exquisitely conserved where the two macromolecules interact. The self-dimerization domain oddly is not particularly conserved within the context of exon 7 conservation. However it may be co-evolving with itself with little net effective change.
Conservation of exon 6 of PRDM11 in Amniota: a sharp break between placentals and marsupials/monotremes/herptiles homSap1 GEKRLQREKSEQVLDNPEDLRGPIHLSVLRQGKSPYKRGFDEGDVHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLVIRKVPKYQ-DDAYSQCATTMTHGVQNIGQTQGEGDWKVPQGVS----KEPGQLEDEEEEPSSFKADSPAEASLAS---DPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPPVLPPQVLELPEFSDPA panTro2 ..........................P...............................................................R......R.............R......................................................................................................... gorGor1 ..........................P......................................................................R.............R......................................................................................................... ponAbe2 -.........................P.................................................P....C..G............R.....A.......R......................................................................................................... rheMac2 ............A.............P......................................................................R.............R......R.................................................................................................. calJac1 ............A.............P.................IQ...................................................R-R..I..S...VCR......................P...........-...............A...................................................... tarSyr1 ............A..........TQ.P...P.................................................................GR.V...A..M.SV.R...D........A.........P...D..........................................................................P... micMur1 ............A..K........Q.P..............................................................H.......R..VA.AR.A..A.R........I.........R...P.................................................................................. tupBel1 ............A.............P.......S.........M....................................................R...A....T..VSR.....N..M.H...........P...........Q......................................................E............... musMus7 .........A..A.E........TQFP..K..R.....S.....I.......................GN...R....................D.-RA.L.------Q.CR.P.........R.A......L.P........T........................................................SL............... ratNor7 .........A..A..........S.FP..K..R.....S.....L........................NA.GR....................D.-RA.L.------Q.CR.P.........K.A......L.P.......HT....................................................V....I..Q............ dipOrd1 .........C..A..S.........IP......RLF................................S...T.......................NP.TA..V..G..VSR........I....A........P.................................................................................. cavPor3 .....P......A.............P.....................T.............................................T.GR..A.......TVSR........T...LA........P....D...AP........................................................................ oryCun2 ............A..ST......TQ.P............L....A.........................G..R...................G...P..AAV....PSVSR........A.A.GP........P........T..............................................L...............G.......... ochPri2 .........A..A..S......SNQ.P...........SL....MQ.H...........................PV.T..............G...H....VM..M..LSR..R..H.RM.P.A.........P......................................................................PG.......... turTru1 ...K........A..........LQ.P.................................................................N.......M.VS.....VTR........I...........A.P.............................................N..............T.....T..A............ bosTau4 ......K.....A..........LQ.P..............................................................R..N...G...M..S.....ASR...D........A.......A.P............................P...............................T.....T..A............ equCab2 .....P......A..........LP.P......................................................................R..M..S.....ASR.......ELS..A.........A.......R.....H..................................................A.I..A....V....... felCat3 -...........A..........LP.P...........................................A......................E...R..M.VS.....V.R........I.............A.....................................................................A.A.....L.... canFam2 ............A...........Q.P.................A....................................................R..M..S.....VSR........I...A.........P.....................................................................A............ myoLuc1 ............A..DL.........P..K....S.............S..................T.....R..................G.......M..SR....VSR..R.....T...A.....T...P.D...................................................................A............ pteVam1 ............A..D..........P..................................................................E...R..M..S.....VSR..E.....T...A.....A...P.....................................................................A............ eriEur1 ............A...SD.....LQ.PM...........L....L.......................T.A..R....................I.GR.PMV....M.SVSR...D....M...GA......T.P...........................S...................................D.....A............ sorAra1 ...Q.....A..A........C.LQ.P...........S.....A...T.....................A..R.......................R..VPVA..I..VSR...D.A..MAP.A.....Q...PM.......A.......V....................................................A......G..... loxAfr3 ............A.........S.Q.P.................L.................................................T..R.....S.....VSR........I...A.........PT....................................................................A............ proCap1 ............G.........S.Q.P...................................................................T..R.....S.....VSR........I....A........P.........P....................P.................................T...T............. dasNov2 ............A...........Q.P.................T.................................................T.G...A.GP....SVSRA..A.A..R...T.........P...........Q................R..................................................... macEug1 .D.K.....P.KA..HQ......LQ.T...H...A.......V.A..PP..................TS.I.E.P..PG.P..L..A..F..E...D..GMAVP..P.DLSRN...KE..A...A......DS.L..............................................I...................L.....P........T monDom .D.K.....P.KA..HQ......LQ.T...H...A.......V.A..PP..................TS.I.E.P..PG.P..L..A..FHTE.V.D..GMAIS..P.DLSRN...KE..A...A.YGPS.DTSL...........................L..................I...................L.....P........T ornAna ...K.L...TDKAPESQ......LQ.T..K........SC....A...T.....................IDE....T..P.AFK.M..F.AE.VFER.GAILP..T.SF.R.HS.....LGH.TPYGPS..K.L..EDQG...PI.V...T....T-..G.SQ.................I.............Q.....LV....P......... allMis .D.K..K....KNA..Q..T.A.L.FTT.K........SY...ES...T.....................F.EK...T..P.ST.ATS...AEEIFER.SSA.Q..SL.LSRNRS.EE..A.H.S.FSSA..V.V........L.L.....T.L....AEGNS..I...............I..........R..............P..Q.....T melGal .D.K..K....KNT..Q..T...LQFTM.K........SY...ES...T.....................F.EK...T..P.ST.ATS...AEEIFER.SGA.Q.----LSRNRS.EE..A.H.S.LSSA..V.V........L.L.....T.L....AEGNS..I...............I..........R.......S......P..Q.....T anaPla .D.........KNVE.Q..M...LQ.TT.K........SC...ES...T.....................F.EN...T..P.S..TAS...AE.IFER.G.A.Q..SL.LSRNRS.EE..I.H.S.FSSA..V.I.D.D....L.L.....T.L....AQGNS..I...............I..........R........E.....P..Q.....T taeGut .D.M..N.N..KNVE.Q..A..ALQFTT.K........SC...ES...T.....................F.EN...T..P.SL.RAS...AE.IFE..GNA.QRSSLSLSRN.S.SE.R..HSS.FISA..MSI........L.L.....T.L....AQGNS..I.S.............I..........R......A.......P..Q.....T anoCar .D.KSL..R..RNTE.QMEMLY.LE.TISK........CSE..VSQ......................T.M.E.KVTRNSAPST..SSRFREQ..SES.G.G.Q.NSPTHSGSRN.DE....H.P.FSVS..T.L....G...L...LN..TDLT..PIDDEALG.....L..........I.......N..R....V...L.....P.YQA..Y.T
Conservation of exon 7 of PRDM11 in Sarcopterygii: high conservation but only partly corresponding to Pfam domains
PRDM11_homSap MTENMKECLAQTNAAVGDMVTVVKTEVCSPLRDQEYGQPC SRRP-DSSAMEVEPKKLKGKRDLIVPKSFQQVDFW FCESCQEYFVDECPNHGPPVFVSDTPVPVGIPDRAALTIPQGMEVVKDTSGESDVRCVNEVIPKGHIFGPYEGQISTQDKSAGFFSWL IVDKNNRYKSIDGSDETKANWMR YVVISREEREQNLLAFQHSERIYFRACRDIRPGEWLRVWYSEDYMKRLHSMSQETIHRNLAR exons 1-5
PRDM11_musMus ..........H.K........................... ...L.EP.S................T......... ................................................AG.......I.............................. ....................... ..................................R...........................
PRDM11_canFam ............K..............Y............ ......P.T............E..M.......... ...............................................EA...N....I.............................. ....................... ..................................R......................H....
PRDM11_loxAfr ..........K.K.......P..........H........ ......P...D.............M.......... ...............................................EA...N....IS............................. ..........V............ .........................V........R......Q....................
PRDM11_monDom ....L.A...H.Q.SM.E....-.......R........W .G....PPS.......P....E..MT......... ............................M...........P......EA..Q.....M.............................. ......H......T......... .............M......K....V........R.....................Q...T.
PRDM11_ornAna ....L.D.....Q.SM.E....-.......H........W .G....P.S...............MS......... ........................................P.I....EA...N....M.............................. .............T......... .I.......................V........R.........................T.
PRDM11_allMis .S..L.D..I..QTSL.E...I-...A...H......... .G....PQS..I............MT......... ...................................S....P......EPN..N....M..................S........... .............T......... ..I..........M...........T........R.....................N...T.
PRDM11_galGal .S..L.D..N..Q.SL.E...I-...A...H......... .G.L..PQS.D.............MT......... ........................................P.I....EP...N....M..................S........... .............T......... ..I..........M....................K.....................N...TT
PRDM11_melGal .S..L.D..N..Q.SL.E...I-...A...HQ........ .G....PQS.D.............MT......... ........................................P.I....EP...N....M..................S........... .............T......... ..I..........M....................K.....................N...TT
PRDM11_anaPla .S..L.D..N..Q.SL.E...I-...A...H......... .G....PQS.D..............T......... ........................................P......EP...N....M..................S........... .............T......... ..I..........M....................K.....................N...T.
PRDM11_taeGut .S..LRD..I..Q.SLRE...I-...A...H......... .G....PQS..M..........V.MT......... ........................................P......EP...N....M..................S..R........ .............T......... ..I..........M................H...K....................MN.SFTS
PRDM11_anoCar .S.KLND..-------.E...I-...P...C.EE....LW .S.KV..QSVD.........Q...MS......... ...................M.L..A...I.........V.P......EAN..R.....G.I....R.Y.....KL.S........... .............T...TS.... ..A..........M...........T........R.....................N...T.
PRDM11_xenTro .S.IS...RVAFSPSL..I.R.-.R..G..VEE.G..HFR .SVGPN.RCLDM...R..E..ESTMS..L...... ..............S....IL.P..L..I.M.E....SV.C.I.....S....E............M........CS....S...... ...N.........T..AY..... .............M......K....T....Q...K.....................N...TQ
PRDM11_latCha ---------------------------------------- .TGAAETPRI.G...RT...LET.MA.TL...... ...E...H..V...T......TM.......M.E.....A.P.IHI..GSN..I..E..D..VQ..R.........T............ .............T......... ......D.............K.....S.NLH...R......DE.............D...TA
PRDM11_homSap GEKRLQREKSEQVLDNPEDLRGPIHLSVLRQGKSPYKRGFDEGDVHPQAKKKKIDLIFKDVLEASLESAKVEAHQLALSTSLV-IRKVPKYQ-DDAYSQCATTMTHGVQNIGQTQGEGDWKVPQGV----SKEPGQLEDEEEEPSSFKADSPAEASLA---SDPHELPTTSFCPNCIRLKKKVRELQAELDMLKSGKLPEPPVLPPQVLELPEFSDPA exon 6
PRDM11_musMus .........A..A.E........TQFP..K..R.....S.....I.......................GN...R.....................D.GRA.L.-----.G.CR.P.........R.....A..L.P........T........................................................SL...............
PRDM11_canFam ............A...........Q.P.................A.....................................................R..M..S.....VSR........I...A.........P.....................................................................A............
PRDM11_loxAfr ............A.........S.Q.P.................L..................................................T..R.....S.....VSR........I...A.........PT....................................................................A............
PRDM11_monDom .D.K.....P.KA..HQ......LQ.T...H...A.......V.A..PP..................TS.I.E.P..PG.P...L..A..FHTE.V.D..GMAIS..P.DLSRN...KE..A...ASYGP..DTSL...........................L..................I...................L.....P........T
PRDM11_ornAna ...K.L...TDKAPESQ......LQ.T..K........SC....A...T.....................IDE....T..P.A.FK.M..F.AE.VFER.GAILP..T.SF.R.HS.....LGH.TPYGP...K.L..EDQG...PI.V...T....T...G.SQ.................I.............Q.....LV....P.........
PRDM11_allMis .D.KS......KNME.Q..M...LQ.TT.K........SCE.AES...T.....................L.E...TT..P.S....AS...TE.VFER.G..IQ.SSP.LSRNRS..E....HSSSFSTA..M.L........L.L.....T.P...STQGNS..I...............I..........R......Q.A.A...P..Q.....T
PRDM11_galGal .D.K..K....KNA..Q..T.A.L.FTT.K........SY...ES...T.....................F.EK...T..P.S.T.ATS...AEEIFER.SSA.Q..SL.LSRNRS.EE..A.H.SSFSSA..V.V........L.L.....T.L...SAEGNS..I...............I..........R..............P..Q.....T
PRDM11_melGal .D.K..K....KNT..Q..T...LQFTM.K........SY...ES...T.....................F.EK...T..P.S.T.ATS...AEEIFER.SGA.Q.----LSRNRS.EE..A.H.SSLSSA..V.V........L.L.....T.L...SAEGNS..I...............I..........R.......S......P..Q.....T
PRDM11_anaPla .D.........KNVE.Q..M...LQ.TT.K........SC...ES...T.....................F.EN...T..P.S...TAS...AE.IFER.G.A.Q..SL.LSRNRS.EE..I.H.SSFSSA..V.I.D.D....L.L.....T.L...SAQGNS..I...............I..........R........E.....P..Q.....T
PRDM11_taeGut .D.M..N.N..KNVE.Q..A..ALQFTT.K........SC...ES...T.....................F.EN...T..P.S.L.RAS...AE.IFE..GNA.QRSSLSLSRN.S.SE.R..HSSSFISA..MSI........L.L.....T.L...SAQGNS..I.S.............I..........R......A.......P..Q.....T
PRDM11_anoCar .D.KSL..R..RNTE.QMEMLY.LE.TISK........CSE..VSQ......................T.M.E.KVTRNSAPS.T..SSRFREQ..SES.G.G.Q.NSPTHSGSRN.DE....H.PSFSV...T.L....G...L...LN..TDLT..PIDDEALG.....L..........I.......N..R....V...L.....P.YQA..Y.T
PRDM11_xenTro .D...L..NN.RL.E.Q..VK.TFP.AT.K....L...SCE.V.L...T..................T.RIDEYH.VT.SP.SGQK.N...LYENHGDR.RMNRQCSSPQ-N.IRNMR...AKHVSASGLNRQASFP...V.DH..V..E..T.S.AI...GNVD.I...............I.......E..R.E.MA.TSQMTN.IN.I...A.AS
PRDM11_latCha .NLK....N..EGW.AQ.N...ML----.K....S....N..AES.Q.P.R.................S.L.GNS..T.SP.P.LK.PI.F.LE.VLQKPEYYYK.VS.LL.R--..VE..S----....HQKS.CSLPDN.DSDRI.EE..D.VPGNSTEE..EDV...............I....E...R.R..Q----.PAS..LPQVQ.LLE.P
<---------------------------------- ZnF_TTF domain ------------------------------->
PRDM11_homSap ASESMVSGPAIMEDDDQEVDSADESVSNDMMTATDEPSKMSSATGRRIRRFKQEWLKKFWFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEMCRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALREDLVERIRQSPCLSVILDGQSDDLLADTVAVYVQYTSSDGPPATEFLSLQE exon 7
PRDM11_musMus .............................V.............................................................................................................................................................................I....................................
PRDM11_canFam ..............................L..A.........................................................................................................................................................................I....................................
PRDM11_loxAfr ............................I..............................................................................................................................................................................I....................................
PRDM11_monDom ....V..V.TML................E.I.......................................................................................................................F.................................................F..I........I...........................
PRDM11_ornAna ....L..I.T.L....P.............IA........................................................................................................................................................................F..I....................................
PRDM11_allMis ....II.V.T....................IA..........V...........................................................................................................Y.................................................F..I....................................
PRDM11_galGal ....II.V.T..................E.IA......................................................................................................................Y.................................................F..I........V...........................
PRDM11_melGal ....II.V.T..................E.IA......................................................................................................................Y.................................................F..I........V...........................
PRDM11_anaPla ....II.V.T....................IA......................................................................................................................Y.................................................F..I........V...........................
PRDM11_taeGut ....II.V.T....................IA......................................................................................................................Y.................................................F..I....................................
PRDM11_anoCar ...TIM.V.T....................I..........AV.-........................................................................R..........................M.....C............................................V....F..I....................I...............
PRDM11_xenTro .P.GVAIATTMID..E...............A.........AGS.....................S..............................................................................M.....Y..............R..................................F..I......E.............................
PRDM11_latCha GVQEAHPSVSL...................IA.S..T..ITVGAS.........................................................................................V.....................SD..............................Q...I.......F........T..I........I...T..............
PRDM11_homSap LGFSSTESYLQALDRAFSALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTAATLCEETEFLGDIRAVRWIIGEQNVLNALIKDYLEVVAHLKEVSSQTQRADASAIALALLQFLMDYQSIKLIYFLLDVIAVLSRLAYIFQGEYLLVSQVDDKIEEAIQEISRLADSPGEYLQEFEEN exon 7
PRDM11_musMus .................A...............................................................................................S........................................D..........................M..........................................................
PRDM11_canFam ..................................I....V.........................................................................S........................................D.............................................V.......................................
PRDM11_loxAfr ....T.D...........S...........I....... ...........................................................................S...............K........................DI............................................V......................................
PRDM11_monDom ....T.D...........S...........I...............................................................................L..S........................................D..G..................................................................................
PRDM11_ornAna ....T.D..........A...V.................V......G....V..........................................................L...........................................D..G..........................................V.......................................
PRDM11_allMis ....T.D...........S...........I...............NL.......................................................K......V..................K........................D..G..........................................V.......................................
PRDM11_galGal ....T.D...........S...........I...............NL..............................................................V..S...............K........................D..G..........................................V.......................................
PRDM11_melGal ....T.D...........S...........I...............NL..............................................................V..S...............K........................D..G..........................................V.......................................
PRDM11_anaPla ....T.D...........S...........I...............NL..............................................................V..................K........................D..G..........................................V.......................................
PRDM11_taeGut ....T.D...........S...........I...............NL..............................................................V..................K........................D..G..........................................V.......................................
PRDM11_anoCar ....A.D..I........S........R.S....I...........N.Y..............L.I.K....V.....................................V..................K........................D..G.........................................F........................................
PRDM11_xenTro ..LPT......GI..............R..............G...NLY.............................................................L..................K........................D..G......T...................................V............E.V..........T.............
PRDM11_latCha ..CVT.D..V..V....AVF.L....QRNV.......TCL..G...NL...................K....V.....................................V..................K........................D..G.......A.................................FV........C................S.......M.....
<-------------------------------------- hATC dimerization domain --------------------------------->
PRDM11_homSap FRESFNGIAMKNLRVAEAKFQSIREKICQKTQVILAQRFDSRSRIFVKACQVFDLAAWPRSSEELMSYGKEDMVQIFDHLEAIPTFSRDVCREGLDPRGSLLMEWRELKADYYTKNGFKDLISHICKYKQRFPLLNKIIQVLKVLPTSTACCEKGRNALQRVRKNHRSRLTLEQLSDLLTIAVNGPPITNFDAKRALDSWFEEKSGNSYALSAEVLSRMSALEQKPALQTMDHGTEFYPDI exon 7
PRDM11_musMus .........V..................................V...............N....L.F................A.........T...........D..............L..................................S...............................A....................T................M.HVV...S.....M
PRDM11_canFam ......................V...................................................................................................G.................................................................A.....................................V...V...S......
PRDM11_loxAfr .................................................................V..........................................................VG..............I........V......................................A.................H..T................V..AI..........
PRDM11_monDom .......V.V..................................T................T...........I.......T..S...EI....M.............................................I...............V........N......................A....................T............D...M.P.....S...S.L
PRDM11_ornAna .......V.V....................A.............T....................V...R......LE......S...E.....A....A..T...................G.VG...........V..I...............S...............................AH..............................S.D...M..SV...S.....M
PRDM11_allMis .........V..................................T................T...............E...TV.S...E.....M.I............V...........LG...............V.I.......S................N....................A.A...C.....................M.....S.D...M..S....S......
PRDM11_galGal .........V..................................T................TD..............E...TV.S...E.....M.TQ...........V...........L................V.I.......S................N......................A...C.....................M.....S.D...M..SV...S......
PRDM11_melGal .........V..................................T................TD..............E...TV.S...E.....M.TQ...........V...........L................V.I.......S................N......................A...C.....................M.....S.D...M..SV...S......
PRDM11_anaPla .........V..................................T................TD..............E...TV.S...E.....M.T............V...........L................V.I.......S................N......................A...C.....................M.....S.D...M..S....S......
PRDM11_taeGut .........V..................................T................TD..............E...TV.S...E.....M.T............V...........L................V.I.......S.......S........N......................A...C.....................M.....S.D...M..S....S......
PRDM11_anoCar .......V.V.............................EP.T.A...........M....A.......R............V....T.II...M.T............V..................R...........I........................N......................A..E.............S........M.....S.DH..M..S....S......
PRDM11_xenTro .........L.......................T..........M..........ST...TT...IN..E...L..YEL..T..N.LH.LG..VA.T..N............C.........G.........LF....V.I....................L...N......D.M....A.....A..A....................S....M..K..S.D...L..P.E..S.Y.Q..
PRDM11_latCha .........L..............................N...P.I....ML.I.T....TDD.KNF.E.EIMV.YEQ..LV...A.E.....T.N....V.........F.S........G.....R....I..RVL.I.....S.A.......S....I...N......D..N......I...S.A...........................N....-D...M..G..FVSD.....
PRDM11 and amniote phylogeny
A recent study searched through UCSC genomic multi-alignment files for the most informative regions for resolving the long-controversial phylogenetic relationships among amniotes. Among the most suitable coding regions was the distal region of the last exon of we now know to be PRDM11. This region was sequenced in various snakes, caecilians, turtles, birds and frogs that are poorly represented in sequence databases. That data is supplemented below with sequences from genome projects in the difference alignment (relative to human) to illustrate the evolution of this highly conserved region.
Note the three sites colored magenta associate turtles with birds + crocodilians to the exclusion of lizards in the overall tree topology: (amphibians,((lizards,(turtles,(birds,crocodilians))),(monotremes,(marsupials,placentals)))). This requires, as do all amniote topologies, various morphological convergences and reversals. The short explanation is that a 1903 choice of anatomical character (number of skull openings -- temporal fenestrae) was -- in retrospect -- a poor choice. This has left an unfortunate legacy of terms such as diapsid, anapsid, synapsid and indeed 'reptile' that do not correctly reflect the evolutionary history of amniotes.
Difference Alignment of Final Exon Region of PRDM11 Used in Establishing Amniote Phylogenetic Tree homSap WFLRYSPTLNEMWCHVCRQYTVQSSRTSAFIIGSKQFKIHTIKLHSQSNLHKKCLQLYKLRMHPEKTEEMCRNMTLLFNTAYHLALEGRPYLDFRPLAELLRKCELKVVDQYMNEGDCQILIHHIARALmagentaLVERIRQSPCLSVILDGQSDDLLA Homo sapiens (human) musMus ................................................................................................................................................I............... Mus musculus (mouse) canFam ................................................................................................................................................I............... Canis familiaris (dog) loxAfr ................................................................................................................................................I............... Loxodonta africana (elephant) monDom ...........................................................................................F.................................................F..I........I...... Monodelphis domestica (opossum) ornAna .............................................................................................................................................F..I............... Ornithorhynchus anatinus (platypus) galGal ...........................................................................................Y.................................................F..I........V...... Gallus gallus (chicken) melGal ...........................................................................................Y.................................................F..I........V...... Meleagris gallopavo (turkey) anaPla ...........................................................................................Y.................................................F..I........V...... Anas platyrhynchos (duck) taeGut ...........................................................................................Y.................................................F..I............... taeGut Taeniopygia guttata (finch) strCam ...........................................................................................Y.................................................F..I............... strCam Struthio camelus (ostrich) allMis ...........................................................................................Y.................................................F..I............... allMis Alligator mississippiensis (alligator) allSin ...........................................................................................Y.................................................F..I............... allSin Alligator sinensis (alligator) croSia ...........................................................................................Y.................................................F..I............... croSia Crocodylus siamensis (crocodile) carIns ...........................................................................................F.................................................F..I............... Carettochelys insculpta (turtle) podUni ...........................................................................................F.................................................F..I............... Podocnemis unifilis (turtle) traScr ...........................................................................................F.................................................F..I............... Trachemys scripta (turtle) pelSin ...Q.......................................................................................F...................................S.............F..I........W...... Pelodiscus sinensis (turtle) anoCar ..........................................................R..........................M.....C............................................V....F..I............... Anolis carolinensis (lizard) hemBow ..........................................................R..........................M.....C...S.............................................F..I............... Hemidactylus bowringii (gecko) sciRee ..........................................................R..........................M.....CE...........................................V....F..I............... Scincella reevesii (skink) dibBou ..........................................................R..........................M.....C.................................................F..I............... Dibamus bourreti (skink) najAtr ..........................................................R..........................I.....C.........................................I..V....F..I............... Naja atra (cobra) batYen ......S.................................................................S....L.......V.....Y..............R..................................F..I......EY....... Batrachuperus yenyuanensis (salamander) ichBan ......S....................................................................................F..............R..................................F..I......E..I..... Ichthyophis bannanicus (caecilian) xenTro ......S..............................................................................M.....Y..............R..................................F..I......E........ Xenopus tropicalis (frog) ranNig ......S..............................................................................M.....Y..............R..........................I.......F..I......E........ Rana nigromaculata (dark-spotted frog) homSap DTVAVYVQYTSSDGPPATEFLSLQELGFSSTESYLQALDRAFSALGIRLQDEKPTVGLGVDGANITASLRASMFMTIRKTLPWLLCLPFMVHRPHLEILDAISGKELPCLEELENNLKQLLSFYRYSPRLMCELRSTAATLCEETEFLGDIRAVRW Homo sapiens (human) musMus ..........................................A...............................................................................................S................. Mus musculus (mouse) canFam ...........................................................I....V.........................................................................S................. Canis familiaris (dog) loxAfr .............................G.............T..............................................................................................S...............K. Loxodonta africana (elephant) monDom .............................T.D...........S...........I...............................................................................L..S................. Monodelphis domestica (opossum) ornAna .............................T.D..........A...V.................V......G....V..........................................................L.................... Ornithorhynchus anatinus (platypus) galGal .............................T.D...........S...........I...............NL..............................................................V..S...............K. Gallus gallus (chicken) melGal .............................T.D...........S...........I...............NL..............................................................V..S...............K. Meleagris gallopavo (turkey) anaPla .............................T.D...........S...........I...............NL..............................................................V..................K. Anas platyrhynchos (duck) taeGut .............................T.D...........S...........I...............NL..............................................................V..................K. taeGut Taeniopygia guttata (finch) strCam .............................T.D...........S...........I...............NL..............................................................V..................K. strCam Struthio camelus (ostrich) allMis .............................T.D...........S...........I...............NL.......................................................K......V..................K. allMis Alligator mississippiensis (alligator) allSin .............................T.D...........S...........I...............DL......A................................................K......V..................K. allSin Alligator sinensis (alligator) croSia .............................T.D...........S...........I...............NL.......................................................K......V..................K. croSia Crocodylus siamensis (crocodile) carIns ............................CT.D...........S...........I...............NL..............................................................V..................K. Carettochelys insculpta (turtle) podUni ............................CT.D...........S...........I...............NL..............................................................V..................K. Podocnemis unifilis (turtle) traScr ............................CT.D...........S...........I...............NL..............................................................V..................K. Trachemys scripta (turtle) pelSin ............................CT.D...........S...........I...............NL..............................................................V..................K. Pelodiscus sinensis (turtle) anoCar .........I...................A.D..I........S........R.S....I...........N.Y..............L.I.K....V.....................................V..................K. Anolis carolinensis (lizard) hemBow .........I...................TAD..I........S..........S....M...........N.Y..............L...K..........................................V..................K. Hemidactylus bowringii (gecko) sciRee .........I...................T.D..I........S..........S....I...........N.Y..............L...K..........................................V..................K. Scincella reevesii (skink) dibBou .........I...................A.D..I........S.............................Y..............L...K..........................................V..................K. Dibamus bourreti (skink) najAtr .........V.C.................T.D..V........S..M.......S....I...........NIY..............L...K..........................................V.................IK. Naja atra (cobra) batYen ...........N...............VPT.......I........................F....G...N.Y.............................................................V................G... Batrachuperus yenyuanensis (salamander) ichBan ...........C.............I.L.TA.....GI.....................I.......G...N.Y.................Y...........................................T..S................. Ichthyophis bannanicus (caecilian) xenTro ...........................LPT......GI..............R..............G...NLY.............................................................L..................K. Xenopus tropicalis (frog) ranNig ...........N..............ALPT......GI..............R.S....I..V....G...NLY.............................................................L..................K. Rana nigromaculata (dark-spotted frog)
PRDM11 relationship to ZNF862 and ZMYM1
ZNF862 is otherwise the most closely related human protein to PRDM11, aligning along almost all of the terminal exon of PRDM11 with low percent identity 195/697 (28%)and gaps totaling 34/697 (5%). The two Pfam domains are not noticably better conserved than bulk exon. ZNF862 too has a very long terminal exon containing the uncommon transcription factor zinc finger type (ZNF_TFF) and a dimerization domain (hATC) often paired with it.
The structure of ZNF862 suggests a partial internal duplication at some point in its history. Its doubled KRAB domain is also found undoubled in PRDM7/9 though percent identity is negligible. Still, the commonality in domains suggests related functionality. ZNF862 reportedly has a pseudogene (NG_024481) as well.
Another human gene, ZMYM1, also has a long terminal exon containing the transcription factor zinc finger type and dimerization domain. This exon is phase 0 unlike the phase 2 terminal exon of PRDM11 and ZNF862. ZMYM1 contains four zinc fingers of MYM-type in its earlier exons somewhat reminiscent of C2H2 domains of PRDM9. The alignment below shows how poorly these domains are conserved between the two proteins.
It follows that PRDM11 shares a distant domain shuffling history with these two proteins as well (which are all quite diverged today). ZNF862 is well-conserved back to monotreme but hard to locate earlier; ZMYM1 only has clear full-length counterparts within placental mammals. Rapid divergence could make it difficult to find earlier orthologs if they existed. At face value, this suggests a major expansion of this overall class of domain-shuffled proteins in early mammals, with only a few members -- like PRDM11 -- having a history back into early vertebrates.
>ZNF862_homSap Homo sapiens (human) aka KIAA0543 KRAB ZNF_TFF hATC dimerization dimerization 0 MEPRESGK 0 0 APVTFDDITVYLLQEEWVLLSQQQKELCGSNKLVAPL 1 2 GPTVANPELFRKFGRGPEPWLGSVQGQRSLLEHHP 1 2 GKKQMGYMGEMEVQGPTRESGQSLPPQKKAYLSHLSTGSGHIEGDWAGRNRKLLKPRSIQKSWFVQFPWLIMNEEQTALFCSACREYPSIRDKRSRLIEGYTGPFKVETLKYHAKSKAHMFCVNALAARDPIWAARFR SIRDPPGDVLASPEPLFTADCPIFYPPGPLGGFDSMAELLPSSRAELEDPGGDGAIPAMYLDCISDLRQKEITDGIHSSSDINILYNDAVESCIQ 0 0 DPSAEGLSEEVPVVFEELPVVFEDVAVYFTREEWGMLDKRQKELYRDVMRMNYELLASL 1 2 GPAAAKPDLISKLERRAAPWIKDPNGPKWGKGRPP 1 2 GNKKMVAVREADTQASAADSALLPGSPVEARASCCSSSICEEGDGPRRIKRTYRPRSIQRSWFGQFPWLVIDPKETKLFCSACIERPNLHDKSSRLVRGYTGPFKVETLKYHEVSKAHRLCVNTVEIKEDTPHTALV PEISSDLMANMEHFFNAAYSIAYHSRPLNDFEKILQLLQSTGTVILGKYRNRTACTQFIKYISETLKREILEDVRNSPCVSVLLDSSTDASEQACVGIYIRYFKQMEVKESYITLAPLYSETADGYFETIVSALDELDI PFRKPGWVVGLGTDGSAMLSCRGGLVEKFQEVIPQLLPVHCVAHRLHLAVVDACGSIDLVKKCDRHIRTVFKFYQSSNKRLNELQEGAAPLEQEIIRLKDLNAVRWVASRRRTLHALLVSWPALARHLQRVAEAGGQIG HRAKGMLKLMRGFHFVKFCHFLLDFLSIYRPLSEVCQKEIVLITEVNATLGRAYVALESLRHQAGPKEEEFNASFKDGRLHGICLDKLEVAEQRFQADRERTVLTGIEYLQQRFDADRPPQLKNMEVFDTMAWPSGIEL ASFGNDDILNLARYFECSLPTGYSEEALLEEWLGLKTIAQHLPFSMLCKNALAQHCRFPLLSKLMAVVVCVPISTSCCERGFKAMNRIRTDERTKLSNEVLNMLMMTAVNGVAVTEYDPQPAIQHWYLTSSGRRFSHVYTCAQVPARSPA 1 2 SARLRKEEMGALYVEEPRTQKPPILPSREAAEVLKDCIMEPPERLLYPHTSQEAPGMS* 0 >ZMYM1_homSap Homo sapiens (human) Q5SVZ6 Zinc finger MYM-type ZnF_TTF hATC_dimerization 0 MKEPLLGGECDKAVASQLGLLDEIKTEPDNAQ 0 0 EYCHRQQSRTQENELKINAVFSES 1 2 ASQLTAGIQLSLASSGVNKMLPSVSTTAIQVSCAGCKKILQKGQTAYQRKGSAQLFCSIPCITEYISSASSPVPSKRTCSNCSK 2 1 DILNPKDVISVQLEDTTSCKTFCSLSCLSSYEEKRKPFVTICTNSILTKCSMCQKTAI 0 0 IQYEVKYQNVKHNLCSNACLSKFHSANNFIMNCCENCGTYCYTSSSLSHILQMEGQSHYFNSSKSITAYKQ 0 0 KPAKPLISVPCKPLKPSDEMIETTSDLGKTELFCSINCFSAYSKAKMESSS 1 2 VSVVSVVHDTSTELLSPKKDTTPVISNIVSLADTDVALPIMNTDVLQ 1 2 DTVSSVTATADVIVD 0 0 LSKSSPSEPSNAVASSSTEQPSVSPSSSVFSQHAIGSSTEVQKDNMKSMKISDELCHPKCTSKVQKVKGKSRSIKKSCCADFECLENSKKDVAFCYSC QLFCQKYFSCGRESFATHGTSNWKKTLEKFRKHEKSEMHLKSLEFWREYQFCDGAVSDDLSIHSKQIEGNKKYLKLIIENILFLGKQCLPLRGNDQSVSSVNKGNFLELLEMRAKDKGEE TFRLMNSQVDFYNSTQIQSDIIEIIKTEMLQDIVNEINDSSAFSIICDETINSAMKEQLSICVRYPQKSSKAILIKERFLGFVDTEEMTGTHLHRTIKTYLQQIGVDMDKIHGQAYDSTT NLKIKFNKIAAEFKKEEPRALYIHCYAHFLDLSIIRFCKEVKELRSALKTLSSLFNTICMSGEMLANFRNIYRLSQNKTCKKHISQSCWTVHDRTLLSVIDSLPEIIETLEVIASHSSNT SFADELSHLLTLVSKFEFVFCLKFLYRVLSVTGILSKELQNKTIDIFSLSSKIEAILECLSSERNDVYFKTIWDGTEEICQKITCKGFKVEKPSLQKRRKIQKSVDLGNSDNMFFPTSTE EQYKINIYYQGLDTILQNLKLCFSEFDYCKIKQISELLFKWNEPLNETTAKHVQEFYKLDEDIIPELRFYRHYAKLNFVIDDSCINFVSLGCLFIQHGLHSNIPCLSKLLYIALSWPITS ASTENSFSTLPRLKTYLCNTMGQEKLTGPALMAVEQELVNKLMEPERLNEIVEKFISQMKEI* 0