Opsin evolution: update blog: Difference between revisions
From genomewiki
Jump to navigationJump to search
Tomemerald (talk | contribs) No edit summary |
Tomemerald (talk | contribs) No edit summary |
||
| (149 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
This | This large collection of opsin evolution articles is updated almost daily. To locate changes, consult the article directory and update blog below. | ||
* [ | * [[Opsin_evolution|Opsin classification tool and curated sequence collection]] | ||
* [ | * [[Opsin_evolution:_orgins_of_opsins|Opsin origins]] | ||
* [ | * [[Opsin_evolution:_key_critters_(deuterostomes)|Key critters with genomics: deuterostomes]] | ||
* [ | * [[Opsin_evolution:_key_critters_%28ecdysozoa%29|Key critters with genomics: ecdysozoa]] | ||
* [ | * [[Opsin_evolution:_key_critters_%28lophotrochozoa%29|Key critters with genomics: lophotrochozoa]] | ||
* [ | * [[Opsin_evolution:_key_critters_(cnidaria)|Key critters with genomics: cnidaria and porifera]] | ||
* [ | * [[Opsin_evolution:_alignment|230 opsins aligned: consensus-colored image and motif searchable text]] | ||
* [ | * [[Opsin_evolution:_ancestral_introns|Ancestral intron reconstruction for ciliary opsins]] | ||
* [ | * [[Opsin_evolution:_informative_indels|Ancestral status of informative coding indels]] | ||
* [[Opsin_evolution:_ancestral_sequences|Ancestral sequence reconstruction for ciliary opsins]] | |||
* [[Opsin_evolution:_LWS_PhyloSNPs|LWS: generating locus for imaging opsins]] | |||
* [[Opsin_evolution:_Encephalopsin_gene_loss|Encephalopsin and TMT: massive gene loss in mammalian clades]] | |||
* [[Opsin_evolution:_Melanopsin_gene_loss|Melanopsins show gene and exon loss in mammals]] | |||
* [[Opsin_evolution:_RGR_phyloSNPs|RGR opsin: abrupt change in DRY motif in boreoeutheres]] | |||
* [[Opsin_evolution:_Peropsin_phyloSNPs|Peropsin: phyloSNPs (synapomorphies) in persopsin evolution]] | |||
* [[Opsin_evolution:_Neuropsin_phyloSNPs|Neuropsin (OPN5): 52 deuterostome genes and a novel paralog, Newropsin]] | |||
* [[Opsin_evolution:_trichromatic_ancestral_mammal|Color vision in the platypus and ancestral mammal]] | |||
* [[Opsins_underground|Opsins and cryptochromes in a subterranean mammal]] | |||
* [[Opsin_evolution:_Cytoplasmic_face|The cytoplasmic face of opsins and GPCR specificity]] | |||
* [[Opsin_evolution:_RBP3_(IRBP)|IRBP Interstitial retinol-binding protein modules]] | |||
* [[Opsin_evolution:_RPE65|RPE65 rate-limiting step in ciliary opsin regeneration]] | |||
* [[USH2A_SNPs|Usherin USH2A roles in vision and hearing]] | |||
* [[CDH23_SNPs|Cadherin CDH23 roles in vision and hearing]] | |||
* [[Opsin_evolution:_transducins|Transducins: comparative genomics of opsin interactions]] | |||
* [[Opsin_evolution:_annotation_tricks|Annotation tricks: genes from trace reads, intron patterns, resolving indels, trimming]] | |||
* Tracking significant updates (this page) | |||
* The photoreceptor fossil record (planned) | * The photoreceptor fossil record (planned) | ||
* <span style="color: #990099;">Opsin evolution: the bottom line (in progress)</span> | * <span style="color: #990099;">Opsin evolution: the bottom line (in progress)</span> | ||
[[Image:TMTsurvivors.png]] | |||
Reverse Chronological Update Blog | |||
14 Apr 12: released major article on [[Cryptochrome_evolution|cryptochrome evolution]], relationships with opsins, with 275 curated [[Cryptochrome_refSeqs|reference sequences]] | |||
15 Feb 12: started new article on opsins of an [[Opsins_underground|underground mammal]], naked mole-rat (Heterocephalus glaber) | |||
30 Dec 11: first nematode opsins located in [[Opsin_evolution:_key_critters_(ecdysozoa)#Nematodes_..._3_opsins|Strongyloides]] | |||
30 Sep 11: the earliest opsins -- added two full-length [[Opsin_evolution:_key_critters_(cnidaria)|ctenophore opsins]] with '''introns''' from Mnemiopsis assembly | |||
13 Aug 11: [[Opsin_evolution:_key_critters_(cnidaria)#Anthozoa:_Acropora_digitifera_.28stony_coral.29_.._13_opsins|first introns]] found in cnidarian opsins (new coral genome, Acropora digitifera) | |||
06 Mar 11: RH7 transcripts found in [[Opsin_evolution:_key_critters_%28ecdysozoa%29#Hexapoda:_Drosophila_melanogaster_.28fruitfly.29_.._7_opsins|butterfly antenna]] -- new anatomical structure? | |||
02 Mar 11: First [[Opsin_evolution:_key_critters_%28deuterostomes%29#Snakes:_Python_molarus_.28python.29_.._11_opsins|snake genome]] has 11 opsins but 8 are lost | |||
06 Oct 10: [[Opsin_evolution:_Neuropsin_phyloSNPs#NEUR4:_11_vertebrate_newwopsins|new opsin]] surfaces in non-amniote vertebrates -- a neuropsin related to NEUR4 in zebrafish and frog | |||
26 Aug 10: first ctenophore opsin now [[Opsin_evolution:_key_critters_%28cnidaria%29#Ctenophora:_Pleurobrachia_pileus_.28sea_gooseberry.29_.._2_opsins|extendable to full length]]; second ctenophore opsin surfaces | |||
03 Jul 10: analyzed the five opsins in the new [[Opsin_evolution:_key_critters_%28ecdysozoa%29#Hexapoda:_Atta_cephalotes_.28ant.29_.._5_opsins|leafcutter ant genome]], Atta cephalotes | |||
06 Jun 10: the [[Opsin_evolution:_key_critters_%28cnidaria%29#Porifera:_Amphimedon_queenslandica_.28sponge.29_.._1_opsin|first sponge opsin]] in unassembled traces proves to be a Lottia contaminant | |||
05 Apr 10: collected assembled salamander transcripts for [[Opsin_evolution:_trichromatic_ancestral_mammal#Opsin_454_transcripts_of_salamander_Ambystoma_tigrinum|12 opsin loci]] from a 454 project; amphibians remain poorly sampled | |||
29 Mar 10: analyzed and classified the 41 'opsins' in the new Hydra genome -- all have lysine at 296 but lack full DRY | |||
24 Mar 10: spellchecked the entire suite of articles (503 pages), fixing many many typos | |||
02 Feb 10: analyzed comparative genomics of the F CWxP [[Opsin_evolution:_ancestral_sequences#The_aromatic_rotameric_lock_of_TM5-TM6_in_opsin_evolution|aromatic lock]] between TM6-TM7 | |||
27 Jan 10: added section on [[Opsin_evolution:_key_critters_%28ecdysozoa%29|Bombyx genome]] and its five opsins | |||
21 Jan 10: greatly improved fasta header lines in the 456 reference sequences using [[Opsin_evolution#Curated_opsin_reference_sequences|space-delimited database format]], facilitating subset selection. | |||
18 Jan 10: queried [http://www.sciencemag.org/cgi/content/full/327/5963/343 new Nasonia genomes] but these were uninformative -- too close to existing Nasonia vitripennis | |||
17 Jan 10: queried [[Opsin_evolution:_key_critters_%28ecdysozoa%29#Chelicerata:_Varroa_destructor_.28bee_mite.29_1_opsin|new chelicerate genome]] (Varroa bee mite) for opsins, finding only a peropsin-like fragment | |||
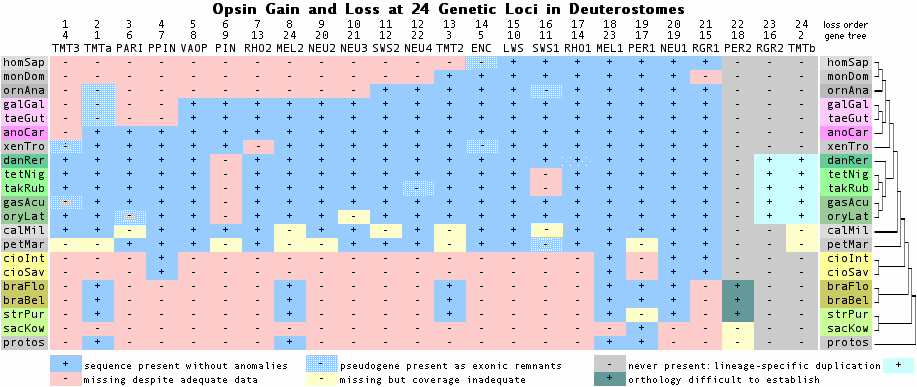
14 Jan 10: snazzy new graphic shows gain and loss at 24 opsin loci during deuterostome history, vindicating 1942 GL Walls hypothesis | |||
13 Jan 10: added section about 'managing' [[Opsin_evolution:_ancestral_introns#Managing_homoplasy_in_intron_data|homoplasy]] of indels, introns and diagnostic residues | |||
12 Jan 10: defined the [[Opsin_evolution:_ancestral_introns#Ancestral_introns|intronation required]] of a GPCR gene to be parental to an opsin family | |||
11 Jan 10: showed longwave opsin introns in arthropods form a [[Opsin_evolution:_ancestral_introns#Ancestral_melanopsin_intronation|distinct yet early coalescing]] melanopsin class | |||
10 Jan 10: established using introns that peropsin, rgropsin and neuropsins [[Opsin_evolution:_ancestral_introns#Ancestral_peropsin.2C_neuropsin_and_RGRopsin_intronation|form a family]] that excludes cilopsins and melanopsins | |||
09 Jan 10: proved arthropod UV opsins are [[Opsin_evolution:_ancestral_introns#Ancestral_melanopsin_intronation|orthologous]] to vertebrate melanopsin | |||
08 Jan 10: improved methods and big update to [[Opsin_evolution:_ancestral_introns#Ancestral_ciliary_opsin_intronation|ancestral introns]] of ciliary opsins | |||
29 Dec 09: discovered a new ur-opsin locus TMTc ranging from fish to frog but lacking transcripts and experiments | |||
27 Dec 09: provided page crosslinks to allow faster navigation between related topics | |||
26 Dec 09: added [[Opsin_evolution:_orgins_of_opsins#Vertebrate_ciliary_opsins|flanking synteny]] to ciliary opsins to sort out gene duplication origins | |||
23 Dec 09: sought informative indels by aligning GPCR and proxy opsins in the [[Opsin_evolution:_informative_indels#Indels_in_the_TM4-EC2-TM5_region|retinal lid]] region (second extracellular loop) | |||
21 Dec 09: extended ur-opsin TMTa into frog and lizard but found [[Opsin_evolution:_Encephalopsin_gene_loss|decayed syntenic pseudogenes]] in chicken, finch and platypus | |||
19 Dec 09: began new section on [[Opsin_evolution:_orgins_of_opsins|opsin origins]] from earlier GPCR | |||
16 Dec 09: located a new moth ciliary opsin and fixed others; confirmed multiple lineage losses for ciliary opsins | |||
15 Dec 09: located a second chelicerate peropsin in Ixodes; gene lost in crustacea and insects | |||
09 Dec 09: reviewed first reported [[Opsin_evolution:_Peropsin_phyloSNPs#Invertebrate_peropsins|peropsin from ecdysozoa]]; reclassified peropsin gene family | |||
08 Dec 09: located [[Opsin_evolution:_informative_indels#Indels_in_melanopsins:_TM2_region|ciliary ur-opsin]] of deuterostomes, new orthologs in frog and lizard, not birds or mammals | |||
07 Dec 09: aligned 411 opsins in [[Opsin_evolution:_informative_indels#411_Opsins_aligned_in_TM2_region|rosetta stone region]] in TM2 | |||
02 Dec 09: major improvements to [[Opsin_evolution:_informative_indels|informative opsin indel]] section | |||
30 Nov 09: alignment-based [[Opsin_evolution|refinement of reference sequence collection]] fixed annotation errors arising from assembly frameshifts | |||
29 Nov 09: [[Opsin_evolution|added]] two new RGR sequences from tunicates and a new amphioxus melanopsin | |||
02 Nov 09: a ciliary opsin from a [[Opsin_evolution:_key_critters_%28cnidaria%29#Anthozoa:_Anemonia_viridis_.28symbiotic_anemone.29_.._1_opsin|new cnidarian]]; revisited [[Opsin_evolution:_key_critters_%28deuterostomes%29#Hemichordata:_Saccoglossus_kowalevskii_.28acornworm.29_.._2_peropsins|acorn worm opsins]] using newly released assembly | |||
05 Sep 09: comparative genomics applied to potential disease SNPs of [[USH2A_SNPs|USH2A]] and [[CDH23_SNPs|CDH25]] | |||
20 Aug 09: new section on molecular commonalities of hearing and vision genes such as [[CDH23_SNPs|PCDH15 and CDH25]] | |||
05 Aug 09: added section on fibronectin domains of [[USH2A_SNPs|Usher Syndrome]] components USH2A and GPR98 | |||
23 Jul 09: [[Opsin_evolution:_key_critters_%28lophotrochozoa%29#Platyhelminthes:_Schistosoma_mansoni.2Fjaponicum_.28trematode.29_.._3_opsins|revisited]] platyhelminthes opsins in view of newly published Schistosoma genome articles | |||
29 Jun 09: found and characterized the [[Opsin_evolution:_key_critters_%28cnidaria%29#Ctenophora:_Pleurobrachia_pileus_.28sea_gooseberry.29_.._1_opsin|first ctenophore opsin]] | |||
28 Jun 09: separated [[Opsin_evolution:_key_critters_%28ecdysozoa%29|ecdysozoa]] and [[Opsin_evolution:_key_critters_%28lophotrochozoa%29|lophotrochozoa]] articles | |||
27 Jun 09: interaction of opsin cytoplasmic loop C2 and C3 [[Opsin_evolution:_transducins#Structure.2Ffunction_of_signalling:_the_G.CE.B1_interaction_with_opsins|co-evolution]] with transducin GNAT1 | |||
25 Jun 09: updated [[Opsin_evolution:_key_critters_%28protostomes%29#Ecdysozoa:_79_opsins|arthropod melanopsins]] and their opsin diagnostics; data gap in myriapods, onychophorans, tardigrades | |||
21 Jun 09: updated section on Ciona opsins and their photoreceptors | |||
19 Jun 09: located 6 opsins in [[Opsin_evolution:_key_critters_%28protostomes%29#Hexapoda:_Acyrthosiphon_pisum_.28pea_aphid.29_.._6_opsins|aphid genome]] including Rh7 ortholog | |||
18 Jun 09: recovered complete ciliary opsin and second UV opsin from [[Opsin_evolution:_key_critters_%28protostomes%29#Panarthropoda:_Rhodnius_prolixus_.28kissing_bug.29_.._2_opsins|new Rhodnia assembly]] | |||
17 Jun 09: comparative genomics of [[Opsin_evolution:_Cytoplasmic_face#The_third_cytoplasmic_loop_in_83_melanopsins|3rd cytoplasmic loop]] in melanopsins | |||
16 Jun 09: new section on drosophila opsins focused on [[Opsin_evolution:_key_critters_%28protostomes%29#Panarthropoda:_Drosophila_melanogaster_.28fruitfly.29_.._7_opsins|'anatomical orphan' RH7]] | |||
14 Jun 09: updated [[Opsin_evolution:_key_critters_%28protostomes%29#Chelicerata:_Ixodes_scapularis_.28tick.29_2_opsins|Chelicerate opsins]], fixed Daphnia gene models, sought Pycnogonida and Onychophora data | |||
10 Jun 09: new section on opsins of [[Opsin_evolution:_key_critters_%28protostomes%29#Ecdysozoa_.._opsin_repertoire_of_the_last_common_ancestor|ancestral Ecdysozoan]], K90 diagnostic of arthropod ultraviolet vision | |||
19 May 09: analyzed [[Opsin_evolution:_key_critters_%28cnidaria%29#Anthozoa:_Acropora_millepora_.28stony_coral.29_.._1_opsin|new melanopsin]] from Acropora (coral) transcriptome project | |||
17 May 09: found [[Opsin_evolution:_key_critters_%28deuterostomes%29#Cephalochordata:_Branchiostoma_.28amphioxus.29_.._11_opsins|new ciliary opsin]] in Branchiostoma, improved and reclassified the 11 opsins of cephalochordates | |||
09 May 09: hagfish opsins: analyzed their [[Opsin_evolution:_key_critters_%28deuterostomes%29#Chordate:_Eptatretus_burgeri_.28hagfish.29_.._0_opsins|potential impacts]] on understanding vertebrate vision evolution | |||
08 May 09: documented a [[Opsin_evolution:_key_critters_%28deuterostomes%29#Urochordata:_Ciona_intestinalis_.28tunicate.29_.._5_opsins|diverged tunicate melanopsin]] with transcript support using K-rhodopsin searching | |||
26 Mar 09: evolutionary analysis of lamprey and amphioxus [[Opsin_evolution:_RBP3_%28IRBP%29|IRBP modules]] (interstitial retinol-binding protein RBP3) | |||
07 Feb 09: integrated sitewide important [[Opsin_evolution:_key_critters_%28deuterostomes%29#Chondrichthyes:_Callorhinchus_milii_.28elephantshark.29_.._17_opsins|new chondricthyean sequences]] for RHO2, LWS1, and LWS2 | |||
05 Feb 09: finished massive [[Opsin_evolution:_Cytoplasmic_face#The_carboxy-terminal_tail_and_VxPx_motif|comparison of cytoplasmic tails]] of all opsin loci | |||
29 Jan 09: discovered [[Opsin_evolution:_Neuropsin_phyloSNPs#NEUR4:_a_fourth_neuropsin_from_lamprey_to_platypus|yet another neuropsin]] NEUR4 that ranges from lamprey to platypus | |||
18 Jan 09: began massive comparative genomic page on structure of [[Opsin_evolution:_Cytoplasmic_face|opsin cytoplasmic faces and Galpha determinants]] | |||
06 Jan 09: began new page on [[Opsin_evolution:_RPE65|origin of RPE65]] and its two paralogs BCMO1 and BCO2 (carotenoid processing) | |||
21 Nov 08: added structural comparison of A2a adenosine receptor binding and DRY region | |||
07 Nov 08: considered implication for opsins of new KH Ciona assembly which corrected 5,109 genes | |||
06 Nov 08: added second hemichordate peropsin using new EST release | |||
08 Sep 08: added duplication history and reference sequences for transducins (alpha subunits of heteromeric G proteins) | |||
27 Aug 08: added choanoflagellate genome, no opsins but Gi and Gq class heteromeric G proteins | |||
26 Aug 08: compiled 16 Nematostella non-opsins yet with lysine in retinal position | |||
25 Aug 08: reviewed placozoan genome project and 4 opsins asserted for it | |||
20 Aug 08: added opsins from Rhodnius genome project (traces only) | |||
12 Aug 08: reviewed exciting new developments in cnidarian (Tripedalia) ciliary opsins and transducin binding site | |||
08 Aug 08: recovered very old SWS2 unprocessed pseudogene in Petromyzon (lamprey) and updated key critter article | |||
01 Aug 08: recovered SWS1 unprocessed pseudogene from Petromyzon marinus (lamprey) genome | |||
30 Jul 08: expanded RHO2 collection to 8 teleost fish and one exon from RHO2 of chondrichthyes; lamprey RHO1/2 still muddled | |||
29 Jul 08: evolution of upstream RORB and CRX control regions for LWS and SWS1 | |||
28 Jul 08: control region section for LWS; recovered LCR_LWS orthologs for 35 species spanning lamprey to human | |||
27 Jul 08: new echidna data for monotreme opsin evolution; gene tree x species tree showing regressive mammalian opsin evolution | |||
20 Apr 08: revisited LWS in many more species; expanded ancestral SWS2+ LWS+ linkage species | |||
13 Apr 08: special section on melanopsins in vertebrates: gene loss of MEL2 and gene shrinkage in MEL1 | |||
06 Apr 08: expanded TMT class of encephalopsins to 14 species, shark to marsupial, showed lost in all placentals | |||
05 Apr 08: demonstrated massive gene loss of mammalian encephalopsins and placental TMT opsins | |||
27 Mar 08: upgraded lamprey larval section and brought opsin collection to 17 (in 11 orthology classes) | |||
23 Mar 08: new section aligning 94 LWS and analyzing its PhyloSNPs | |||
17 Mar 08: discovery of a novel opsin occuring from sharks to birds, lost in mammals: newropsin | |||
14 Mar 08: new section on neuropsin comparative genomics | |||
07 Mar 08: new section on peropsin synapomorphies | |||
20 Feb 08: new section on RGR: clade-defining changes (phyoSNPs), gene loss in marsupial | |||
10 Jan 08: added Tribolium critter section on opsin loss, ommitidia cell gain, non-opsin GPCR content. | |||
09 Jan 08: posted paraphyletic gene tree explanation for 'too many' opsins in Daphnia, Nematostella, and Cladonema: | |||
opsins duplicated, diverged, neofunctionalized to non-opsin non-photoreceptors nesting within opsins. | |||
08 Jan 08: added key critter section on hydrozoan Cladonema putative opsins. | |||
07 Jan 08: added ancient intron discussion for melanopsins. | |||
05 Jan 08: finished ancient intron section for peropsin group. | |||
04 Jan 08: added new key critters section for hydrozoan Cladonema radiatum, | |||
03 Jan 08: made comparative conservation profile for 54 new cnidarian opsins proposed by Gehring group. | |||
02 Jan 08: added comparative genomics of RGR because of curious loss of DRY motif in boreoeutheres. | |||
31 Dec 07: posted fullscale comparison of ciliary opsin introns | |||
29 Dec 07: extracted scattered elements of the binding pocket for comparative genomic covariance study. | |||
28 Dec 07: added new discussion on peropsins as more than mere photoisomerases and in need of subfamily refinement. | |||
27 Dec 07: recovered new Aplysia and Lottia peropsins orthologous to experimentally studied squid 'retinochrome'. | |||
26 Dec 07: started new page on coding indels and completely rewrote main introductory page. | |||
25 Dec 07: added key critters section for human body louse and its 3 rhabdomeric opsins. | |||
24 Dec 07: fixed arrestin quasi-cognate section and proposed bioinformatic approach to unstudied opsin/arrestin pairs | |||
23 Dec 07: added alignment of C-terminus and comparative genomics of phosphorylation sites vis-a-vis arrestins | |||
22 Dec 07: upgraded alignment of all opsin sequences, trimmed at one or both ends, color consensus and motif searchable. | |||
21 Dec 07: added an annotation trick that allows trimming and residue selection from the reference opsin set. | |||
20 Dec 07: added comparative genomics discussion of counterion and its switching. | |||
19 Dec 07: added greatly to opsin structure/function in ancient sequence section to explain conserved diagnostic motifs. | |||
18 Dec 07: began new page on ancestral ciliary opsin sequence reconstruction. | |||
17 Dec 07: added new ciliary opsins from Daphnia, Tribolium, Aedes, and Culex genomes. | |||
16 Dec 07; began new page on ancestral ciliary opsin intron pattern. | |||
14 Dec 07: upgraded sponge photoreceptor account, found final exon of new Platynereis ciliary exon. | |||
13 Dec 07: extended JGI staff models of 3 intronless ciliary encephalopsins from Nematostella to full length. | |||
12 Dec 07: added Nematostella and Hydra sections awaiting clarification. | |||
11 Dec 07: finished larval and adult photoreception in Ciona. | |||
10 Dec 07: expanded section on Ciona to include possible otolith paralogous photoreceptor. | |||
09 Dec 07: added a treatment of two tunicate pinopsin-like opsins and their ancient introns. | |||
08 Dec 07: added a section on intron cross-comparison to the annotation tutorial. | |||
08 Dec 07: added treatment of two platyhelminthes to Lophotrochozoa area. | |||
07 Dec 07: added 2 trematode and 1 planaria opsins, exon pattern matches melanopsin. | |||
06 Dec 07: added cnidarian larval rhabdomeric single-cell eye and adult rhopalium imagery. | |||
05 Dec 07: added Capitella literature survey and a new melanopsin from genome assembly. | |||
04 Dec 07: added Helobedella literature survey and new opsins from genome assembly. | |||
03 Dec 07: added Lottia, reviewed genome assembly, found an opsin but could not locate anything on ocelli. | |||
02 Dec 07: added Aplysia eye anatomy and apparent melanopsin ortholog to key critters. | |||
29 Nov 07: added platynereis story to critters, described second new ciliary opsin. | |||
29 Nov 07: added graphics comparing eye systems in deuts and lophs, illustrated everse and inverse eyes. | |||
29 Nov 07: added sea urchin opsins to critter discussion, graphic of expression in larva and adult. | |||
29 Nov 07: added 6 sea urchin reference opsins by repairing gene models, adding introns, and reclassifying. | |||
28 Nov 07: added Xenoturbella and Convoluta section to basal key critters, no opsins available. | |||
28 Nov 07: upgraded key critters with treatment of Saccoglossus. | |||
28 Nov 07: found first hemichordate opsin, put methods into detailed tutorial. | |||
28 Nov 07: received copywrite permissions for use of handdrawn invertebrate eyes illustrations. | |||
28 Nov 07: added jump navigation to key critters as article is getting fairly long. | |||
27 Nov 07: added scallop Go opsins from an important historic study. | |||
27 Nov 07: reorganized curated sequences into deuterostome, ecdysozoa, lophotrochozoa blocks sorted by receptor type. | |||
site visits: 2008 2009 2010 2011 2012 2014 | |||
#pages topic 22 Apr 12 May 29 Jul 4 Sep 4 Nov 3 Jan 8 Feb 26 Mar 2 Jun 5 Sep 8 Dec 6 Mar 6 Jun 13 Sep 21 Jan 22 May 7 Sep 2 Jan 18 May 18 Apr | |||
47 main 814 896 1076 1227 1706 1919 2123 2380 2693 3128 3561 4181 4570 4938 5418 6091 6798 7510 9171 23391 | |||
14 key.deuterostomes 0 0 107 170 250 385 447 631 907 1104 1460 1680 1835 2073 2377 2720 3222 3718 4942 18253 | |||
32 key.ecdysozoa 1106 1207 1279 1351 1492 1685 1766 2009 2324 98 245 430 603 855 1121 1471 1877 2242 3398 9232 | |||
11 key.lophotrochozoa 0 0 0 0 0 0 0 0 0 121 305 432 621 794 1071 1376 1721 2020 2783 5582 | |||
25 key.cnidaria 0 0 0 120 209 320 391 515 731 974 1304 1562 1961 2316 2835 3505 4246 5210 6994 25284 | |||
04 opsin.origins 0 0 0 0 0 0 0 0 0 0 0 294 524 761 1056 1395 1691 2085 3094 8306 | |||
08 alignment 231 256 304 324 380 429 475 555 636 731 872 966 1086 1237 1423 1593 1811 1963 2341 3696 | |||
10 introns 180 202 252 274 340 396 455 548 637 777 980 1203 1435 1674 1891 2188 2485 2708 3556 8080 | |||
02 indels 97 118 157 179 227 284 322 405 521 634 829 1070 1315 1505 1719 2060 2317 2615 3145 5949 | |||
11 ancestral 247 269 318 351 414 476 520 628 765 901 1109 1267 1452 1649 1853 2139 2420 2683 3167 5789 | |||
24 LWS 68 104 169 218 291 338 409 548 665 825 1045 1194 1330 1559 1791 2098 2571 2968 3758 8390 | |||
12 encephalopsin 67 110 181 216 286 350 454 625 842 1030 1289 1468 1762 1997 2303 2594 2867 3100 3655 5761 | |||
13 melanopsin 24 70 132 160 207 244 317 424 540 651 802 913 1055 1214 1398 1637 1913 2091 2659 4790 | |||
13 RGR.phyloSNPs 98 120 181 200 259 299 528 662 822 983 1178 1339 1532 1728 1995 2275 2639 2915 3810 7457 | |||
16 peropsin 89 117 174 195 254 332 381 461 573 710 891 1072 1244 1444 1691 1965 2216 2493 3045 6830 | |||
25 | 15 neuropsin 83 102 164 188 225 264 390 471 558 672 903 1097 1227 1467 1759 1985 2247 2471 3192 9972 | ||
08 platypus 290 338 488 574 743 868 978 1123 1324 1484 1743 1962 2300 2527 2889 3342 3790 4234 4990 8138 | |||
06 tricks 169 191 304 343 396 468 530 631 769 938 1253 1438 1622 1854 2113 2365 2697 3072 3950 7907 | |||
27 cytoplasmic.face 0 0 0 0 0 0 91 132 277 445 670 814 977 1150 1346 1572 1861 2137 2864 6526 | |||
41 RPE.BCO2.BCMO1 0 0 0 0 0 0 85 163 261 447 592 691 869 993 1197 1461 1687 1839 2436 4978 | |||
20 IRBP.(RBP3) 0 0 0 0 0 0 0 339 772 968 1202 1310 1493 1643 1840 2053 2368 2679 3164 5183 | |||
20 CDH23 0 0 0 0 0 0 0 0 0 162 326 474 709 945 1211 1741 2085 2490 3296 11970 | |||
28 USH2A 0 0 0 0 0 0 0 0 0 222 364 480 702 911 1178 1537 1830 2133 2900 5880 | |||
17 transducins 0 0 0 0 148 468 378 509 674 918 1164 1345 1538 1802 2063 2402 2722 2976 3437 5455 | |||
31 underground 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 782 7069 | |||
03 blog 1178 1329 1798 2158 2610 3179 3412 3960 4599 5436 6211 7002 7860 9107 10354 11987 13540 15422 18726 36975 | |||
532 total 4741 5429 7084 8248 10437 12579 14219 17719 21890 26683 30298 35684 41640 48143 55892 65552 75621 85774 109255 275661 | |||
... visits.per.day 31 34 22 31 36 39 49 74 61 50yr 38 61 65 66 69 80 102 87yr 173 ... | |||
13 | |||
03 | |||
[[Category:Comparative Genomics]] | [[Category:Comparative Genomics]] | ||
Latest revision as of 20:49, 19 April 2014
This large collection of opsin evolution articles is updated almost daily. To locate changes, consult the article directory and update blog below.
- Opsin classification tool and curated sequence collection
- Opsin origins
- Key critters with genomics: deuterostomes
- Key critters with genomics: ecdysozoa
- Key critters with genomics: lophotrochozoa
- Key critters with genomics: cnidaria and porifera
- 230 opsins aligned: consensus-colored image and motif searchable text
- Ancestral intron reconstruction for ciliary opsins
- Ancestral status of informative coding indels
- Ancestral sequence reconstruction for ciliary opsins
- LWS: generating locus for imaging opsins
- Encephalopsin and TMT: massive gene loss in mammalian clades
- Melanopsins show gene and exon loss in mammals
- RGR opsin: abrupt change in DRY motif in boreoeutheres
- Peropsin: phyloSNPs (synapomorphies) in persopsin evolution
- Neuropsin (OPN5): 52 deuterostome genes and a novel paralog, Newropsin
- Color vision in the platypus and ancestral mammal
- Opsins and cryptochromes in a subterranean mammal
- The cytoplasmic face of opsins and GPCR specificity
- IRBP Interstitial retinol-binding protein modules
- RPE65 rate-limiting step in ciliary opsin regeneration
- Usherin USH2A roles in vision and hearing
- Cadherin CDH23 roles in vision and hearing
- Transducins: comparative genomics of opsin interactions
- Annotation tricks: genes from trace reads, intron patterns, resolving indels, trimming
- Tracking significant updates (this page)
- The photoreceptor fossil record (planned)
- Opsin evolution: the bottom line (in progress)
Reverse Chronological Update Blog 14 Apr 12: released major article on cryptochrome evolution, relationships with opsins, with 275 curated reference sequences 15 Feb 12: started new article on opsins of an underground mammal, naked mole-rat (Heterocephalus glaber) 30 Dec 11: first nematode opsins located in Strongyloides 30 Sep 11: the earliest opsins -- added two full-length ctenophore opsins with introns from Mnemiopsis assembly 13 Aug 11: first introns found in cnidarian opsins (new coral genome, Acropora digitifera) 06 Mar 11: RH7 transcripts found in butterfly antenna -- new anatomical structure? 02 Mar 11: First snake genome has 11 opsins but 8 are lost 06 Oct 10: new opsin surfaces in non-amniote vertebrates -- a neuropsin related to NEUR4 in zebrafish and frog 26 Aug 10: first ctenophore opsin now extendable to full length; second ctenophore opsin surfaces 03 Jul 10: analyzed the five opsins in the new leafcutter ant genome, Atta cephalotes 06 Jun 10: the first sponge opsin in unassembled traces proves to be a Lottia contaminant 05 Apr 10: collected assembled salamander transcripts for 12 opsin loci from a 454 project; amphibians remain poorly sampled 29 Mar 10: analyzed and classified the 41 'opsins' in the new Hydra genome -- all have lysine at 296 but lack full DRY 24 Mar 10: spellchecked the entire suite of articles (503 pages), fixing many many typos 02 Feb 10: analyzed comparative genomics of the F CWxP aromatic lock between TM6-TM7 27 Jan 10: added section on Bombyx genome and its five opsins 21 Jan 10: greatly improved fasta header lines in the 456 reference sequences using space-delimited database format, facilitating subset selection. 18 Jan 10: queried new Nasonia genomes but these were uninformative -- too close to existing Nasonia vitripennis 17 Jan 10: queried new chelicerate genome (Varroa bee mite) for opsins, finding only a peropsin-like fragment 14 Jan 10: snazzy new graphic shows gain and loss at 24 opsin loci during deuterostome history, vindicating 1942 GL Walls hypothesis 13 Jan 10: added section about 'managing' homoplasy of indels, introns and diagnostic residues 12 Jan 10: defined the intronation required of a GPCR gene to be parental to an opsin family 11 Jan 10: showed longwave opsin introns in arthropods form a distinct yet early coalescing melanopsin class 10 Jan 10: established using introns that peropsin, rgropsin and neuropsins form a family that excludes cilopsins and melanopsins 09 Jan 10: proved arthropod UV opsins are orthologous to vertebrate melanopsin 08 Jan 10: improved methods and big update to ancestral introns of ciliary opsins 29 Dec 09: discovered a new ur-opsin locus TMTc ranging from fish to frog but lacking transcripts and experiments 27 Dec 09: provided page crosslinks to allow faster navigation between related topics 26 Dec 09: added flanking synteny to ciliary opsins to sort out gene duplication origins 23 Dec 09: sought informative indels by aligning GPCR and proxy opsins in the retinal lid region (second extracellular loop) 21 Dec 09: extended ur-opsin TMTa into frog and lizard but found decayed syntenic pseudogenes in chicken, finch and platypus 19 Dec 09: began new section on opsin origins from earlier GPCR 16 Dec 09: located a new moth ciliary opsin and fixed others; confirmed multiple lineage losses for ciliary opsins 15 Dec 09: located a second chelicerate peropsin in Ixodes; gene lost in crustacea and insects 09 Dec 09: reviewed first reported peropsin from ecdysozoa; reclassified peropsin gene family 08 Dec 09: located ciliary ur-opsin of deuterostomes, new orthologs in frog and lizard, not birds or mammals 07 Dec 09: aligned 411 opsins in rosetta stone region in TM2 02 Dec 09: major improvements to informative opsin indel section 30 Nov 09: alignment-based refinement of reference sequence collection fixed annotation errors arising from assembly frameshifts 29 Nov 09: added two new RGR sequences from tunicates and a new amphioxus melanopsin 02 Nov 09: a ciliary opsin from a new cnidarian; revisited acorn worm opsins using newly released assembly 05 Sep 09: comparative genomics applied to potential disease SNPs of USH2A and CDH25 20 Aug 09: new section on molecular commonalities of hearing and vision genes such as PCDH15 and CDH25 05 Aug 09: added section on fibronectin domains of Usher Syndrome components USH2A and GPR98 23 Jul 09: revisited platyhelminthes opsins in view of newly published Schistosoma genome articles 29 Jun 09: found and characterized the first ctenophore opsin 28 Jun 09: separated ecdysozoa and lophotrochozoa articles 27 Jun 09: interaction of opsin cytoplasmic loop C2 and C3 co-evolution with transducin GNAT1 25 Jun 09: updated arthropod melanopsins and their opsin diagnostics; data gap in myriapods, onychophorans, tardigrades 21 Jun 09: updated section on Ciona opsins and their photoreceptors 19 Jun 09: located 6 opsins in aphid genome including Rh7 ortholog 18 Jun 09: recovered complete ciliary opsin and second UV opsin from new Rhodnia assembly 17 Jun 09: comparative genomics of 3rd cytoplasmic loop in melanopsins 16 Jun 09: new section on drosophila opsins focused on 'anatomical orphan' RH7 14 Jun 09: updated Chelicerate opsins, fixed Daphnia gene models, sought Pycnogonida and Onychophora data 10 Jun 09: new section on opsins of ancestral Ecdysozoan, K90 diagnostic of arthropod ultraviolet vision 19 May 09: analyzed new melanopsin from Acropora (coral) transcriptome project 17 May 09: found new ciliary opsin in Branchiostoma, improved and reclassified the 11 opsins of cephalochordates 09 May 09: hagfish opsins: analyzed their potential impacts on understanding vertebrate vision evolution 08 May 09: documented a diverged tunicate melanopsin with transcript support using K-rhodopsin searching 26 Mar 09: evolutionary analysis of lamprey and amphioxus IRBP modules (interstitial retinol-binding protein RBP3) 07 Feb 09: integrated sitewide important new chondricthyean sequences for RHO2, LWS1, and LWS2 05 Feb 09: finished massive comparison of cytoplasmic tails of all opsin loci 29 Jan 09: discovered yet another neuropsin NEUR4 that ranges from lamprey to platypus 18 Jan 09: began massive comparative genomic page on structure of opsin cytoplasmic faces and Galpha determinants 06 Jan 09: began new page on origin of RPE65 and its two paralogs BCMO1 and BCO2 (carotenoid processing) 21 Nov 08: added structural comparison of A2a adenosine receptor binding and DRY region 07 Nov 08: considered implication for opsins of new KH Ciona assembly which corrected 5,109 genes 06 Nov 08: added second hemichordate peropsin using new EST release 08 Sep 08: added duplication history and reference sequences for transducins (alpha subunits of heteromeric G proteins) 27 Aug 08: added choanoflagellate genome, no opsins but Gi and Gq class heteromeric G proteins 26 Aug 08: compiled 16 Nematostella non-opsins yet with lysine in retinal position 25 Aug 08: reviewed placozoan genome project and 4 opsins asserted for it 20 Aug 08: added opsins from Rhodnius genome project (traces only) 12 Aug 08: reviewed exciting new developments in cnidarian (Tripedalia) ciliary opsins and transducin binding site 08 Aug 08: recovered very old SWS2 unprocessed pseudogene in Petromyzon (lamprey) and updated key critter article 01 Aug 08: recovered SWS1 unprocessed pseudogene from Petromyzon marinus (lamprey) genome 30 Jul 08: expanded RHO2 collection to 8 teleost fish and one exon from RHO2 of chondrichthyes; lamprey RHO1/2 still muddled 29 Jul 08: evolution of upstream RORB and CRX control regions for LWS and SWS1 28 Jul 08: control region section for LWS; recovered LCR_LWS orthologs for 35 species spanning lamprey to human 27 Jul 08: new echidna data for monotreme opsin evolution; gene tree x species tree showing regressive mammalian opsin evolution 20 Apr 08: revisited LWS in many more species; expanded ancestral SWS2+ LWS+ linkage species 13 Apr 08: special section on melanopsins in vertebrates: gene loss of MEL2 and gene shrinkage in MEL1 06 Apr 08: expanded TMT class of encephalopsins to 14 species, shark to marsupial, showed lost in all placentals 05 Apr 08: demonstrated massive gene loss of mammalian encephalopsins and placental TMT opsins 27 Mar 08: upgraded lamprey larval section and brought opsin collection to 17 (in 11 orthology classes) 23 Mar 08: new section aligning 94 LWS and analyzing its PhyloSNPs 17 Mar 08: discovery of a novel opsin occuring from sharks to birds, lost in mammals: newropsin 14 Mar 08: new section on neuropsin comparative genomics 07 Mar 08: new section on peropsin synapomorphies 20 Feb 08: new section on RGR: clade-defining changes (phyoSNPs), gene loss in marsupial 10 Jan 08: added Tribolium critter section on opsin loss, ommitidia cell gain, non-opsin GPCR content. 09 Jan 08: posted paraphyletic gene tree explanation for 'too many' opsins in Daphnia, Nematostella, and Cladonema: opsins duplicated, diverged, neofunctionalized to non-opsin non-photoreceptors nesting within opsins. 08 Jan 08: added key critter section on hydrozoan Cladonema putative opsins. 07 Jan 08: added ancient intron discussion for melanopsins. 05 Jan 08: finished ancient intron section for peropsin group. 04 Jan 08: added new key critters section for hydrozoan Cladonema radiatum, 03 Jan 08: made comparative conservation profile for 54 new cnidarian opsins proposed by Gehring group. 02 Jan 08: added comparative genomics of RGR because of curious loss of DRY motif in boreoeutheres. 31 Dec 07: posted fullscale comparison of ciliary opsin introns 29 Dec 07: extracted scattered elements of the binding pocket for comparative genomic covariance study. 28 Dec 07: added new discussion on peropsins as more than mere photoisomerases and in need of subfamily refinement. 27 Dec 07: recovered new Aplysia and Lottia peropsins orthologous to experimentally studied squid 'retinochrome'. 26 Dec 07: started new page on coding indels and completely rewrote main introductory page. 25 Dec 07: added key critters section for human body louse and its 3 rhabdomeric opsins. 24 Dec 07: fixed arrestin quasi-cognate section and proposed bioinformatic approach to unstudied opsin/arrestin pairs 23 Dec 07: added alignment of C-terminus and comparative genomics of phosphorylation sites vis-a-vis arrestins 22 Dec 07: upgraded alignment of all opsin sequences, trimmed at one or both ends, color consensus and motif searchable. 21 Dec 07: added an annotation trick that allows trimming and residue selection from the reference opsin set. 20 Dec 07: added comparative genomics discussion of counterion and its switching. 19 Dec 07: added greatly to opsin structure/function in ancient sequence section to explain conserved diagnostic motifs. 18 Dec 07: began new page on ancestral ciliary opsin sequence reconstruction. 17 Dec 07: added new ciliary opsins from Daphnia, Tribolium, Aedes, and Culex genomes. 16 Dec 07; began new page on ancestral ciliary opsin intron pattern. 14 Dec 07: upgraded sponge photoreceptor account, found final exon of new Platynereis ciliary exon. 13 Dec 07: extended JGI staff models of 3 intronless ciliary encephalopsins from Nematostella to full length. 12 Dec 07: added Nematostella and Hydra sections awaiting clarification. 11 Dec 07: finished larval and adult photoreception in Ciona. 10 Dec 07: expanded section on Ciona to include possible otolith paralogous photoreceptor. 09 Dec 07: added a treatment of two tunicate pinopsin-like opsins and their ancient introns. 08 Dec 07: added a section on intron cross-comparison to the annotation tutorial. 08 Dec 07: added treatment of two platyhelminthes to Lophotrochozoa area. 07 Dec 07: added 2 trematode and 1 planaria opsins, exon pattern matches melanopsin. 06 Dec 07: added cnidarian larval rhabdomeric single-cell eye and adult rhopalium imagery. 05 Dec 07: added Capitella literature survey and a new melanopsin from genome assembly. 04 Dec 07: added Helobedella literature survey and new opsins from genome assembly. 03 Dec 07: added Lottia, reviewed genome assembly, found an opsin but could not locate anything on ocelli. 02 Dec 07: added Aplysia eye anatomy and apparent melanopsin ortholog to key critters. 29 Nov 07: added platynereis story to critters, described second new ciliary opsin. 29 Nov 07: added graphics comparing eye systems in deuts and lophs, illustrated everse and inverse eyes. 29 Nov 07: added sea urchin opsins to critter discussion, graphic of expression in larva and adult. 29 Nov 07: added 6 sea urchin reference opsins by repairing gene models, adding introns, and reclassifying. 28 Nov 07: added Xenoturbella and Convoluta section to basal key critters, no opsins available. 28 Nov 07: upgraded key critters with treatment of Saccoglossus. 28 Nov 07: found first hemichordate opsin, put methods into detailed tutorial. 28 Nov 07: received copywrite permissions for use of handdrawn invertebrate eyes illustrations. 28 Nov 07: added jump navigation to key critters as article is getting fairly long. 27 Nov 07: added scallop Go opsins from an important historic study. 27 Nov 07: reorganized curated sequences into deuterostome, ecdysozoa, lophotrochozoa blocks sorted by receptor type.
site visits: 2008 2009 2010 2011 2012 2014 #pages topic 22 Apr 12 May 29 Jul 4 Sep 4 Nov 3 Jan 8 Feb 26 Mar 2 Jun 5 Sep 8 Dec 6 Mar 6 Jun 13 Sep 21 Jan 22 May 7 Sep 2 Jan 18 May 18 Apr 47 main 814 896 1076 1227 1706 1919 2123 2380 2693 3128 3561 4181 4570 4938 5418 6091 6798 7510 9171 23391 14 key.deuterostomes 0 0 107 170 250 385 447 631 907 1104 1460 1680 1835 2073 2377 2720 3222 3718 4942 18253 32 key.ecdysozoa 1106 1207 1279 1351 1492 1685 1766 2009 2324 98 245 430 603 855 1121 1471 1877 2242 3398 9232 11 key.lophotrochozoa 0 0 0 0 0 0 0 0 0 121 305 432 621 794 1071 1376 1721 2020 2783 5582 25 key.cnidaria 0 0 0 120 209 320 391 515 731 974 1304 1562 1961 2316 2835 3505 4246 5210 6994 25284 04 opsin.origins 0 0 0 0 0 0 0 0 0 0 0 294 524 761 1056 1395 1691 2085 3094 8306 08 alignment 231 256 304 324 380 429 475 555 636 731 872 966 1086 1237 1423 1593 1811 1963 2341 3696 10 introns 180 202 252 274 340 396 455 548 637 777 980 1203 1435 1674 1891 2188 2485 2708 3556 8080 02 indels 97 118 157 179 227 284 322 405 521 634 829 1070 1315 1505 1719 2060 2317 2615 3145 5949 11 ancestral 247 269 318 351 414 476 520 628 765 901 1109 1267 1452 1649 1853 2139 2420 2683 3167 5789 24 LWS 68 104 169 218 291 338 409 548 665 825 1045 1194 1330 1559 1791 2098 2571 2968 3758 8390 12 encephalopsin 67 110 181 216 286 350 454 625 842 1030 1289 1468 1762 1997 2303 2594 2867 3100 3655 5761 13 melanopsin 24 70 132 160 207 244 317 424 540 651 802 913 1055 1214 1398 1637 1913 2091 2659 4790 13 RGR.phyloSNPs 98 120 181 200 259 299 528 662 822 983 1178 1339 1532 1728 1995 2275 2639 2915 3810 7457 16 peropsin 89 117 174 195 254 332 381 461 573 710 891 1072 1244 1444 1691 1965 2216 2493 3045 6830 15 neuropsin 83 102 164 188 225 264 390 471 558 672 903 1097 1227 1467 1759 1985 2247 2471 3192 9972 08 platypus 290 338 488 574 743 868 978 1123 1324 1484 1743 1962 2300 2527 2889 3342 3790 4234 4990 8138 06 tricks 169 191 304 343 396 468 530 631 769 938 1253 1438 1622 1854 2113 2365 2697 3072 3950 7907 27 cytoplasmic.face 0 0 0 0 0 0 91 132 277 445 670 814 977 1150 1346 1572 1861 2137 2864 6526 41 RPE.BCO2.BCMO1 0 0 0 0 0 0 85 163 261 447 592 691 869 993 1197 1461 1687 1839 2436 4978 20 IRBP.(RBP3) 0 0 0 0 0 0 0 339 772 968 1202 1310 1493 1643 1840 2053 2368 2679 3164 5183 20 CDH23 0 0 0 0 0 0 0 0 0 162 326 474 709 945 1211 1741 2085 2490 3296 11970 28 USH2A 0 0 0 0 0 0 0 0 0 222 364 480 702 911 1178 1537 1830 2133 2900 5880 17 transducins 0 0 0 0 148 468 378 509 674 918 1164 1345 1538 1802 2063 2402 2722 2976 3437 5455 31 underground 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 782 7069 03 blog 1178 1329 1798 2158 2610 3179 3412 3960 4599 5436 6211 7002 7860 9107 10354 11987 13540 15422 18726 36975 532 total 4741 5429 7084 8248 10437 12579 14219 17719 21890 26683 30298 35684 41640 48143 55892 65552 75621 85774 109255 275661 ... visits.per.day 31 34 22 31 36 39 49 74 61 50yr 38 61 65 66 69 80 102 87yr 173 ...