LOXHD1 SNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 338: | Line 338: | ||
2 ELKPVKYEVTVVTGDEKGAGTDANVSVILYGDNGDTGPRPLKKKFVNLFERNQHDKFTIEALDLGKLTKLHIEHDNKGWGASWLLDRVEVHNVDSNETIIFPCKQWLDKKKGDGQIAKDLLPES* 0 | 2 ELKPVKYEVTVVTGDEKGAGTDANVSVILYGDNGDTGPRPLKKKFVNLFERNQHDKFTIEALDLGKLTKLHIEHDNKGWGASWLLDRVEVHNVDSNETIIFPCKQWLDKKKGDGQIAKDLLPES* 0 | ||
>Monosiga brevicollis (monosiga) Eukaryota|Choanoflagellates XM_001742822 17 PLAT domains [http://www.ncbi.nlm.nih.gov//genomes/geblast.cgi?bact=off&gi=5881 blast] exons colored; PLAT domains numbered | <span style="color: #0066CC;">>Trichoplax adhaerens (trichoplax) Metazoa XM_002107971 17 PLAT domains [http://www.ncbi.nlm.nih.gov//genomes/geblast.cgi?bact=off&gi=6008 blast] | ||
MSASLAMQELMSDPSGHFAPKPPPGRSKINRKRPQT<span style="color: #996633;">APGRQITTASKAPKLWRSSTYNEPNLKRRKFIVTLNHKKDISSISRNAPYANRAYTTAMATAARTYSNALNKGQQKEIIPIYNPLCDPHLNDYYARKFGLLNSRD</span>ESRKNRKQ<span style="color: #996633;">AG | |||
01 VAYQFGVKTGDKKGSDTDA</span>VYIQVIGTKDKIPKKRLFKKQETEKTERGNLFKFDKSTVEKFAVQHRDIGDPVKLIVE<span style="color: #996633;">HDGNEKRHGWFLEEITLTNIQSKKSWLFPCHKWLSKYEGDRKLCYE LKPLAKAGK | |||
02 A</span>VYEVSVLTGDKRGAGTDANVSVTLFGKHTSSPKIQLLKS<span style="color: #996633;">SKHKNPFERNNTDEFKIRTRDVGKLSKIRIEHDNAGFGPGWFLDK</span>VIICNLEKPNVKYYCPCNQWLAKDVGDKSISRD LTAYTDPNAAPS<span style="color: #996633;">A | |||
03 YVYIVHTFTGNKRGAGTDANVYAVIFGDSGDTGEKRLDNSKNNFEKSR</span>KDTFKLSCSCVGKLERLRIRHDNTGLFAGWYLDKV<span style="color: #996633;">VVEDPQEQQSYTFYCRRWLSKTEDDGEICRDLI VSASGDDGDDSVAPK</span>G | |||
04 YPYHIHVTTSDVKNAGTDAEVYVVMHGEGKKSKELNSGKLVLANSEKKKNTFERAMTDIFHMECAEMLSPLTKLTVGHDNKGLAAGWHLDRa<span style="color: #996633;">IVIDCPTTGIEQTFLCQQWLDRKAGDGLTERELVEA FDMRKTRRP</span>K | |||
05 QLWFAWIWTSDIRGAGTDANVSMQIYGDKGKSQEIKLGNNTDNFEQATLDKFK<span style="color: #996633;">LEIDQVGVPYKLRIGHDNSNAFPGWHLDKVKLENMNDKEQYLFNCNRWLSRSEEDNEIIRELPAS GPNCPNYP</span>I | |||
06 VIYEVSVHTGNKMGGGTDANVFIKIYGELGDSGYRPLKSSKSHNNKFERNQVDVFHIEAVTLKALKKIKIGHDGNNP<span style="color: #996633;">GAGWFLDKVVIKELNGEASNEFPCNR</span>WLSKSEDDGQIVRELFLK SDTPLLK<span style="color: #996633;">T | |||
07 TSYHISVKTGDVRNAGTDANVFIQIFGAKDDTGRVRLKQSLNTSNKFERNRIDKFIIEAAQIGK</span>IEKIIIGHDGKGLGSGWFLDYIELDVPSVGRLYRFSCHQWFDSTEGDRKVERELYPS ECIKSAA<span style="color: #996633;">K | |||
08 IPYQISVHTGDIRHAGTDSNVFAVIYGENGKTEELKLRNKSDNFERGQVDVFK</span>VECEDVGKLRKLRIGHDSAGMGSAWFLDK<span style="color: #996633;">VYVRRLPPKSGKKSKETDEREEETAKKDADEPEKLDENNYLFVANRWLSKEEGDRQTVIEIS PVGVDGALA</span>E | |||
09 MTYTIRVITGNKFGCGTNANVFINMYGEEGDSGERQLKKSETHTDKFERNQ<span style="color: #996633;">EDVFKISCLSLGELKKIKIRHDNSGFRPAWFLDKVIIEVGESKYQFMCDRWLAKDEDDGQISRELLPQ SDEQSRAEAIGASKDLQKK</span>VAS | |||
10 TTYNVSVTTGDIKGAGTDANVHIVLYGEKDDTGLIHLKNSTTHSNKFERNQEDRFVVEAIDIGELKKIK<span style="color: #996633;">IGHDNKGGMAGWFLNKVEIDIPSLGRRLLFPCGRWIDKGKDDGALER ELYPLNEAEETYRP</span>H | |||
11 IPYEVTVYTTDKRGASTGANVYVVIYGEENQTEQASLEPDKKRRKQYFKNGAIDKFVLE<span style="color: #996633;">LDDVGEEITKLRIGHDGKGWGAGWHLDKVEICRLLDGGKASKKFTFQCNRWLASDEDDGAIVRELVPS EIVEKSSKDGGQVKTKVTKPTDGLK</span>V | |||
12 KPYTIHVFTGDVDGAGTNANVFLTIFGESGDSGERKLAKSDTHYDKFERNQ<span style="color: #996633;">EDIFHIEAADLGRLFKVKIRHDNTGSLFSPAWFLNRIEIVDDENEETTAFPCERWLAKKKDDGKIDRTLFVK GWEGDTSSVATTRSK</span>ASFRSNSTDLGEMVSASGRRASSRKGPVMAPIDEK<span style="color: #996633;">C | |||
13 VPYNIKVTVGEESSKNFQETLHLELFGQIEEEKSGPIELSPEKKSDKTFYPGKITTFYVSAAEVNIIEKIQVS</span>HNSYMPDSGIYLKEIEVDVPTIGNKYIFPCNRWLAKDKDDSKTSRIFTAA | |||
14 NAQVTSYTA<span style="color: #996633;">GTDSNIFVILFGTKGRTPEISLEKNEDRFERAKVDIIP</span>LELDDVGTIKKIRIGHDGKGSRTDWYLEK<span style="color: #996633;">ASIQRMDTLDMYMFRANQWFSKKIDDKKLVREI PAETSKEGATTIK</span>K | |||
15 INYVLSTHTSDKRGSGTDANVFVIIFGENGDSGEIALKKSETNWNKFEKGQTDVFLINDRLSLGRLQKLRIWHDNA<span style="color: #996633;">GFGASWHLASVDIVDESTGVKYTFPCDKWLSKSNGDKLILREL PCAETAGNTAASKATKQESKSGK</span> | |||
16 AEYEIAFTTGTEKRAGTNQDVAIVLKGKSEKSREFLIENNEDKKYFSKGKTNKFTYTCKPLGDITKAIVSHRESAIGEEPDSKNSSWYLKVVTVVHKASGTT<span style="color: #996633;">YKFPCNKWIDLDDDEENNSSVTLKC KSADVAASSKAKPV</span>ELKP | |||
17 VKYEVTVVTGDEKGAGTDANVSVILYGDNGDTGPRPLKKKFVNLFERNQHDKFTIEALDLGKLTKLHIEHDNKGWGASWLLDRVEVHNVDSNETIIFPCKQWLDKKKGDGQIAKDLLPES* </span> | |||
<span style="color: #0066CC;">>Monosiga brevicollis (monosiga) Eukaryota|Choanoflagellates XM_001742822 17 PLAT domains [http://www.ncbi.nlm.nih.gov//genomes/geblast.cgi?bact=off&gi=5881 blast] exons colored; PLAT domains numbered | |||
MADLPVPTTALQRLQLERSRYSFAAA<span style="color: #996633;">DPPPSFVQRYTSADVLSH</span>RLYAQGLASDVGPGSHSSL<span style="color: #996633;">GMSRRVELPPYNPLNDPALANYFARKFEWNSTRSVGAGSGSRRSASASLTRTRQPVRGASAHGHTKSRTKSSHRK | |||
01 GHASLEIFTSKRSNPVSKSEKYITVVGTKGRSDAIQLAAPGVHFRAGNKDVFQVNLTGIGKPT</span>KVILENTGTKRTDGWCVSKVVLVKKTDKGTRRYRFSGPVWL<span style="color: #996633;">SKHHDEMKLKR VLHIDDEDIGSG | 01 GHASLEIFTSKRSNPVSKSEKYITVVGTKGRSDAIQLAAPGVHFRAGNKDVFQVNLTGIGKPT</span>KVILENTGTKRTDGWCVSKVVLVKKTDKGTRRYRFSGPVWL<span style="color: #996633;">SKHHDEMKLKR VLHIDDEDIGSG | ||
02 NG</span>YRIDCYTGDVANAGTDAVATIQLFGSKGQSPMVELRRSDGQAFQRARVATFTLD<span style="color: #996633;">DLANLGKLKKLVISHNGHGMASGWFLDK</span>IIVTSLSSNKATVFPCDAWLDRKNGRSK<span style="color: #996633;">ELVAR TAAAGAEGM | 02 NG</span>YRIDCYTGDVANAGTDAVATIQLFGSKGQSPMVELRRSDGQAFQRARVATFTLD<span style="color: #996633;">DLANLGKLKKLVISHNGHGMASGWFLDK</span>IIVTSLSSNKATVFPCDAWLDRKNGRSK<span style="color: #996633;">ELVAR TAAAGAEGM | ||
Revision as of 22:15, 12 November 2009
Introduction
LOXHD1, a large mis-annotated gene on chromosome 18 consisting solely of concatenated 120-residue PLAT domains (also seen in polycystin/lipoxygenase/alpha-toxin proteins), recently surfaced as a causative gene for presbycusis or age-progressive postlingual non-syndromic hearing loss type DFNB77. MYO3 (myosin IIIA) and DFNB59 (protein product pejvakin -- echo in Persian) are the only other genes to date with this autosomal recessive progressive phenotype.
There is no evidence suggesting that the three encoded proteins physically interact within auditory hair cells or could cause digenic disease (as perhapss seen in the Usher Syndrome complex). Perhaps with larger numbers of pedigrees, these conditions might exhibit syndromic effects (eg on vision or kidney function as well). LOXDH1 is specific to hair cell stereocilia within the cochlea but is also expressed in testis and placenta and other cell types (not including retina or RPE to date).
Effects are not seen during development but rather in mature stereocilia which degenerate over time. LOXDH1 binds to the inner plasma membrane of stereocilia, perhaps organizing protein binding there. As with other concatenated domain proteins such as CDC23 and USH2A, subtle dysfunction in one of many internally repeated domains evidently cannot be compensated for by the others. Here the mouse I1342N disrupts the hydrophobic pocket between two beta sheets of the tenth PLAT domain but the protein is still made and localized properly. The first morphological change is observed at day 21: fused stereocilia and membrane ruffling at the apical hair cell surface in the basal cochlear turn. Secondary effects arise later and include damage to spiral ganglion neurons.
The one known human allele R670* would eliminate all but the first five PLAT domains (of 16 total), possibly producing a normal but shortened protein in contrast to mouse, but might not produce protein at all due to nonsense-mediated decay. These individuals do not experience tinnitus, balance problems or vertigo, suggesting normal vestibular function even though LOXHD1 is expressed there too.
One difference between cochlear and vestibular hair cells is retention of the apical kinocilium in the latter. This is lost in developing cochlear hair cells as stereocilia mature. This correlates with peak LOXHD1 expression, suggesting to Grillet et al that LOXDHY might help stabilize the stereociliary bundle in the absence of kinocilium.
The kinocilium is a non-motile cilium-like structure present in the crista ampullaris of the semicircular ducts and the sensory maculae of the utricle and saccule but not in the cochlear duct organ of Corti (where they degenerate following maturation of the stereociliary bundle).
Since retinal receptor cells are modified cilia and LOXDH1 acts in hair cells after the kinocilium is gone, a syndromic effect is not expected based on kinocilium/photoreceptor homologization.
Phylogenetic occurence
The PLAT domain is quite ancient, readily tracing back to prokaryotes. However in other proteins it occurs only as single copy, often in conjunction with one or more catalytic domains. A common denominator of PLAT domain proteins is their association with lipid or membrane metabolism. This is perhaps mediated by extruded tryptophans which extend out from the standard beta barrel surface.
LOXHD1 itself has highly conserved orthologs throughout vertebrates, indeed deuterostomes (though the Branchiostoma gene model XM_002606291 contains other domains). These species all contain mechanosensory cells with ciliary F-actin based protrusions. No counterparts of the gene can be found in Ecdysozoa (strictly non-cilary mechanosensation) yet lophotrochozoa (Schistosoma, and perhaps Aplysia) can have full length counterparts, as can cnidrians (Hydra, Nematostella) though gene models here seem fragmentary.
This establishes gene loss in the ancestral ecdysozoan, rather than gene innovation in Bilatera. However the overall impression is phylogenetically spotty occurence outside of deuterostomes. It's unclear whether some other gene product can compensate for lost LOXHD1 or whether it does not play a central role.
Most surprisingly, full length orthologs of LOXHD1 with 16 PLAT domains and very similar intronation patterns are readily recoverable from early eukaryote genomes, notably Trichoplax and the choanflagellate Monosiga. This seems to rule out an obligatory association with F-actin-based mechanosensory organelles as the basal metazoan Trichoplax exhibits only 4 distinct cell types and the early animal Monosiga is unicellular. This rules out LOXDH1 as a metazoan innovation. Lateral gene transfer makes no sense in this context and is contradicted by the gene tree. The extraordinary conservation Monosiga to human -- surely in the 95th percentile of all genes -- suggests conservation of some fundamental intracellular role.
In the plant world, Chlamydomonas and Micromonas have proteins (XM_001703369, XM_002502455) with 20 PLAT domains and numerous WD40 domains (ie are not necessarily orthologous to animal pure poly-PLAT proteins). A curious note on the genbank entry states "identified by comparative genomics as being present only in organisms having motile cilia."
Within deuterostomes, the large length of the gene does not sit well with gappy, incomplete and sometimes chaotic assemblies. Chicken, lamprey, amphioxus, tunicate and sea urchin are among the many that do not yield satisfactory full length gene models, in part because transcript support is spotty. However these species do contain long poly-PLAT proteins no doubt orthologous to human LOXHD1. Some of these unreliable sequences are provided in the reference sequence compilation below. Ironically, the obscure Monosiga and Trichoplax have better assemblies and more accurate gene models presumably because their genomes are much smaller.
Thus within vertebrates it is only feasible to compare selected PLAT motifs that fall with an exon or two. Here there is a risk of using best-blast because many cross-ove matches will occur to other PLAT domains within the same protein, so some additional signature must be used.
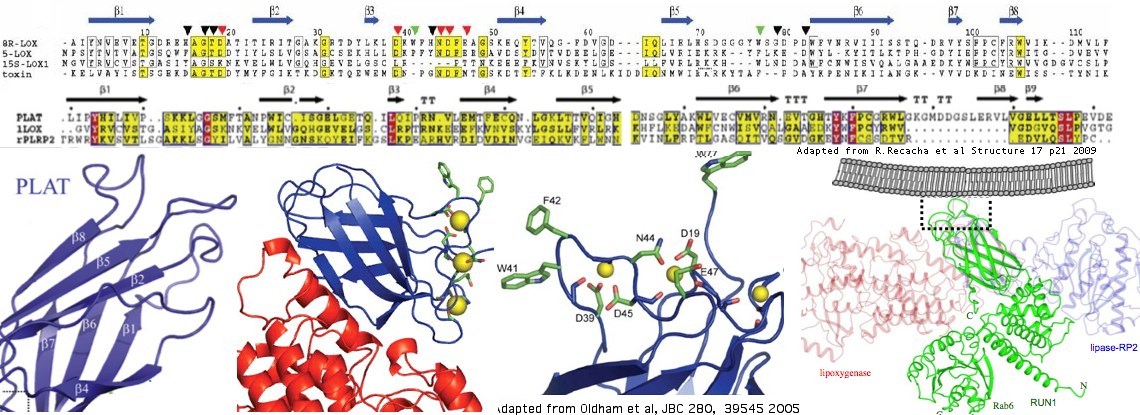
PLAT domain structure
The structure of three proteins containing single PLAT domains have been determined. The best blastp of human LOXHD1 to these is a coral lipoxygenase/allene oxidase natural fusion protein with PDB accession 3DY5. Studies of that protein have called the PLAT region a C2-like domain in conflict with SMART/Pfam/InterPro. Lower quality but still useful matches occur with RAB6 (accession 3CWZ) and rabbit reticulocyte lipoxygenase (accession 2P0MA).
The basic structure of a PLAT domain consists of a sandwich of two 2-stranded beta sheets (beta barrel). The lipoxygenases are calcium-binding via extended pairs of acid residues and have conserved hydrophobic residues likely inserting into the plasma membrane but neither these nor the hydrophobic residues are conserved in LOXHD1.
The independently folding PLAT domain is sometimes denoted as LH2 or 'C2-like' depending on the structural publication and domain tool (eg Pfam or SMART). The secondary structure is so generic that convergent evolution (non-homologous origins) must be considered, as with TIM proteins. Here however significant blastp matches at the primary sequence level (that unify PLAT and LH2 sequences to the exclusion of all C2) cannot have originated repeatedly by chance given the vast combinatorial space available to amino acid sequences. Further, all PLAT/LH2 domains share characteristic conserved residues. Thus PLAT and LH2 domains are in fact all homologous (descended from a single source) and so multiplicity of domain names is inappropriate. PLAT is used here because it is an acronym of representative protein classes containing the domain.
C2 domains bear an uncertain relationship to PLAT domains because a resemblance occurs only at the secondary and tertiary structural levels. The lack of convincing blastp of HMM profile matches could be due either to extreme divergence of primary sequence or to convergent evolution resulting in the same basic fold (and perhaps a similarly convergent hydrophobic intermediating function). The name implies calcium binding yet evem in this class some domains function independently of calcium. It's not clear whether calcium binding is ancestral or derived. If ancestral, it's not been resolved whether binding has been lost once or multiple times in different wings of the domain tree. PLAT domains can be structurally aligned to C2 domains but that is unnecessary and potentially misleading given three determined structures for bona fide PLAT domains.
LOXHD1, which tracks back to the earliest unicellular animals, lacks capacity for calcium binding in all 16 of its domains in all extant descendant clades based on non-conservation of acidic liganding residues relative to coral lipoxygenase, human LOX5 and Clostridium alpha-toxin PLAT domains.
The 20 other human proteins with significant blastp matching to the PLAT domain of LOXHD1 are shown below along with flanking domain context. After adjusting for gene expansions (paralogy), only 4 classes occur: polycystins, lipoxygenases, lipases, and RAB6-interacting. These all contain a single PLAT domain. The final column shows its best-blastp to LOXHD1; no strong pattern emerges though perhaps PLAT:09 may conserve the most ancestral features and PLAT:15 the least. The first two columns at bottom shows the blastp score of a given human LOXHD1 PLAT domain averaged over it match to the 12 other genes; the second two columns show the average match of other genes over the 16 LOXHD1 PLAT domains.
The domain tree thus looks like ((((LOXHD1,RAB6s),polycystins),lipoxygenases),lipases). One evolutionary scenario envisions a single standalone PLAT domain that served as an auxillary protein to various enzymes with hydrophobic substrates. Later copies fused with these in separate events under selection for efficiency. The standalone gene subsequently expanded to a poly-PLAT protein; today the standalone domain itself is gone (no longer represented in extant species).
PKD1L1 GPS-TM-LH2-TM chr07:47780815 polycystin 1-like 1 pdb:3CWZ PLAT:14 36% PKD1 GPS-TM-LH2-TM chr16: 2078712 polycystin 1 pdb:3CWZ PLAT:01 38% PKD1L2 GPS-TM-LH2-TM chr16:79691985 polycystin 1-like 2 signal pdb:3CWZ PLAT:11 35% PKD1L3 GPS-TM-LH2-TM chr16:70556160 polycystic kidney disease pdb:3CWZ PLAT:01 40% PKDREJ GPS-TM-LH2-TM chr22:45030224 receptor egg jelly signal pdb:3CWZ PLAT:11 32% DENND5A RUN-LH2-RUN chr11: 9116949 RAB6 interacting 1 pdb:3CWZ PLAT:09 42% DENND5B RUN-LH2-RUN chr12:31426424 RAB6 interacting 2 pdb:3CWZ PLAT:09 37% ALOX5 NH2-LH2-lipox chr10:45189635 arachidonate 5-lipoxygenase pdb:2P0M PLAT:01 37% ALOX15 NH2-LH2-lipox chr17: 4480963 arachidonate 15-lipoxygenase pdb:2P0M PLAT:03 26% ALOX12 NH2-LH2-lipox chr17: 6840108 arachidonate 12-lipoxygenase pdb:2P0M PLAT:09 30% ALOX15B NH2-LH2-lipox chr17: 7888129 arachidonate 15-lipoxygenase pdb:2P0M PLAT:02 34% ALOX12B NH2-LH2-lipox chr17: 7916679 arachidonate 12R-lipoxygenase pdb:2P0M PLAT:09 29% ALOXE3 NH2-LH2-lipox chr17: 7939943 arachidonate lipoxygenase 3 pdb:2P0M PLAT:16 32% LIPC LIP-PLAT chr15:56511467 lipase C pdb:2P0M PLAT:11 30% LIPG LIP-PLAT chr18:45342425 endothelial lipase pdb:2P0M PLAT:09 32% LPL LIP-PLAT chr08:19840862 lipoprotein lipase pdb:2P0M PLAT:11 24% PNLIP LIP-PLAT chr10:118295418 pancreatic lipase pdb:2P0M PLAT:16 24% PNLIPRP1 LIP-PLAT chr10:118340480 pancreatic lipase-related 1 pdb:2P0M PLAT:16 27% PNLIPRP2 LIP-PLAT chr10:118370455 pancreatic lipase-related 2 pdb:2P0M PLAT:01 31% PNLIPRP3 LIP-PLAT chr10:118177414 pancreatic lipase-related 3 pdb:2P0M PLAT:02 27%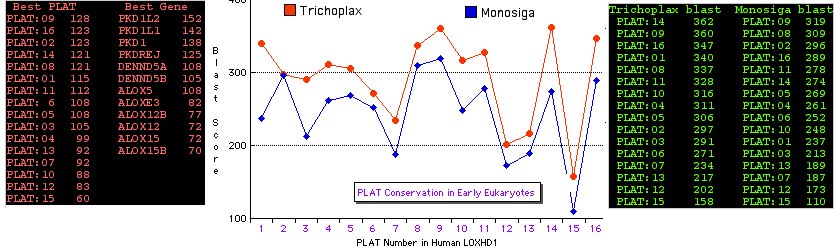
Below all PLAT domains found in human proteins are aligned with the 16 domains of human LOXHD1. The coral lipoxygenase is included because it provides the template for beta strand assignment and locating the problematic calcium binding and hydrophobic extended residues. The lower alignment is just with human LOXHD1 domains and uses an optimized seventh PLAT domain (the KE expansion is removed).
Individual PLAT domains can be quite conserved. That is illustrated by a difference alignment of the fifth PLAT domain in 40 vertebrates (human to lamprey) below. Note columns where human sequence represents a unique change -- such as H->R in column 5 -- may be minor alleles or disease variants rather than the predominate residue in the overall human population.
01.hg18_6 MARYHVTVCTGELEGAGTDANVYLCLFGDVGDTGERLLYNCRNNTDLFEKGNADEFTIESVTMRNVRRVRIRHDGKGSGSGWYLDRVLVREEGQPESDNVEFPCLRWLDKDKDDGQLVRELLPSDSSATLK 02.panTro ....R.............................................................................................................................. 03.gorGor S..R...........................................................................--------------------------..................G...... 04.ponAbe ....R..............T.....................................................................................-....................N.... 05.rheMac ....R.........................................................................................................................N.... 06.calJac ....R...........................................................K.............C...............................................N.... 07.micMur ....R...........................................................M.............................................................N.... 08.otoGar ....R. ............K.................-..........H.P.V.......T....................N.... 09.tupBel ...R...........................................................K............A................................................N.... 10.mm9_6_ ....R...........................................................K.....V.......................................................N.... 11.rn4_6_ ....R...........................................................K.....V........................................Q................... 12.dipOrd ....R.....................Y..................E..................K.....V.......S.S.............................................N.... 13.cavPor ....R.......F.............Y...................................L.K.............................................................N.... 14.speTri .V..R.....................Y......-.....-------------............K............P......E...L.....................................N.... 15.oryCun ..........S.............K..........................................................TG.N.... 16.ochPri ....R...........................................................K.............S....V..........................................N.... 17.vicPac ...R.......V.............................I.....................K.................F.E.........................................N.... 18.turTru ....R...........................................V...............K.....V...........F.......................---------...........N.... 19.bosTau ....R.....................Y.....................................K.................F.E....................-....................N.... 20.equCab ....R...........................................................K............A....F.E.........................................N.... 21.felCat ....R...........................................................K............G....F.E.........................................N.... 22.canFam ....R...........................................................K............G....F.E.........................................N.... 23.myoLuc ...HR......D.R......S.....Y............T.K..DEM.Q...............K........T.V.G....F..................Y........................N.... 24.pteVam ...R.........................................M.................K..........R.G....F.E.........................................N.... 25.eriEur ....R.....................Y.....................................K............G....F.A..............M..........................N.... 26.loxAfr ..Y.......-..M.....................A..L.K............G......E.........................................N.... 27.proCap ....R.....................Y..A.......M...........R.........A..L.K............A......E..............M...---...................NN.... 28.echTel ....R.....................Y..........MF.....V..............A..L.K......M.....A......-...LKK.........K..G.G....................N...R 29.choHof .V..R.....................Y.........W.........M....S...Y...A..L.K.K.I.VG.....GN...F.E..............M....N...AR.Q.....I......G.N.... 30.monDom ..Q.R..L...DI.....N.QAFV.....A......M.......MEICQ..........A..V.K......G.....K....F.VK..................N..F...Q..............NSR.. 31.ornAna IVK.R...V..D.N......R.FI..I.........I.........T.........FV.A..LKQ......G.....GS.....AK.I.........EA.....Y.....NE....I....V.AGE.PL.. 32.galGal VIK.R......MVS.S......FV..I..Q....D.V..K.I..VNK.........F..A..LKQ......G.....GS.....AK.I.........EA.....Y.....NE....I....V.AGE.PL.. 33.anoCar IK.R...H..NVS.S....H.FA..I..Q....D.V.Q.SV.TVNK....S....I..A.NLKQ......G.....GS.....GK.I...D.....EAQ....Y.....NE....II...V.AGR.SL.E 34.xenTro ...F.IE.K..MNK.......D.I..A..LKQ.K....G.....G.C...M.K.V...D.K..TEAI....N..F.RNE...HII...V.AGD.QN.R 35.tetNig LK.RI.I...NVG.G....S.F.NII..L......SMITSK..VNK.....H...L....CLGR.S...VG...R.G.C..F..K.T.........SAI....F....HNE....I....V.AGDGRLF 36.fr2_6_ LIK.R..I...NVS.G....S.F.NVI..L.......MIMSK..VNK.....H...L....SLGQ.....VG...R.G.C..F..K.M.........SAI....F....HNE....I....V.A 37.gasAcu LIK.R..I...NVS.S....S.F.NVI..L.......MFMSK..VNK.....H...L....SLGQ.....VG...R.G.C..F..K.M...D.....LAT....F....RNE....I....V.AGDGRLF 38.oryLat IK.R..I...TVS.S....S.F.NVI..L.......M.MSK..VNK.....H...L....WLGQ.....VG...R.G.C..F..K.M.........AAI....N....QNE....II...V.AGEG 39.danRer VK.R...Y..DVS.S....H.F...I..L....D.S.I..K..INK..R......I..A.SIGP......G...R.G.C.....K.TI.....A..QA.....S..F.RNE....I....Q.FADGRLY 40.petMar VK.L..IH..KHG.G......FV..Y.EQ....D.M..TS...VNK..R..V...V..C..LKR.T....G...R.GS...F..KIM.K-D.RRQCEE.....N....D.E....I....VAGGCQMLK Consensus ...yrvt..tg...g.gtda.v..cl.gd.gdtger.ly.crnn...fekgna...t..s.t$rk.r...!r...k.ggs..y$d.!lvre#.qpes.nv...c.r.l.k#k....l!...l.sgsnatlk
Expansion of the PLAT:07 domain
The seventh PLAT domain (human numbering) is unique domains in having a large compositionally simple expansion in the middle of the domain. It is a unique signature of the LOXHD1 orthology class that would distinguish it from other poly-PLAT domain concatenates of independent origin. That issue is completely hypothetical because no other protein in any vertebrate genome contains more than a single PLAT domain. Indeed, the LOXHD1 gene is single-copy in all species that retain it (including teleost fish). Thus even with very ancient eukaryotic divergencs, the corresponding genes can be taken as straightforward human orthologs.
Presumably a loop out of the beta barrel, the expansion consists of an extremely high AG content at the dna level and encodes charged residues, primarily lysine K and glutamate E residues. These are asymmetrically distributed with the lysines N-terminal and the glutamates distal. The region will be called the KE loop of PLAT:07 here.
Despite minimal net charge, this region may not be able to interact with (hydrophobic) membranes. Yet the split PLAT domain retains the potential to form a standard beta barrel. The seventh PLAT domain is quite variable even within mammals and must be hand-annotated to ensure accuracy (because its quasi-repetitive nature wreaks havoc with alignment). This section uses comparative genomics within eukaryotes to answer the following questions:
- when did the high GA section arise in phylogenetic time? [early eukaryotes]
- what is its source? [internal expansion, replication slippage]
- did it arise from intron or retroposon retention? [species lacking the repeat also have a single exon]
- can the boundaries of 7th PLAT domain be better located? [conservation on both sides of the KE region allow definition of the PLAT domain segments]
- is the 44-species alignment accurate in this region? [corrections are needed when longer than human, eg platypus]
- does the KE domain disrupts PLAT function? [evidently not: the split domain is highly conserved and can form a normal beta barrel]
- is it a mammalian innovation or an unwanted intrusion? [this ancient expansion has variable expansion and contraction in different clades]
homSap LEAADVGEVYKLRLGHTGEGFGPSWFVDTVWLRHLVVREVDLTPEEEARKKKEKDKLRQLLKKERLKAKLQRKKKKRKGSDEEDEGEEEESSSSEESSSEEEEMEEEEEEEE FGPGMQEVIEQHKFEAHRWLARGKEDNELVVELVPAGKPGPE 1 ornAna LEATDVGEIYKVRLGHSGEGFGSGWFIESLVLKRLVLKEVEPNPEEEKRKAKERERAREQRRKERLKAKQQRKKKKKMKKSSDDEDSEAEDSEEEEGSSEEESSSSSSEEEVEEEFGPGIKEVIDVYKFEAHRWLARDEDDKELIVELEPANRPGPE 1 galGal VEAADVGKIYKIRIGHDGKGIGDGWFLESVTLKRLATKMDETD KKKKKKKKKSEEEEEEEE TKVEEVMDVYTFVAHRWLAKDEGDKELVVELVPDGESELE 1 taeGut REAADVGKIYKIRIGHDGTGIGDGWFLESVTLKRLATKTEGSD KKKKKKKKSEEEETKE EEGMDVYTFVAHRWLAKDEGDKELVVELVPDGESDLE 1 anoCar VEAVDVGKVYKIRIGHDGKGFGDGWFLDSVVVKKLPTKVP KKKKKKKKKKTPEEEEAEE GPGIMEVYNFTPCRWLASDEEDKELVVELVPDEGSELE 1 oryLat IEALDVGKIYKIRIYHDGSGIGDGWFLETVDIKRLTMALVQVEV KKEEAPKKDKKKDKKKKKKEEEEVEIIEE MQEVVETFTFTCNRWLARDEEDGEIVVELLTEENEDLE 1 gasAcu IEAKDVGKIFKIRIGHDGSGIGSGWFLETVDVKRLILALVPKEK KKEDKKKKKKKKEDVDEEGGEE MQEVVLTYSFPCSRWLAGGEEDGELVVELLPDDAKELE 1 takRub IEAKDVGKIFKIRIGHDGLGIGSGWFLEKVYVKHLIMALVPREN KKDDKKKKKKKKKDKEDEEEVGGEE MQEVVVTYHFPCSRWLASGEDDDDLVVELLPEDAEELE 1 petMar VEAMDVGKVVKLRVGHDNSGMGSGWFLDSIVIRRLRQSSPHRPQPVDA EEDEDEEDDEEAEDED VQTYTFPCKRWLARDEDDGEIVRELLPQDCAEME 1 sacKow IEAADVAMLTKIRIGHDNSGRSAGWYLERVIIERFPPKRKM KRKRSGTPRRRGEYDEEDYDD IPETNVVNFVCNRWFAKDEEDHQIVRELLPTDEEALKGH 1 triAdh VECEDVGKLRKLRIGHDSAGMGSAWFLDKVYVRRLPPKSGKKSKET DEREEETAKKDADEPEKLDE NNYLFVANRWLSKEEGDRQTVIEISPVGVDGALA 1 monBre LACKPVGRPSKIRLSAHGGGMSADWHLEKIEVHELGQAR IYTFEHNDWLRKGTKAKPFMVELPLRRIETVDDN 1
Introns relative to PLAT domains
Here it emerges that PLAT domains correspond to 2-3 consecutive exons with conserved phasing, allowing for potential alternative splicing that reduced domain count while not introducing premature stop codons. This may reflect the origin of the 16 PLAT domains by iterated inhomogenous recombination. (to be continued).
Reference sequences (intronated)
The sequences below are broken into individual exons with reading phase (codon overhang) indicated by 012. Alternating colors show odd and even numbered PLAT domains. The 7th domain (which contains the charged KE expansion loop in mammals) is shown in red. In some cases, the PLAT domains are numbered and exon phases are not indicated. In the case of poor quality but phylogenetically important sequences, an approximate sequence is shown.
>LOXHD1_homSap_full 41 exons|2273 aa|chr18|span 179,559 bp|no signal peptide|no conserved cysteines|16 PLAT domains|EIW --> WNW mouse deafness 0 MMPQKKRRRKKDIDFLALYEAELLNYASEDDEGELEHEYYKAR 1 2 VYEVVTATGDVRGAGTDANVFITLFGENGLSPKLQLTSK 2 1 SKSAFEKGNVDVFRVRTNNVGLIYKVR 2 1 IEHDNTGLNASWYLDHVIVTDMKRPHLRYYFNCNNWLSKVEGDRQWCRDLLASFNPMDMPR 1 2 GNKYEVKVYTGDVIGAGTDADVFINIFGEYGDT 1 2 GERRLENEKDNFEKGAEDRFILDAPDLGQLMKINVGHNNKGGSAGWFLSQ 0 0 IVIEDIGNKRKYDFPLNRWLALDEDDGKIQRDILVGGAETT 1 2 AITYIVTVFTGDVRGAGTKSKIYLVMYGARGNKNSGKIFLEGGVFDRGRTDIFHIELAVLLSPLSRVSVGHGNVGVNRGWFCEK 0 0 VVILCPFTGIQQTFPCSNWLDEKKADGLIERQLYEMVSLRKKRLK 1 2 KFPWSLWVWTTDLKKAGTNSPIFIQIYGQKGRTDEILLNPNNKWFKPGIIEKFR 0 0 IELPDLGRFYKIRVWHDKRSSGSGWHLER 0 0 MTLMNTLNKDKYNFNCNRWLDANEDDNEIVREMTAEGPTVRRIMG 1 2 MARYHVTVCTGELEGAGTDANVYLCLFGDVGDTGERLLYNCRNNTDLFEKGN 0 0 ADEFTIESVTMRNVRRVRIRHDGKGSGSGWYLDRVLVREEGQPESDNVEFPCLR 2 1 WLDKDKDDGQLVRELLPSDSSATLK 1 2 NFRYHISLKTGDVSGASTDSRVYIKLYGDKSDTIKQVLLVSDNNLKDYFERGRVDEFTLETLNIGN 0 0 INRLVIGHDSTGMHASWFLGSVQIRVPRQGKQYTFPANRWLDKNQADGRLEVELYPSEVVEIQK 1 2 LVHYEVEIWTGDVGGAGTSARVYMQIYGEKGKTEVLFLSSRSKVFERASKDTFQ 0 0 LEAADVGEVYKLRLGHTGEGFGPSWFVDTVWLRHLVVREVDLTPEEEARKKKEKDKLRQLLKKERLKAKLQRKKKKRKGSDEEDEGEEEESSSSEESSSEEEEMEEEEEEEEFGPGMQEVIEQHKFEAHRWLARGKEDNELVVELVPAGKPGPE 1 2 RNTYEVQVVTGNVPKAGTDANVYLTIYGEEYGDTGERPLKKSDKSNKFEQGQ 0 0 TDTFTIYAIDLGALTKIRIRHDNTGNRAGWFLDRIDITDMNNEIT 2 1 YYFPCQRWLAVEEDDGQLSRELLPVDESYVLPQSEEGRGGGDNNPLDNLALEQK 1 2 DKSTTFSVTIKTGVKKNAGTDANVFITLFGTQDDT 1 2 GMTLLKSSKTNSDKFERDSIEIFTVETLDLGDLWKVRLGHDNT 1 2 GKAPGWFVDWVEVDAPSLGKCMTFPCGRWLAKNEDDGSIIRDLFHAELQTRLYTP 1 2 FVPYEITLYTSDVFAAGTDANIFIIIYGCDAVCTQQKYLCTNKREQKQFFERKSASRFIVE 0 0 LEDVGEIIEKIRIGHNNTGMNPGWHCSHVDIRRLLPDKD 0 0 GAETLTFPCDRWLATSEDDKKTIRELVPYDIFTEKYMKDGSLRQVYKEVEEPLD 1 2 IVLYSVQIFTGNIPGAGTDAKVYITIYGDLGDTGERYLGKSENRTNKFERGT 0 0 ADTFIIEAADLGVIYKIKLRHDNSKWCADWYVEKVEIWNDTNEDEFLFLCGRWLSLKKEDGRLERLFYEK 0 0 EYTGDRSSNCSSPADFWEIALSSKMADVDISTVTGPMADYVQEGP 1 2 IIPYYVSVTTGKHKDAATDSRAFIFLIGEDDERSKRIWLDYPRGKRGFSRGSVEEFYVAGLDVGIIKKIE 0 0 LGHDGASPESCWLVEELCLAVPTQGTKYMLNCNCWLAKDRGDGITSRVFDLLDAMVVNIGVK 0 0 VLYEMTVWTGDVVGGGTDSNIFMTLYGINGSTEEMQLDKKKAR 2 1 FEREQNDTFIMEILDIAPFTKMRIRIDGLGSRPEWFLER 0 0 ILLKNMNTGDLTMFYYGDWLSQRKGKKTLVCEMCAVIDEEEMMEWTSYTVAVKTSDIL 1 2 GAGTDANVFIIIFGENGDSGTLALKQSANWNKFERNNTDTFNFPDMLSLGHLCKLRVWHDNK 1 2 GIFPGWHLSYVDVKDNSRDETFHFQCDCWLSKSEGDGQTVRDFACANNKICDELEET 1 2 TYEIVIETGNGGETRENVWLILEGRKNRSKEFLMENSSRQRAFRK 2 1 GTTDTFEFDSIYLGDIASLCVGHLAREDRFIPKRELAWHVKTITITEMEYGNV 2 1 YFFNCDCLIPLKRKRKYFKVFEVTKTTESFASKVQSLVPVKYEVIVTTGYEPGAGTDANVFVTIFGANGDTGKRELKQKMRNLFERGSTDRFFLETLELGELRKVRLEHDSSGYCSGWLVEKVEVTNTSTGVATIFNCGRWLDKKRGDGLTWRDLFPSV* 0 >Gallus gallus gappy genome,flawed XM_425221 15 PLAT domains MKETKKNKAEEEEEEEEEEEEENEEVVEDVVEEAVEEDGEDESQQKKSKKKSKSEDSEDDGEVKKKKKKKKKTKRKEYSSEEEDDYERRKKKKKKGKKSK EKKKKKGDKSKKGKKEDNFLELYEQELRDYHSDSSNATEDEYNKKKVYEVVTVTGDVRGAGTDANVFVTLFGEFGITPKTHLTSKSSTAFERSKTDVFRV KTNNVGQIKKIRIEHDNTGLNAGWFLDRVIVTDMNRPHLRFYFPCNNWLSKEDGDGLYVRDLIGSLNPMDVPKINKYVVRVFTGEVSGSGTDADVFINIF GEKGDTGVRKLDNDKDNFEKGAEDKFTLDAPNLGRLRKINIGHNNKGGSAGWFLAKVIIEDIGNKCVYQFPVGRWFALDEDDGKIQRDILVGGTEATGIV YNVAVVTGDIRGAGTNSKIHVILHGSKGLKNSGTIFLEGGEFERARTDLFNVEIASLLSPLSRVTIGHDNCGVSSGWYCEKVVVYCPFTGIEQTFPCGKW LDEDEGDGLIERELYEMVSLRQRRLKKNPWSLWIWTSDIKNAGTDATIFFQIYGDKGKSDEMKLDNNSDNFEAGQTDKFMIELPDLGTFYKLRIWHEKRN PFAGWHLDKVTLLKTLTKDKYSFNCGRWLDINEDDNEIVRELPAEGSLVTEVMPVIKYRVTVCTGMVSGSGTDANVFVCLIGDQGDTGDRVLYKCINNVN KFEKGNADEFFVEAVTLKQVRRVRIGHDGKGGSSGWYLAKVIVREEGQPESEAVEFPCYRWLDKNEDDGQIVRELVPAGESPLLKNVSYHISVKTGDIPG ASSDSKVFIKLYGEKADTSKETLLVSDNDLGNYFERGRVDEFTIDTMDIGKINRILIGHDNVGLRSGWFLASVQITVPVQGRQYMFPCNRWLDKDEADGR VEVEVYPSEILPIEKLINYEVSVVTGDVRAAGTNAKVFMQIYGETGKTELIILENRSNNFERGATDIFEVEAADVGKIYKIRIGHDGKGIGDGWFLESVT LKRLATKMDETDKKKKKKKKKSEEEEEEEETKVEEVMDVYTFVAHRWLAKDEGDKELVVELVPDGESELEENTYEVHVLTGSVWGSGTDANVFLSIYGIE RGDTGERQLKRSNNLNKFEKGQVDVFTIKAIDLGELKKLRIRHDNSGSSPSWFLERVEIVDLKESTTYYFPCQRWLAVEEDDGQIVRELVPVDEAFVKKD SENDGQSLATLGLEQKAKSTTYIVKVKTGDKKNAGTDANVFITLYGSKDDTGIVSLKASKLNKNKFERGKIDEFTVESVDIGDLKKIKIGHDNAGNSNGW FLEWVEIDAPSLGQCLKFPCGRWLDKSEDDGAIERFIFPAELQTTEYIPFVPYEITVYTSDIFGAGTDADVFIVLYGSDGICTQQKSLCLNKREQRMYFE RNSVNQFIVELEDVGDIIEKIRIGHNGGGLNSGWHLDRVAIRRLLPNGKGSETITFPCERWLAKSEDDGEIIRELVPSDIFTEKLMKDGTLKQIEEEVED PLEVHTYKISVFTGDIYGAGTDANVFLNIYGDLGDTGERKLSKSETNFNKFERGQSYDGERRSLSESSVSLSSLDSSGNPKEKKSRLVKSAEEGLLIPYH ITVTTGTEYDSSTDSRVFIIIMGPQKVRTERLWLDLPEGKDEFADGSVEKFSVWGLDVGEIKKVEVGHDGATPESCWLMEELTIVVPTKGVMYNFVCKCW LARDKGDGLTSRILNILDADCVNVGIKILYEVTVVTGDIESGGTDAGIFMTVFGSNGNTEEMQLDKNGDRFERGQEDSFIMEIADIAPLRKMRIRTDAKG TRPDWFLERIVMRNLTNQEVATFTYGDWLSKVKNAKGSLVCEMPAMVNDEQMMEDTTYTIQVKTSDIGGAGTDANVSLILFGENGDSGTLALKESNKSNK FERNQMDEFNFPNMLSLGDLCKVRIWHDNKAYEIVTVTSNREDAETKENIWIILEGKLGRSKEFLMENSSKKRRFERRGSTDTFQFSSKNLGDIAAICVG HCPKDGKKSSAKADVYWHVKEIIITEMELCNKYFFRCNGKIPLRYKRRDYKVFECAKVIESFASKARSLVPVKYETIVVTGFEKGAGTDANVFITIFGLN GDSGKRALKQKFRNLFERGKTNRFYLETLDMGELKKVRIEHDNSGLAPGWLVERVEITNSATGVTTIFPCGKWLDENRGDGLTWRELFPRY* >LOXHD1_cioSav Ciona savigny approx gene model 15 PLAT domains 0 EVIVEDTKYDRTYTFPCDRWLSLYKEDCQLTRHLMAVSGRQS 1 KRTTFEITTVTGNGEKRGAGTDANVFLTMRGSKGVSPKLQLKSEYCSRRNTFERGQSDVFRVKSVNVGNLKRIRMEIDDTGFAPAWFLELQVVVVDLANPSQRIYFPCGQWLSDDEQLFRDL VGSSNPLAR 2 PKSVNAYVVHTFTGDVRGAGTDATVKITLFGDHGDSGQLVLDDSKNNFERARKDTFSLQCPHLGKIKKIRIGKGHDNGGVSPGWYLDKVVVDDTMMDCVYSFPCQRWFAK 3 DEDDGRITRELVAGVGEAGIPYQVQVFTGRFTSSCMGGRRVKSVQGKSILDGKFQRNRVDICDIESTTMLSPLDHIEIGHDNSGTGPGWYLEKVVITCPTNGCEQVFSCRKWLATDEGDGLISR QLYESKSLRK 4 QVVQSAWNCLIWTSDVRNAGTDANVFIQVYGENGKSDELPLDNETDNFETGQKDKFKINLPEIGCIYKLRVWHDDSNPFSGWHLDKIELEPIGGKKGDLYTFKCGKWLDTNEGDGEVIREIPST GPGVKR 5 PQPLVRYSVSVVTGDKRGAGTDANVSCCLYGRQGDTGNRPLTASKNNRNKFERGQLHLMLIIQTQHHRWLATDEDDGAIVRELTSEGSP 6 QLLNTSYHVAVKTGDIRGAGTDADVYIQIYGSDGDTGRLKLTRGENIRNKFERGRTDKFKLEATDIGRLERIYIGHSDSGIGAGWFLDGIEIDVPSSGMHYTFKANRWLASDEGDGKTEIELYPT QTR 7 QQDKLPYEITVATGDTLNAGTDAQVVLQLYGEDGKSEVMKLRNKTDNFERKAIDKFKIEAPDIGPITKIRIGHDGRGVGSGWFLDSVKIQRFVHKTSKRKKRRRGQNQSEVETYWFVCKRWFDKSEDDGQ 8 IIRELEYTVDVYTGDKYGAGTDSNVFLTLYGENGDSGERKLDKSVNINKFERNQVDSFKLKAADLSTLKKLKIRHDNSEIGSSWFVDRVEVKVKGETWVFPCQRWLSTKEDDGQIERTLVPV DATYQRLQQQKQGRKASIAVRDAIALET 9 KAQLTTYEVSVKTGDVWGSGTDANVYLTLFGEKDDSGINLKTSKTYRDKFERNHIDVFTVESADLGVLKKIKIGHDNAGLSPGWYLEYVEVDAASLGMKWRFPCGRWIAKDKDDGKLEKELHLAT DETVG 10 YAAKVPYEITVYTSEISGAGTDADVYVVLYGREGIATEQTSLCSSKDERKSRFNKKGIDTFVLELDDIGEVIEKLRIGHDGKGWGAGWHLDKVEVRRLLEGVKGSETSTFTCGRWLARDEDDGEIVRELVPT KIVKEQVTKSGKLKRSESKLD 11 DALKQQYKVHVFTGDVFRGGTDANVFITIFGENGDTGERKLLKSETNTDKFERGKEDIFKLDAVDLGKLFKVHIRHDNALIQPAWFLDRVEIEDTDTGSIYVFHCERWLAKNKEDGKIARSLYVV GYDGETSTRRSLATTKSGGSLGKKSILSDTIP 12 EGPAIPYTVKIETGSEEGSSCKAPAFIKIFGPSKKGTVSRRIPLEPYSGKFKRGTIETFNIEAIDVGSIKKIEIGHSSVDEEWFVESVMVNQPTLGKSYTFKCHKWFARSKDDGLVSRIFTTD DAEKLS 13 FKAKIPYEITLHTGNVDNAGTDCTITLSVYGSKGNSPDITVKKSEEDGVLLERGSIDTINVELDEIAPLKMVRVGHDGRGQRADWFCDKKTLSRELSASVGGRS 14 VLKKTNYKINVKTSNLRSAGTNANVSCIVFGTFGETGELKLKVSETHKNKFERDAMDTFTFQDKLSVGEPQKLRVWHDSKGFSSGWHLDHIEVCDESTQQKYMFPCGRWLADDEDDKQTIREL TCSNLDQ 15 AKEGGQYTATVRTSDKRDAAPNSDGWVILHGKMGKSRRMKLREGKLGRGKASTFKLDSNDLGALQKITLGLEGDNMGRWHVDDVVIENPSGGQFKFDCHAWVSNEDGTNTRGSNFDC TATKGRTISMS 16 SLLPVKYEVTVVTADEKGSGTDSNVSLTIYGTNGDSGKRPLKQRFRDLFERKQTDKFTLEILDLGDLTKVQIRLEHDDAGFNADWLCDNVTIRNPVSGQSWTFICREWIGLKKANQLFREVLPR >LOXHD1_cioInt Ciona intestinalis XM_002122635 defective assembly; frag gene model 16 PLAT domains EVIVEDTKYDRTYTFPCDRWLSLYKEDCQLTRHLMAVSGRQSKRTTFEITTVTGNF GEKRGAGTDANVFITLRGSKGVSPKLQLKSD 21 RHNTFERGNSDVFRLKSANVGNLKRVRVEIDENGFAPAWFLERVVVVDMSKPSQRIFFPCGQWLSDDEQLFRDLVGSTDPLARPKM NAYVVHVFTGDMRGAGTDAEVFITLYGGKKGQLSSGKIVLDGKFQRNRVDICDVESASMLSPLDHIEIGHDNSGSGPGWFLDKVVVTCPTNGCEQVFSCRKWFATDEGDGLTSRELYESKSLRKQVMQK SAWNCLIWTSDVRNAGTDANVFIQVYGENGKSDEIALDNETDNFETGQKDKFKINIPEVGRMYKLRVWHDDSNPFSGWHLDKIVLEPVGGKKGSSYTFNCQKWLDTNEGDGQIIREIPATGPGVKRPQPL VRYQVTVVTGDKRNAGTDANVFCCLYGRQGDTGNRPLEASKSNRNKFERGQSDEFTIEAVDLKSVNKVKIGHDDSGSGWHLKEIIVRDETNGESTVFPCDRWLATDEDDGAIVRELTSEGSPQLLNT TSYHVSVKTGDMRGAGTDADVYIQIYGSDGDGRLKLHQGENIRNKFERGRTDRFKLEATDIGRIERIYIGHNDSGVGAGWFLDGVEIDVPSSGMHYTFKSNRWLASDEGDGKTEVELYPTQTRQQDKLL PYEITVTTGDIMNGGTDAQVVVQLYGEDGKSEVIRLRNKTDNFERKAVDKFKIEAPDIGPITKIRIGHDGRGVGSGWFLDSVKIQRFVRKTSKRKKRRRESMVSNDQNEVETYWFVCRRWFDKSEDDGQIIRELVPTDESGKPIDEALQE LEYSVDVYTGDKYGAGTDSNVFITLYGENGDSGERKLDKSDNINKFERNQVDGFKLKAADLSNLKKLKIRHDNSELGSSWFVDRVEVKVKGQTWVFPCQRWLSTNEDDGQIERTLVPVDVATYQRLQQKQGSGRKASVAVRDAIALETKAQL TTYEVSVKTGDIRGSGTDANVYLVLFGENDDSGMINLKTSKTHRDKFERNHTDVFTVESADLGNLKKVKIGHDNAGLSPGWYLEYVEVDAPSLGMKWRFPSGRWIAKDKDDGKLEKELFLAMDETVGYAAK VPYEITVYTSEVSGAGTDADVYVVLYGRDGMSTEKTSLCPTKDERKSKFNKKSVDTFVLELDDVGEVIEKLRIGHDGKGWGAGWHLDKVEIRRLLEGAKGSETYSFTCGRWLARDEDDGEIVRELVPTQLVQEKVTKS QQYKVHVYTGDVFNAGTDANVYITIYGENGDTGERKLLKSETNTDKFERGKEDIFKIDAVDLGKLFKVHIRHDNALLNPSWNLNRVEVEDTDTGNRYVFHCERWLAKGKDDGKIVHVYTGDVF IPYTVKIETGSDEGSSSTASAFIKIYGPSKKGKVSRRITLDPPSGKFKKGTVESFNIDAIDVGTIKKIEIGHTGVDDEWFIESVMINQPTLGKSFTFKCHKWLARSKDDGLTTRIFTIDDAEKLTYKAK IPYEVTIHTGNVPNAGTDCLITMSVYGTKGNAPDITIKKSEEEGILLERGSVDVFNVEMEEIAPLKMIRISHDGRGQRQDWFCDKIELRNLNSGKVTVFPCDSWLSKKLGEKTLSRELSASVGGRSVLKK TTYKLNVKTSNMRSAGTNANVSCIVFGTFGDTGELRLKESETHKNKFERDSLDVFTYQDKLSIGQLQKLRVWHDSKGFSSGWHLEYIEVVDESTQQKFMFPCNRWLADDEDDKQTIRELTCSNLDQATEGHTV GQYTATITTGDKRDAASSCDGWIILHGKMGKSRRLKIRSGKMGRGKKSEVKLDAGDVGKMEAITLGLHDMGRWYVEDVTIETGNGGQYKFDCHAWVSNEDGTNTRGRTFECSATKGRTVSMSSLIP IKYEVTVVTADEKGAGTDANVKLTIYGENGDSGSRALKQRFRDLFERKQTDKFTLEILDLGELTKIRLEHDDGGFNADWLCDHVIVRNPVIGQSWTFVCREWIGTKKAGQLFREVMPRSTG* >Schistosoma mansoni (flatworm) XP_002576380 2301aa Bilatera|Platyhelminthes blast 0 MNLEDSTNDDYLKNSKSRFSFHRKEEKEHKGIICCGHNTCHLDRYNLISSVKPFTARPYTLYKVVLTTADVPGAGTSAQ 1 2 IYITLKGEWGSSTRQKLRKEKVPTRNLRFYFYPGSTNTFSVVSPDLGGLHSVFIE 0 0 HDSLRKSDSWLLESVQVFHPLTKKRYMFMCNHWFSLYKEDGLIARELFGIRSAKT 1 2 KYSIVTVTGDQEGSGTNSKVYITIYGRTGITPRIELSQENKSTGKDILCAPFGRGTSTKFIVKAPNVGAITNIRIKQDESGNEPHWFLERVVVTDMSYPQWTYYFHLSCWLSSKYGDGKSCRLVRGYREPTGTGV 1 2 ETEYKLTFYTSDQKDAGTTGEVYVKLYGEKDSSREIWVNSINQKNRRQPTSYHFTRGSTVEVYLPPCPQLGEITKLKVGHNRAGSSPSWFLNK 0 0 VIVDDLRMNRVFEFPCYAWITKPSETIIVCQKSIKKEEKEIIWQ 1 2 KIPLEIRLYTGDVPNAGTTAKVYL 12 HQKTTSENKEDVYTTPYIWLEDGNYDRNGVTLFSIDLPVPKFISPLSQLVIGHDNTGHSPSWFFDK 0 0 VQIYCPLNGVEQTFLYRKWLTSSKPGMKVEQILCEEKSLRKCTDK 1 2 KIPWEISVKTSPIANSYVTAAVSIIIFGSKDKTMKIQLDRSNLEVSSEWTKVSGNEVVEMFRPNVESKFRIYIKDIGIPCKLRIQHDNKGSNPNWHLQE 0 0 IVLTNLRTHEQYEFYCNRWLSTREDDGSILREIPAKGPGITNPLP 1 2 LYHYIIQIFTGNKPNSGTNANIFINIFGEKGDCGERWLGRSVNRNSELFQQNQ 00 MDEFVIEAVQLGSINKICIGHEERSPGYGWYLAKIVLTIKENPKYKLTFECYR 21 WFDVGEDDGQIVRELFAHSSLS 1 2 AIAYNVTVLTGTCRNAGTVANVFVHLYGLQGESKDMQLKHKETEITKFEAGKSEEFILACGKLGE 0 0 IKKIKIWHDSIAPHSGWFLDEVVVIDYFLGRRYVFHVNRWLAKNEADGLISLDLQPTSIINIER 1 2 KSSYEVIVKTGGRKYAGTDSHVYITMFGINSESKEYHLANSKTHNNKFEQNHEDLFN 0 0 LNAVGLGSLKKIRVRHTNTGIAPGWFLDYILIRESIEQKKMEYFFPCYQWLSATRLDGLVIRELSVASENIFERWKSGHDISDDSRLILE 1 2 DQSTIYHVRIHTGSQSNISSDANVQIQLYGDKDFTGNIRLYKAYYGKEDILVNKFQAGQIANFIVKAINIGNIKKIR 2 1 IEHDTAVGSARWFVERVEVEARKLGLLWKFECNRFITPQQPELILYPEPKDISFVKP 1 2 MTNYQIKTFTSDISGAETTSSVYIQLYGNDSLPSSIRRLHQNNDSEQRFQRNKIDTFYVELEELQEPFSKLRIWHNDKGSSTDWHLNKVEIRKIKTNQY 0 0 VFVTYIFPCNKWLSRNMDQAALERELIPSHMLQDQNGVLQFERQLSPKWLMNTYEVRITTGDKAYAGTDASVYITLFGENGDSGERKLTKSLTHRNKFERGQ 0 0 TDVFQLEIVDLGKINKVRIRHDNSGVNPSWYLSTIEVFNISKSQSVHGLKDQPNETHHLVKYIFNCEAWLSTEHDEKVLDRFFSAEIIKPQDLPGIVPKTSKDIVPHKLIQDLSQGKKALE 1 2 TIPYLITVITGSDRKAGTPGPVWISCVDKDKMNSEKFILCDCYNRTMLKRGTTRYFRFAGVKLNDLTEIQ 0 0 VGNDAPESPSMGWYIQSLYLSFPTIGKMYLFDCKEWLSTNRGNKKRTCNLKISNTNIVKFKP 1 2 KITYHLKITTANVKRAGTDCSINLQIFGTNGVTNCYILEKTSNRFAQGITDNISLEMEDVGKLLKMRIGHDNQ 0 0 GKNKHWNLSCVEVTVANTNQLYRFVYDDWLSLTYGKRKSLWADLPAMIGDTVQLK 1 2 ETCLDIFVKTGNMPASSTDANVYVQLFGEYGDSGEILLKQTVSNQKPFQNNS 0 0 IDHFKIPSILKLGNLARCRIWHDNKGSSPNWYCEWLEVKEVLIPGEKNLACNWKFAFNKWLSVSDDNKQLLRDAPCSEVYMNDSKGHRTIDQESIETLLTTADSMNISDLKDNPEGKL 1 2 VYEVVIETGNLKDSGTTCDAWIILEGKHGRSPKLELVNQVGNPILQINQMNTFQ 2 1 LPSFPLGDLETIRLGIQERNINKQTNPNDIQSQKWFCEKVSIKDPVSKRTYIFTIHQWLSVTPTSNLKKDVLVNYSKIIEDPYKKAFNELK 1 2 NKQTVMYKVSIYTGTKGCANTDANIFITMFSTTPGLNSGRIALKRENNNLFDRKQLDEFYVESIDL 1 2 ESIQRIIIEHDNTGVSPDWYLDKVLITNQTNNQIHLFQCYQWISKKKGDCRLWKELLVSN* 0 >Trichoplax adhaerens (trichoplax) Metazoa XM_002107971 17 PLAT domains blast 0 MSASLAMQELMSDPSGHFAPKPPPGRSKINRKRPQT 1 2 APGRQITTASKAPKLWRSSTYNEPNLKRRKFIVTLNHKKDISSISRNAPYANRAYTTAMATAARTYSNALNKGQQKEIIPIYNPLCDPHLNDYYARKFGLLNSRD 0 0 ESRKNRKQ 1 2 AGVAYQFGVKTGDKKGSDTDA 2 1 VYIQVIGTKDKIPKKRLFKKQETEKTERGNLFKFDKSTVEKFAVQHRDIGDPVKLIVE 0 0 HDGNEKRHGWFLEEITLTNIQSKKSWLFPCHKWLSKYEGDRKLCYELKPLAKAGKA 1 2 VYEVSVLTGDKRGAGTDANVSVTLFGKHTSSPKIQLLKS 2 1 SKHKNPFERNNTDEFKIRTRDVGKLSKIRIEHDNAGFGPGWFLDK 0 0 VIICNLEKPNVKYYCPCNQWLAKDVGDKSISRDLTAYTDPNAAPS 1 2 AYVYIVHTFTGNKRGAGTDANVYAVIFGDSGDTGEKRLDNSKNNFEKSR 2 1 KDTFKLSCSCVGKLERLRIRHDNTGLFAGWYLDKV 1 2 VVEDPQEQQSYTFYCRRWLSKTEDDGEICRDLIVSASGDDGDDSVAPK 1 2 GYPYHIHVTTSDVKNAGTDAEVYVVMHGEGKKSKELNSGKLVLANSEKKKNTFERAMTDIFHMECAEMLSPLTKLTVGHDNKGLAAGWHLDR 0 0 IVIDCPTTGIEQTFLCQQWLDRKAGDGLTERELVEAFDMRKTRRP 1 2 KQLWFAWIWTSDIRGAGTDANVSMQIYGDKGKSQEIKLGNNTDNFEQATLDKFK 0 0 LEIDQVGVPYKLRIGHDNSNAFPGWHLDKVKLENMNDKEQYLFNCNRWLSRSEEDNEIIRELPASGPNCPNYP 1 2 IVIYEVSVHTGNKMGGGTDANVFIKIYGELGDSGYRPLKSSKSHNNKFERNQVDVFHIEAVTLKALKKIKIGHDGNNP 1 2 GAGWFLDKVVIKELNGEASNEFPCNR 2 1 WLSKSEDDGQIVRELFLKSDTPLLK 1 2 TTSYHISVKTGDVRNAGTDANVFIQIFGAKDDTGRVRLKQSLNTSNKFERNRIDKFIIEAAQIGK 0 0 IEKIIIGHDGKGLGSGWFLDYIELDVPSVGRLYRFSCHQWFDSTEGDRKVERELYPSECIKSAA 1 2 KIPYQISVHTGDIRHAGTDSNVFAVIYGENGKTEELKLRNKSDNFERGQVDVFK 0 0 VECEDVGKLRKLRIGHDSAGMGSAWFLDK 0 0 VYVRRLPPKSGKKSKETDEREEETAKKDADEPEKLDENNYLFVANRWLSKEEGDRQTVIEISPVGVDGALA 1 2 EMTYTIRVITGNKFGCGTNANVFINMYGEEGDSGERQLKKSETHTDKFERNQ 0 0 EDVFKISCLSLGELKKIKIRHDNSGFRPAWFLDKVIIEVGESKYQFMCDRWLAKDEDDGQISRELLPQSDEQSRAEAIGASKDLQKK 1 2 VASTTYNVSVTTGDIKGAGTDANVHIVLYGEKDDTGLIHLKNSTTHSNKFERNQEDRFVVEAIDIGELKKIK 2 1 IGHDNKGGMAGWFLNKVEIDIPSLGRRLLFPCGRWIDKGKDDGALERELYPLNEAEETYRP 1 2 HIPYEVTVYTTDKRGASTGANVYVVIYGEENQTEQASLEPDKKRRKQYFKNGAIDKFVLE 0 0 LDDVGEEITKLRIGHDGKGWGAGWHLDKVEICRLLDGGKASKKFTFQCNRWLASDEDDGAIVRELVPSEIVEKSSKDGGQVKTKVTKPTDGLK 1 2 VKPYTIHVFTGDVDGAGTNANVFLTIFGESGDSGERKLAKSDTHYDKFERNQ 0 0 EDIFHIEAADLGRLFKVKIRHDNTGSLFSPAWFLNRIEIVDDENEETTAFPCERWLAKKKDDGKIDRTLFVKGWEGDTSSVATTRSK 1 2 ASFRSNSTDLGEMVSASGRRASSRKGPVMAPIDEK 1 2 CVPYNIKVTVGEESSKNFQETLHLELFGQIEEEKSGPIELSPEKKSDKTFYPGKITTFYVSAAEVNIIEKIQVS 1 2 HNSYMPDSGIYLKEIEVDVPTIGNKYIFPCNRWLAKDKDDSKTSRIFTAANAQVTSYTA 1 2 GTDSNIFVILFGTKGRTPEISLEKNEDRFERAKVDIIP 0 0 LELDDVGTIKKIRIGHDGKGSRTDWYLEK 0 0 ASIQRMDTLDMYMFRANQWFSKKIDDKKLVREIPAETSKEGATTIK 1 2 KINYVLSTHTSDKRGSGTDANVFVIIFGENGDSGEIALKKSETNWNKFEKGQTDVFLINDRLSLGRLQKLRIWHDNA 1 2 GFGASWHLASVDIVDESTGVKYTFPCDKWLSKSNGDKLILRELPCAETAGNTAASKATKQESKSGKA 1 2 EYEIAFTTGTEKRAGTNQDVAIVLKGKSEKSREFLIENNEDKKYFSKGKTNKFTYTCKPLGDITKAIVSHRESAIGEEPDSKNSSWYLKVVTVVHKASGTT 2 1 YKFPCNKWIDLDDDEENNSSVTLKCKSADVAASSKAKPV 1 2 ELKPVKYEVTVVTGDEKGAGTDANVSVILYGDNGDTGPRPLKKKFVNLFERNQHDKFTIEALDLGKLTKLHIEHDNKGWGASWLLDRVEVHNVDSNETIIFPCKQWLDKKKGDGQIAKDLLPES* 0 >Trichoplax adhaerens (trichoplax) Metazoa XM_002107971 17 PLAT domains blast MSASLAMQELMSDPSGHFAPKPPPGRSKINRKRPQTAPGRQITTASKAPKLWRSSTYNEPNLKRRKFIVTLNHKKDISSISRNAPYANRAYTTAMATAARTYSNALNKGQQKEIIPIYNPLCDPHLNDYYARKFGLLNSRDESRKNRKQAG 01 VAYQFGVKTGDKKGSDTDAVYIQVIGTKDKIPKKRLFKKQETEKTERGNLFKFDKSTVEKFAVQHRDIGDPVKLIVEHDGNEKRHGWFLEEITLTNIQSKKSWLFPCHKWLSKYEGDRKLCYE LKPLAKAGK 02 AVYEVSVLTGDKRGAGTDANVSVTLFGKHTSSPKIQLLKSSKHKNPFERNNTDEFKIRTRDVGKLSKIRIEHDNAGFGPGWFLDKVIICNLEKPNVKYYCPCNQWLAKDVGDKSISRD LTAYTDPNAAPSA 03 YVYIVHTFTGNKRGAGTDANVYAVIFGDSGDTGEKRLDNSKNNFEKSRKDTFKLSCSCVGKLERLRIRHDNTGLFAGWYLDKVVVEDPQEQQSYTFYCRRWLSKTEDDGEICRDLI VSASGDDGDDSVAPKG 04 YPYHIHVTTSDVKNAGTDAEVYVVMHGEGKKSKELNSGKLVLANSEKKKNTFERAMTDIFHMECAEMLSPLTKLTVGHDNKGLAAGWHLDRaIVIDCPTTGIEQTFLCQQWLDRKAGDGLTERELVEA FDMRKTRRPK 05 QLWFAWIWTSDIRGAGTDANVSMQIYGDKGKSQEIKLGNNTDNFEQATLDKFKLEIDQVGVPYKLRIGHDNSNAFPGWHLDKVKLENMNDKEQYLFNCNRWLSRSEEDNEIIRELPAS GPNCPNYPI 06 VIYEVSVHTGNKMGGGTDANVFIKIYGELGDSGYRPLKSSKSHNNKFERNQVDVFHIEAVTLKALKKIKIGHDGNNPGAGWFLDKVVIKELNGEASNEFPCNRWLSKSEDDGQIVRELFLK SDTPLLKT 07 TSYHISVKTGDVRNAGTDANVFIQIFGAKDDTGRVRLKQSLNTSNKFERNRIDKFIIEAAQIGKIEKIIIGHDGKGLGSGWFLDYIELDVPSVGRLYRFSCHQWFDSTEGDRKVERELYPS ECIKSAAK 08 IPYQISVHTGDIRHAGTDSNVFAVIYGENGKTEELKLRNKSDNFERGQVDVFKVECEDVGKLRKLRIGHDSAGMGSAWFLDKVYVRRLPPKSGKKSKETDEREEETAKKDADEPEKLDENNYLFVANRWLSKEEGDRQTVIEIS PVGVDGALAE 09 MTYTIRVITGNKFGCGTNANVFINMYGEEGDSGERQLKKSETHTDKFERNQEDVFKISCLSLGELKKIKIRHDNSGFRPAWFLDKVIIEVGESKYQFMCDRWLAKDEDDGQISRELLPQ SDEQSRAEAIGASKDLQKKVAS 10 TTYNVSVTTGDIKGAGTDANVHIVLYGEKDDTGLIHLKNSTTHSNKFERNQEDRFVVEAIDIGELKKIKIGHDNKGGMAGWFLNKVEIDIPSLGRRLLFPCGRWIDKGKDDGALER ELYPLNEAEETYRPH 11 IPYEVTVYTTDKRGASTGANVYVVIYGEENQTEQASLEPDKKRRKQYFKNGAIDKFVLELDDVGEEITKLRIGHDGKGWGAGWHLDKVEICRLLDGGKASKKFTFQCNRWLASDEDDGAIVRELVPS EIVEKSSKDGGQVKTKVTKPTDGLKV 12 KPYTIHVFTGDVDGAGTNANVFLTIFGESGDSGERKLAKSDTHYDKFERNQEDIFHIEAADLGRLFKVKIRHDNTGSLFSPAWFLNRIEIVDDENEETTAFPCERWLAKKKDDGKIDRTLFVK GWEGDTSSVATTRSKASFRSNSTDLGEMVSASGRRASSRKGPVMAPIDEKC 13 VPYNIKVTVGEESSKNFQETLHLELFGQIEEEKSGPIELSPEKKSDKTFYPGKITTFYVSAAEVNIIEKIQVSHNSYMPDSGIYLKEIEVDVPTIGNKYIFPCNRWLAKDKDDSKTSRIFTAA 14 NAQVTSYTAGTDSNIFVILFGTKGRTPEISLEKNEDRFERAKVDIIPLELDDVGTIKKIRIGHDGKGSRTDWYLEKASIQRMDTLDMYMFRANQWFSKKIDDKKLVREI PAETSKEGATTIKK 15 INYVLSTHTSDKRGSGTDANVFVIIFGENGDSGEIALKKSETNWNKFEKGQTDVFLINDRLSLGRLQKLRIWHDNAGFGASWHLASVDIVDESTGVKYTFPCDKWLSKSNGDKLILREL PCAETAGNTAASKATKQESKSGK 16 AEYEIAFTTGTEKRAGTNQDVAIVLKGKSEKSREFLIENNEDKKYFSKGKTNKFTYTCKPLGDITKAIVSHRESAIGEEPDSKNSSWYLKVVTVVHKASGTTYKFPCNKWIDLDDDEENNSSVTLKC KSADVAASSKAKPVELKP 17 VKYEVTVVTGDEKGAGTDANVSVILYGDNGDTGPRPLKKKFVNLFERNQHDKFTIEALDLGKLTKLHIEHDNKGWGASWLLDRVEVHNVDSNETIIFPCKQWLDKKKGDGQIAKDLLPES* >Monosiga brevicollis (monosiga) Eukaryota|Choanoflagellates XM_001742822 17 PLAT domains blast exons colored; PLAT domains numbered MADLPVPTTALQRLQLERSRYSFAAADPPPSFVQRYTSADVLSHRLYAQGLASDVGPGSHSSLGMSRRVELPPYNPLNDPALANYFARKFEWNSTRSVGAGSGSRRSASASLTRTRQPVRGASAHGHTKSRTKSSHRK 01 GHASLEIFTSKRSNPVSKSEKYITVVGTKGRSDAIQLAAPGVHFRAGNKDVFQVNLTGIGKPTKVILENTGTKRTDGWCVSKVVLVKKTDKGTRRYRFSGPVWLSKHHDEMKLKR VLHIDDEDIGSG 02 NGYRIDCYTGDVANAGTDAVATIQLFGSKGQSPMVELRRSDGQAFQRARVATFTLDDLANLGKLKKLVISHNGHGMASGWFLDKIIVTSLSSNKATVFPCDAWLDRKNGRSKELVAR TAAAGAEGM 03 TTFTIRVMTGDRRGAGTDANVQCTLFGEDGESGPHTLNTSRNDFRRGHTDVFAVSSRKIGTLKRLRIWHDNGGAGPAWFLDAVEVVDEASGQTYRFECNRWLAKDEDDGQISRELTCN GDSSWGL 04 KSYKLTIFTGDKRNAGTSANVFCKLVGERGASDNVILENSSKNFQRDRTDIFTVEASDLGSLRHIVLGHDNHGMGAGWYVERFSLEVPSEGKLYNVDVKQWFATDMSDGAIERTFRLD NAETLDIAQR 05 LDWKCTIYTSDVANAGTDANVFMQVYGKKGKTDVVPLKNKSDTFERGQTDELRVQLINVGSLRKLRVWHDNKGMASGWHLDRIVLSRDGEEYIFPCAEWLAVSEGDKEIVRELPAT GPNVKKPLQL 06 VEYTVRVATGHARFAGTNADVFVMLTGELGDSGKRALLRSQTNRNKFERGKEDVFTVAAVDLGKLTSVTVGHNNAGTSAGWFLDKIVVLDPRRGEEEEFPCHRWLAVDADDGQIERELVPK MAEHQAAAT 07 TTYIVKIKTGDVRHAGTDANVFVQLFGKTGESTQLKLRNSETYSDAFERNKMDIFKFELLDLGDLSRILVGHDNKGMGAAWFLDYVEVEVPSIRTRWKFPCSRWFSKSQDDGLTER EIYAEKEAGEPMEEDVS 08 APYLFRFYTSDVAFAGTDANVSVVLYGDEGKTEELVVNNQSDNFERGKADDFKLACKPVGRPSKIRLSAHGGGMSADWHLEKIEVHELGQARIYTFEHNDWLRKGTKAKPFMVEL PLRRIETVDDNGREVVEELALDANK 09 RTYRVKVHTGDQKGAGTDANVYVNLHGSLGDSGDRHLKNSLTHTNKFQRKTVDEFDIDAVTLGDINKVKVWHDNAGLGAAWYLEKIEVVDTADDKTYIFPCAQWFAKSMGDGQIAREL GVLEEQKPADFQTKNVG 10 FKYRISVHTSDVKHAGTDANVDIVLYGEKGDTGKIRLAKSETHRDMWERGNCDVFTVSAIELGDLKRVDIMHDGKGVGSGWHLNKVVVDAPQAGKTWTFMCDAWLDKATDDGTMAK TLYASADAIEEYSAH 11 VPYEIIIKTSDVRNAGTDANVFIDLYGRDQEERDLTAHHEFKDAVKAHFERNLEDRFNVELPDVGSIYKIRLGHDGKGMSSSWHVASVVVINQRTHERFEFPCDAWLSKDKDDKKLVREFAVG EVKALEGDKVVRRESILSLQE 12 AIYKIHVFTGDIKHAGTDANIYVQIFGDTGDSGEIKLEKSETYRDKFERGHEDIFTHRCLDLGPLRKIKVRSDGKGLMGGDWYLDRVEVHQENDLSEPPVRFVCQDWFKRGKQEGDTLEREITAQ VDAAVAMKEATALEDEAARLQRKATTLSRKSTKGSPLRKGTGTTETNAELEEAQRQANIARRKADEAKERAGLAGADDTQ 13 LEYKVTVYTGTDTQAGTTANVWLQLFGEKDAPLTPAPPSPSKLSRSGSLFGRRRRASSDAQSVSSSLSAGASGPRETSTGRLQLNNAPADLQSGAKTTFTVTGLDVGELVGLEIGHDDERDKWYLEQVVVEVPKSATHAARRYEFKAGVWLAARADGSTSGSAK GKSKASVRLQPSEIHAGSERI 14 LEYTLKVYTASADGAGCTAVPQVQLFGDKHTTEALPLRAGGDILPASVVETQHRVPDLGALLKVRLMVPRGSSWTVEKVEFGRAGQTPITFVGSDGSAVTLGAERLS FDFLPAAAPASSGGKGKRRGSTTSSVAVAQQ 15 TSYRVYVTTADERGTGTDANVSIILYGAMGDSGEHSLTKSETFDDPFERGNTDVFTLEVPDLGELQRARIWHDGKGMFSSWKLDKIVVVVEATQSRYELPCGQWLSKNKGDKQLTRDLAVASKR VNALT 16 GTYKLEVATDSRAGGGCKGPVRIMLLDAEKNQLPLTLEPPGGEFAPGSVEHLVFDNVMLLGPLTELRIRRKPSGASSRRGAEDDDEDDNEASGASSQSGSAVSPWHLEHIIVKHLQSGQSFVFKGPSKGLSRSRAKLSVHTEA TTEEAVAQATKASV 17 AQYEVAVTTGTERGAGTDSNVFVTLFGKNGDSGERALAKSKTFRNMFESGNTDVFDVECQDLGELTKIEVKSDLKGFGAAWQLDKIKVTRTGSQNSWQFKCDQWFDKKQGAEHTFSVA S*
>LOXHD1_homSap_full dna: each line is an exon ATGATGCCCCAGAAGAAAAGGCGGAGGAAGAAGGACATCGACTTCCTGGCCCTGTACGAAGCGGAGCTGCTGAACTACGCCTCGGAGGACGACGAGGGGGAGCTGGAACACGAGTACTACAAGGCCAGAG TGTATGAAGTGGTCACAGCCACGGGGGATGTTCGCGGTGCAGGGACGGATGCCAATGTCTTCATCACGCTTTTTGGAGAGAATGGGCTCTCTCCCAAGCTCCAGCTCACCAGCAA GAGCAAGTCTGCCTTTGAGAAGGGCAACGTCGATGTGTTCCGGGTGAGAACCAACAATGTGGGCCTCATCTATAAAGTCAG GATTGAGCATGACAACACGGGCTTGAATGCCAGCTGGTACCTGGACCATGTGATTGTGACCGACATGAAGAGGCCTCATCTCCGTTACTACTTCAACTGCAACAACTGGCTGAGCAAGGTGGAAGGTGACCGCCAGTGGTGCCGTGACCTGCTGGCCAGCTTCAACCCCATGGACATGCCCAGAG GTAATAAGTATGAAGTCAAGGTATACACTGGTGATGTAATTGGTGCAGGGACAGATGCTGATGTCTTCATCAATATTTTTGGAGAGTATGGAGACACAG GGGAGCGTAGGCTAGAAAATGAAAAGGACAACTTTGAAAAGGGAGCTGAAGACAGGTTCATCCTGGATGCCCCGGATTTGGGGCAGCTGATGAAGATCAATGTTGGCCACAACAATAAGGGGGGCTCTGCAGGTTGGTTCCTGTCCCAG ATAGTCATTGAAGATATTGGGAACAAAAGAAAATATGACTTCCCCCTTAACCGCTGGCTGGCCTTGGACGAAGACGATGGCAAAATCCAAAGGGATATCTTAGTGGGCGGAGCTGAGACCACAG CTATTACGTATATTGTCACCGTCTTCACTGGGGATGTCCGGGGGGCTGGTACCAAATCCAAAATCTACTTGGTCATGTATGGGGCCAGAGGGAATAAGAACAGTGGGAAAATCTTCCTGGAGGGCGGCGTGTTTGACCGAGGCCGCACGGACATCTTCCACATCGAGCTGGCTGTCCTCCTTAGCCCCCTGAGTCGGGTCTCCGTCGGGCATGGCAATGTGGGTGTCAACAGAGGCTGGTTCTGTGAGAAG GTGGTGATTCTGTGCCCCTTCACTGGTATCCAGCAGACCTTCCCTTGTAGCAACTGGCTGGATGAGAAGAAAGCGGATGGGTTGATTGAGAGGCAGCTCTATGAGATGGTGTCTCTCAGGAAGAAGCGGCTGAAAA AATTCCCTTGGTCCCTGTGGGTCTGGACAACCGACCTAAAGAAAGCTGGTACCAACTCTCCCATCTTCATCCAGATTTATGGGCAGAAGGGGCGGACAGATGAGATTCTCCTGAATCCCAACAACAAGTGGTTCAAACCCGGCATAATCGAGAAGTTTAGG ATTGAGCTCCCGGATCTTGGCAGGTTTTATAAGATTCGAGTATGGCATGATAAAAGGAGTTCTGGTTCTGGATGGCATTTAGAAAGG ATGACCCTGATGAACACTCTGAACAAAGACAAGTACAACTTCAATTGCAACCGCTGGCTGGATGCCAATGAGGATGACAATGAGATAGTGAGGGAAATGACTGCAGAAGGCCCAACAGTGCGCAGGATCATGGGCA TGGCCCGGTACCATGTGACTGTGTGCACAGGTGAACTTGAAGGTGCTGGGACCGATGCCAACGTCTATCTCTGCCTTTTTGGTGATGTGGGGGACACGGGGGAACGGCTGCTCTACAACTGCAGGAATAACACAGACCTGTTTGAAAAGGGCAAT GCTGACGAGTTCACTATCGAGTCTGTCACCATGCGGAATGTGAGGCGGGTGAGGATCAGACACGATGGCAAAGGCTCCGGCAGCGGCTGGTACCTGGACAGAGTGCTGGTGAGAGAGGAGGGGCAGCCTGAGAGCGACAACGTGGAGTTCCCATGTCTCAG GTGGTTGGACAAGGATAAGGATGATGGGCAGCTGGTCCGAGAGTTGCTACCCAGTGACAGCAGCGCGACACTGAAGA ACTTTCGCTATCACATCAGCTTGAAGACTGGGGATGTCTCTGGGGCCAGCACGGATTCTAGAGTCTACATCAAGCTCTATGGGGATAAATCTGACACCATCAAGCAAGTTCTTCTTGTCTCTGACAACAACCTCAAAGACTACTTTGAACGTGGCCGGGTGGATGAGTTCACCCTCGAGACCCTGAACATTGGAAAT ATCAACCGGCTGGTGATTGGGCATGACAGCACTGGCATGCATGCCAGCTGGTTCCTGGGCAGCGTTCAGATCCGTGTGCCCCGTCAAGGCAAGCAGTACACCTTTCCCGCCAACCGCTGGCTGGACAAGAACCAGGCTGACGGGCGCCTGGAGGTGGAGCTGTATCCCAGCGAGGTGGTGGAGATCCAGAAAT TGGTCCACTATGAGGTTGAGATTTGGACAGGAGATGTGGGTGGCGCAGGCACCAGTGCCCGAGTCTACATGCAGATCTATGGAGAGAAAGGCAAGACAGAAGTGCTCTTCCTCTCCAGCCGCTCAAAAGTTTTTGAACGGGCGTCCAAGGACACATTCCAG CTTGAGGCGGCCGACGTGGGCGAGGTCTATAAGCTCCGGCTCGGGCACACGGGCGAGGGCTTTGGGCCCAGCTGGTTCGTGGACACCGTGTGGCTGCGGCACCTGGTGGTGCGGGAGGTGGACCTCACGCCGGAGGAGGAGGCCCGGAAGAAGAAGGAGAAGGACAAGCTGCGGCAGCTGCTCAAGAAGGAGCGGCTGAAGGCCAAGCTGCAGAGGAAGAAGAAGAAGAGGAAGGGCAGCGACGAAGAGGACGAGGGGGAGGAAGAGGAGTCGTCCTCATCAGAGGAGTCCTCGTCAGAGGAGGAGGAGATGGAAGAAGAGGAGGAAGAGGAGGAGTTTGGGCCGGGGATGCAGGAGGTGATTGAGCAGCACAAGTTCGAAGCCCACCGCTGGCTGGCCCGGGGCAAGGAGGACAACGAACTTGTCGTGGAGTTGGTGCCAGCTGGCAAGCCGGGTCCTGAGC GAAACACCTATGAGGTTCAGGTGGTCACGGGGAATGTGCCCAAGGCCGGCACTGATGCTAACGTCTACCTAACCATCTACGGCGAGGAGTATGGAGACACGGGCGAACGACCCCTGAAGAAGTCAGACAAGTCCAACAAATTTGAGCAGGGGCAG ACAGACACCTTCACCATCTATGCCATTGACCTGGGGGCCCTGACCAAGATTCGGATTCGCCACGACAACACAGGCAACAGAGCAGGCTGGTTCCTGGACAGAATAGACATTACTGACATGAACAACGAGATCAC GTACTACTTTCCATGCCAACGTTGGCTGGCAGTGGAGGAAGATGATGGCCAGCTGTCCAGGGAGCTGTTGCCAGTGGATGAGTCCTATGTGCTGCCACAGAGCGAGGAGGGTAGGGGAGGCGGTGACAACAACCCCCTCGACAACCTGGCCCTGGAGCAGAAAG ATAAATCTACCACATTCTCAGTGACCATAAAGACTGGGGTTAAGAAGAATGCGGGCACAGATGCTAATGTCTTCATCACACTCTTTGGCACACAGGATGACACTG GAATGACCCTCCTGAAGTCCTCCAAGACAAACAGCGATAAGTTTGAGAGGGACAGCATTGAAATCTTCACGGTGGAGACGCTGGATCTGGGAGACCTGTGGAAAGTCCGGCTTGGCCATGACAACACAG GCAAGGCCCCAGGCTGGTTTGTAGACTGGGTAGAGGTGGATGCCCCATCTCTTGGGAAGTGCATGACGTTTCCCTGTGGCCGCTGGCTGGCCAAAAACGAAGACGACGGGTCCATCATCAGAGACCTCTTCCATGCAGAGCTTCAGACGAGGCTGTACACACCAT TTGTTCCTTACGAGATCACCCTCTACACCAGTGATGTCTTTGCTGCTGGGACAGATGCCAACATCTTCATCATCATCTATGGCTGCGATGCCGTGTGCACCCAGCAGAAGTATCTGTGTACCAACAAGAGGGAACAGAAGCAGTTCTTTGAGAGGAAGTCTGCCTCCCGCTTCATCGTAGAG TTAGAAGATGTGGGAGAAATCATTGAAAAAATTCGGATTGGCCATAATAACACGGGCATGAATCCTGGGTGGCACTGCTCTCACGTGGACATCCGCAGGCTCCTCCCGGATAAAGAC GGTGCAGAGACCTTGACTTTCCCATGCGATCGGTGGCTTGCCACCTCTGAGGATGACAAAAAGACCATTCGAGAACTGGTTCCATATGACATCTTCACTGAGAAATACATGAAAGATGGGTCCTTACGGCAAGTCTACAAGGAAGTAGAAGAGCCTCTGGACa TTGTGCTGTACTCGGTGCAGATCTTCACAGGGAACATTCCTGGGGCAGGGACGGATGCCAAGGTGTACATCACCATCTATGGAGACCTCGGGGACACTGGGGAGCGATACCTTGGCAAGTCAGAGAACCGGACCAACAAGTTCGAGAGAGGAACG GCTGACACCTTCATCATCGAGGCCGCTGACCTAGGCGTCATCTACAAGATCAAGCTCCGCCATGACAACTCCAAGTGGTGCGCAGACTGGTACGTGGAGAAGGTGGAGATCTGGAATGACACCAACGAGGACGAGTTCCTGTTCCTATGCGGGCGCTGGCTCTCCCTGAAGAAGGAGGATGGGCGACTCGAGAGGCTCTTTTACGAGAAG GAGTACACTGGGGACCGCAGCAGCAACTGCAGCAGCCCTGCTGACTTCTGGGAGATCGCCCTGAGCTCCAAGATGGCCGATGTCGACATCAGCACAGTGACCGGGCCCATGGCTGACTACGTTCAAGAGGGCCCAA TTATTCCCTACTATGTGTCAGTCACCACTGGGAAGCACAAGGACGCGGCCACTGACAGCCGAGCCTTCATCTTTCTCATCGGGGAGGATGATGAACGTAGTAAGCGCATCTGGTTGGACTACCCCCGAGGGAAGAGGGGCTTCAGCCGTGGCTCTGTGGAGGAGTTCTACGTCGCAGGCTTGGATGTGGGCATCATCAAGAAAATAGAG CTGGGCCATGACGGGGCCTCCCCTGAGAGCTGCTGGCTGGTGGAAGAGTTGTGTTTGGCAGTGCCCACCCAGGGCACCAAGTACATGTTGAACTGTAACTGCTGGCTGGCCAAGGACAGAGGCGACGGCATCACCTCCCGTGTCTTCGACCTCTTGGATGCCATGGTGGTGAACATTGGGGTGAAG GTTCTCTATGAAATGACGGTGTGGACAGGGGATGTGGTTGGCGGGGGCACTGACTCCAACATCTTCATGACCCTCTACGGCATCAACGGGAGCACAGAGGAGATGCAGCTGGACAAAAAGAAAGCCAG GTTTGAGCGGGAGCAGAACGACACCTTCATCATGGAGATCCTAGACATTGCTCCATTCACCAAGATGCGGATCCGGATTGATGGCCTGGGCAGTCGGCCGGAGTGGTTCCTGGAGAGG ATCCTACTGAAGAACATGAACACTGGAGACCTGACCATGTTCTACTATGGAGACTGGCTGTCCCAGCGGAAGGGCAAGAAGACCCTGGTGTGTGAAATGTGTGCCGTTATCGATGAGGAAGAAATGATGGAGTGGACCTCCTACACCGTCGCAGTTAAGACCAGCGACATCCTGG GAGCAGGCACTGATGCCAACGTGTTCATCATCATCTTCGGGGAGAACGGGGATAGTGGGACACTGGCCCTGAAGCAGTCGGCAAACTGGAACAAGTTTGAGCGGAACAACACGGACACATTCAACTTCCCTGACATGCTGAGCTTGGGCCACCTCTGCAAGCTGAGGGTCTGGCACGACAACAAAG GGATATTTCCTGGCTGGCATCTGAGCTATGTCGATGTGAAGGACAACTCCCGCGACGAGACCTTCCACTTCCAGTGTGACTGCTGGCTCTCCAAGAGTGAGGGTGACGGGCAGACGGTCCGCGACTTTGCCTGTGCCAACAACAAGATCTGTGATGAGCTGGAAGAGACCA CCTACGAGATCGTCATAGAAACGGGCAACGGAGGCGAAACCAGGGAGAACGTCTGGCTCATCCTGGAGGGCAGGAAGAACCGATCCAAAGAGTTTCTCATGGAAAATTCTTCTAGGCAGCGGGCCTTTAGGAA GGGGACCACAGACACGTTTGAGTTTGACAGCATCTACTTGGGGGACATTGCCTCCCTCTGTGTGGGCCACCTTGCCAGGGAAGACCGGTTTATCCCCAAGAGAGAACTTGCCTGGCATGTCAAGACCATCACCATCACCGAGATGGAGTACGGCAATGT GTACTTCTTTAACTGTGACTGCCTCATCCCCCTCAAGAGGAAGAGGAAGTACTTCAAGGTATTCGAGGTTACCAAGACGACAGAGAGCTTTGCCAGCAAGGTCCAGAGCCTGGTGCCCGTCAAGTACGAAGTCATCGTGACAACAGGCTATGAGCCAGGGGCAGGCACTGATGCCAACGTCTTCGTGACCATCTTTGGGGCCAACGGAGACACAGGCAAGCGGGAGCTGAAGCAGAAAATGCGCAACCTCTTCGAGCGGGGCAGCACAGACCGCTTCTTCCTGGAGACGCTGGAGCTGGGTGAGCTGCGCAAGGTGCGCCTGGAGCACGACAGCAGTGGCTACTGCTCAGGCTGGCTGGTGGAGAAGGTGGAGGTCACCAACACCAGCACCGGCGTGGCCACCATCTTCAACTGTGGCAGGTGGCTGGACAAGAAGCGGGGGGATGGACTCACCTGGAGAGACCTCTTCCCTTCTGTC
Reference PLAT domain sets
>LOXHD1_homSap_PLAT:01 RVYEVVTATGDVRGAGTDANVFITLFGENGLSPKLQLTSKSKSAFEKGNVDVFRVRTNNVGLIYKVRIEHDNTGLNASWYLDHVIVTDMKRPHLRYYFNCNNWLSKVEGDRQWCRD >LOXHD1_homSap_PLAT:02 NKYEVKVYTGDVIGAGTDADVFINIFGEYGDTGERRLENEKDNFEKGAEDRFILDAPDLGQLMKINVGHNNKGGSAGWFLSQIVIEDIGNKRKYDFPLNRWLALDEDDGKIQRDILVGG >LOXHD1_homSap_PLAT:03 ITYIVTVFTGDVRGAGTKSKIYLVMYGARGNKNSGKIFLEGGVFDRGRTDIFHIELAVLLSPLSRVSVGHGNVGVNRGWFCEKVVILCPFTGIQQTFPCSNWLDEKKADGLIER >LOXHD1_homSap_PLAT:04 FPWSLWVWTTDLKKAGTNSPIFIQIYGQKGRTDEILLNPNNKWFKPGIIEKFRIELPDLGRFYKIRVWHDKRSSGSGWHLERMTLMNTLNKDKYNFNCNRWLDANEDDNEIVREM >LOXHD1_homSap_PLAT:05 ARYHVTVCTGELEGAGTDANVYLCLFGDVGDTGERLLYNCRNNTDLFEKGNADEFTIESVTMRNVRRVRIRHDGKGSGSGWYLDRVLVREEGQPESDNVEFPCLRWLDKDKDDGQLVRELLPS >LOXHD1_homSap_PLAT:06 FRYHISLKTGDVSGASTDSRVYIKLYGDKSDTIKQVLLVSDNNLKDYFERGRVDEFTLETLNIGNINRLVIGHDSTGMHASWFLGSVQIRVPRQGKQYTFPANRWLDKNQADGRLEV >LOXHD1_homSap_PLAT:07 VHYEVEIWTGDVGGAGTSARVYMQIYGEKGKTEVLFLSSRSKVFERASKDTFQLEAADVGEVYKLRLGHTGEGFGPSWFVDTVWLRHLVVREVDLTPEEEARKKKEKDKLR >LOXHD1_homSap_PLAT:08 NTYEVQVVTGNVPKAGTDANVYLTIYGEEYGDTGERPLKKSDKSNKFEQGQTDTFTIYAIDLGALTKIRIRHDNTGNRAGWFLDRIDITDMNNEITYYFPCQRWLAVEEDDGQLSRELLPV >LOXHD1_homSap_PLAT:09 TTFSVTIKTGVKKNAGTDANVFITLFGTQDDTGMTLLKSSKTNSDKFERDSIEIFTVETLDLGDLWKVRLGHDNTGKAPGWFVDWVEVDAPSLGKCMTFPCGRWLAKNEDDGSIIRDLFHA >LOXHD1_homSap_PLAT:10 VPYEITLYTSDVFAAGTDANIFIIIYGCDAVCTQQKYLCTNKREQKQFFERKSASRFIVELEDVGEIIEKIRIGHNNTGMNPGWHCSHVDIRRLLPDKDGAETLTFPCDRWLATSEDDKKTIRELVP >LOXHD1_homSap_PLAT:11 VLYSVQIFTGNIPGAGTDAKVYITIYGDLGDTGERYLGKSENRTNKFERGTADTFIIEAADLGVIYKIKLRHDNSKWCADWYVEKVEIWNDTNEDEFLFLCGRWLSLKKEDGRLERLFYEK >LOXHD1_homSap_PLAT:12 IPYYVSVTTGKHKDAATDSRAFIFLIGEDDERSKRIWLDYPRGKRGFSRGSVEEFYVAGLDVGIIKKIELGHDGASPESCWLVEELCLAVPTQGTKYMLNCNCWLAKDRGDGITSRVFD >LOXHD1_homSap_PLAT:13 VLYEMTVWTGDVVGGGTDSNIFMTLYGINGSTEEMQLDKKKARFEREQNDTFIMEILDIAPFTKMRIRIDGLGSRPEWFLERILLKNMNTGDLTMFYYGDWLSQRKGKKTLVC >LOXHD1_homSap_PLAT:14 TSYTVAVKTSDILGAGTDANVFIIIFGENGDSGTLALKQSANWNKFERNNTDTFNFPDMLSLGHLCKLRVWHDNKGIFPGWHLSYVDVKDNSRDETFHFQCDCWLSKSEGDGQTVRDFAC >LOXHD1_homSap_PLAT:15 TTYEIVIETGNGGETRENVWLILEGRKNRSKEFLMENSSRQRAFRKGTTDTFEFDSIYLGDIASLCVGHLAREDRFIPKRELAWHVKTITITEMEYGNVYFFNCDCLIPLKRKRKYFKVF >LOXHD1_homSap_PLAT:16 VKYEVIVTTGYEPGAGTDANVFVTIFGANGDTGKRELKQKMRNLFERGSTDRFFLETLELGELRKVRLEHDSSGYCSGWLVEKVEVTNTSTGVATIFNCGRWLDKKRGDGLTWRDLFPS >PKD1L1_homSap_PLAT chr7: 47780815 polycystin-1L1 GPS-TM-LH2-TM pdb:3CWZ QLYAVVIDTGFRAPARLTSKVYIVLCGDNGLSETKELSCPEKPLFERNSRHTFILSAPAQLGLLRKIRLWHDSRGPSPGWFISHVMVKELHTGQGWFFPAQCWLSAGRHDGRVERELTC >PKD1_homSap_PLAT chr16: 2078712 polycystin 1 GPS-TM-LH2-TM pdb:3CWZ FKYEILVKTGWGRGSGTTAHVGIMLYGVDSRSGHRHLDGDRAFHRNSLDIFRIATPHSLGSVWKIRVWHDNKGLSPAWFLQHVIVRDLQTARSAFFLVNDWLSVETEANGGLVEK >PKD1L2_homSap_PLAT chr16: 79691985 polycystin 1-like 2 signal...GPS-TM-LH2-TM pdb:3CWZ YHYLVTVYTGHRRGAATSSKVTVTLYGLDGEREPHHLADPDTPVFERGAVDAFLLSTLFPLGELRSLRLWHDNSGDRPSWYVSRVLVYDLVMDRKWYFLCNSWLSINVGDCVLDKVFPVA >PKD1L3_homSap chr16:70,556,160 GPS-TM-LH2-TM polycystic kidney disease 1-like 3 FHYLIQVYTGYRRSAATTAKVVITLYGSEGRSEPHHLCDPQKTVFERGGLDVFLLTTWTSLGNLHSLRLWHDNSGVSPSWYVSQVIVCDMAVKRKWHFLCNCWLAVDLGDCELDRVFIPV >PKDREJ_homSap_PLAT chr22: 45030224 receptor for egg jelly-like signal...GPS-TM-LH2-TM pdb:3CWZ CYLVTIFTGSRWGSGTRANVFVQLRGTVSTSDVHCLSHPHFTTLYRGSINTFLLTTKSDLGDIHSIRVWHNNEGRSPSWYLSRIKVENLFSRHIWLFICQKWLSVDTTLDRTFHVT >DENND5A_homSap_PLAT chr11: 9116949 RAB6 interacting 1 RUN-LH2-RUN pdb:3CWZ HILIVPSKKLGGSMFTANPWICISGELGETQIMQIPRNVLEMTFECQNLGKLTTVQIGHDNSGLYAKWLVEYVMVRNEITGHTYKFPCGRWLGKGMDDGSLER >DENND5B_homSap_PLAT chr12: 31426424 RAB6 interacting 2 RUN-LH2-RUN pdb:3CWZ RSVIIPIKKLSNAIITSNPWICVSGELGDTGVMQIPKNLLEMTFECQNLGKLTTVQIGHDNSGLLAKWLVDCVMVRNEITGHTYRFPCGRWLGKGIDDGSLER >ALOX5_homSap_PLAT chr10: 45189635 arachidonate 5-lipoxygenase NH2-LH2-lipox pdb:2P0M PSYTVTVATGSQWFAGTDDYIYLSLVGSAGCSEKHLLDKPFYNDFERGAVDSYDVTVDEELGEIQLVRIEKRKYWLNDDWYLKYITLKTPHGDYIEFPCYRWITGDVEVVLRDG >ALOX15_homSap_PLAT chr17: 4480963 arachidonate 15-lipoxygenase NH2-LH2-lipox pdb:2P0M GLYRIRVSTGASLYAGSNNQVQLWLVGQHGEAALGKRLWPARGKETELKVEVPEYLGPLLFVKLRKRHLLKDDAWFCNWISVQGPGAGDEVRFPCYRWVEGNGVLSLPEG >ALOX12_homSap_PLAT chr17: 6840108 arachidonate 12-lipoxygenase NH2-LH2-lipox pdb:2P0M GRYRIRVATGAWLFSGSYNRVQLWLVGTRGEAELELQLRPARGEEEEFDHDVAEDLGLLQFVRLRKHHWLVDDAWFCDRITVQGPGACAEVAFPCYRWVQGEDILSLPEG >ALOX15B_homSap_PLAT chr17 7,888,129 arachidonate 15-lipoxygenase NH2-LH2-lipox pdb:2P0M AEFRVRVSTGEAFGAGTWDKVSVSIVGTRGESPPLPLDNLGKEFTAGAEEDFQVTLPEDVGRVLLLRVHKAPPVLPLLGPLAPDAWFCRWFQLTPPRGGHLLFPCYQWLEGAGTLVLQEG >ALOX12B_homSap_PLAT chr17: 7916679 arachidonate 12R-lipoxygenase NH2-LH2-lipox pdb:2P0M ATYKVRVATGTDLLSGTRDSISLTIVGTQGESHKQLLNHFGRDFATGAVGQYTVQCPQDLGELIIIRLHKERYAFFPKDPWYCNYVQICAPNGRIYHFPAYQWMDGYETLALREA >ALOXE3_homSap_PLAT chr17: 7939943 arachidonate lipoxygenase 3 NH2-LH2-lipox pdb:2P0M AVYRLCVTTGPYLRAGTLDNISVTLVGTCGESPKQRLDRMGRDFAPGSVQKYKVRCTAELGELLLLRVHKERYAFFRKDSWYCSRICVTEPDGSVSHFPCYQWIEGYCTVELRPG >LIPC_homSap LIP-PLAT chr15:56511467 lipase C YHYQLKIQFINQTETPIQTTFTMSLLGTKEKMQKIPITLGKGIASNKTYSFLITLDVDIGELIMIKFKWENSAVWANVWDTVQTIIPWSTGPRHSGLVLKTIRVKAGETQQRMTFCSENTDDLLLRPTQEKIFVKC >LIPG_homSap LIP-PLAT chr18:45342425 endothelial lipase YHYQMKIHVFSYKNMGEIEPTFYVTLYGTNADSQTLPLEIVERIEQNATNTFLVYTEEDLGDLLKIQLTWEGASQSWYNLWKEFRSYLSQPRNPGRELNIRRIRVKSGETQRKLTFCTEDPENTSISPGRELWFRKC >LPL_homSap LIP-PLAT chr8:19840862 lipoprotein lipase FHYQVKIHFSGTESETHTNQAFEISLYGTVAESENIPFTLPEVSTNKTYSFLIYTEVDIGELLMLKLKWKSDSYFSWSDWWSSPGFAIQKIRVKAGETQKKVIFCSREKVSHLQKGKAPAVFVKC >PNLIP_homSap LIP-PLAT chr10:118295418 pancreatic lipase WRYKVSVTLSGKKVTGHILVSLFGNKGNSKQYEIFKGTLKPDSTHSNEFDSDVDVGDLQMVKFIWYNNVINPTLPRVGASKIIVETNVGKQFNFCSPETVREEVLLTLTPC >PNLIPRP1_homSap LIP-PLAT chr10:118340480 pancreatic lipase-related 1 WRYGVSITLSGRTATGQIKVALFGNKGNTHQYSIFRGILKPGSTHSYEFDAKLDVGTIEKVKFLWNNNVINPTLPKVGATKITVQKGEEKTVYNFCSEDTVREDTLLTLTPC >PNLIPRP2_homSap LIP-PLAT chr10:118370455 pancreatic lipase-related 2 WRYKVSVTLSGKEKVNGYIRIALYGSNENSKQYEIFKGSLKPDASHTCAIDVDFNVGKIQKVKFLWNKRGINLSEPKLGASQITVQSGEDGTEYNFCSSDTVEENVLQSLYPC >PNLIPRP3_homSap LIP-PLAT chr10:118177414 pancreatic lipase-related protein 3 chr16:70520943 WRHKLSVKLSGSEVTQGTVFLRVGGAVRKTGEFAIVSGKLEPGMTYTKLIDADVNVGNITSVQFIWKKHLFEDSQNKLGAEMVINTSGKYGYKSTFCSQDIMGPNILQNLKPC >LOXHD1_schMan_PLAT:01 Schistosoma mansoni (flatworm) Bilatera; Platyhelminthes; Trematoda XP_002576380 from contig NS_000200 TLYKVVLTTADVPGAGTSAQIYITLKGEWGSSTRQKLRKEKVPTRNLRFYFYPGSTNTFSVVSPDLGGLHSVFIEHDSLRKSDSWLLESVQVFHPLTKKRYMFMCNHWFSLYKEDGLIARELF >LOXHD1_schMan_PLAT:02 TKYSIVTVTGDQEGSGTNSKVYITIYGRTGITPRIELSQENKSTGKDILCAPFGRGTSTKFIVKAPNVGAITNIRIKQDESGNEPHWFLERVVVTDMSYPQWTYYFHLSCWLSSKYGDGKSCR >LOXHD1_schMan_PLAT:03 TEYKLTFYTSDQKDAGTTGEVYVKLYGEKDSSREIWVNSINQKNRRQPTSYHFTRGSTVEVYLPPCPQLGEITKLKVGHNRAGSSPSWFLNKVIVDDLRMNRVFEFPCYAWITK >LOXHD1_schMan_PLAT:04 IPLEIRLYTGDVPNAGTTAKVYLHQKTTSENKEDVYTTPYIWLEDGNYDRNGVTLFSIDLPVPKFISPLSQLVIGHDNTGHSPSWFFDKVQIYCPLNGVEQTFLYRKWLTSSKPGMKVEQILCEE >LOXHD1_schMan_PLAT:05 IPWEISVKTSPIANSYVTAAVSIIIFGSKDKTMKIQLDRSNLEVSSEWTKVSGNEVVEMFRPNVESKFRIYIKDIGIPCKLRIQHDNKGSNPNWHLQEIVLTNLRTHEQYEFYCNRWLSTREDDGSILREIPAK >LOXHD1_schMan_PLAT:06 YIIQIFTGNKPNSGTNANIFINIFGEKGDCGERWLGRSVNRNSELFQQNQMDEFVIEAVQLGSINKICIGHEERSPGYGWYLAKIVLTIKENPKYKLTFECYRWFDVGEDDGQIVRELFA >LOXHD1_schMan_PLAT:07 IAYNVTVLTGTCRNAGTVANVFVHLYGLQGESKDMQLKHKETEITKFEAGKSEEFILACGKLGEIKKIKIWHDSIAPHSGWFLDEVVVIDYFLGRRYVFHVNRWLAKNEADGLISLDLQPT >LOXHD1_schMan_PLAT:08 SYEVIVKTGGRKYAGTDSHVYITMFGINSESKEYHLANSKTHNNKFEQNHEDLFNLNAVGLGSLKKIRVRHTNTGIAPGWFLDYILIRESIEQKKMEYFFPCYQWLSATRLDGLVIREL >LOXHD1_schMan_PLAT:09 TIYHVRIHTGSQSNISSDANVQIQLYGDKDFTGNIRLYKAYYGKEDILVNKFQAGQIANFIVKAINIGNIKKIRIEHDTAVGSARWFVERVEVEARKLGLLWKFECNR >LOXHD1_schMan_PLAT:10 TNYQIKTFTSDISGAETTSSVYIQLYGNDSLPSSIRRLHQNNDSEQRFQRNKIDTFYVELEELQEPFSKLRIWHNDKGSSTDWHLNKVEIRKIKTNQYVFVTYIFPCNKWLSRNMDQAALERELIPS >LOXHD1_schMan_PLAT:11 NTYEVRITTGDKAYAGTDASVYITLFGENGDSGERKLTKSLTHRNKFERGQTDVFQLEIVDLGKINKVRIRHDNSGVNPSWYLSTIEVFNISKSQSVHGLKDQPNETHHLVKYIFNCEAWLSTEHDEKVLDR >LOXHD1_schMan_PLAT:12 IPYLITVITGSDRKAGTPGPVWISCVDKDKMNSEKFILCDCYNRTMLKRGTTRYFRFAGVKLNDLTEIQVGNDAPESPSMGWYIQSLYLSFPTIGKMYLFDCKEWLSTNRGNKK >LOXHD1_schMan_PLAT:13 ITYHLKITTANVKRAGTDCSINLQIFGTNGVTNCYILEKTSNRFAQGITDNISLEMEDVGKLLKMRIGHDNQGKNKHWNLSCVEVTVANTNQLYRFVYDDWLSLTYGKRKSLWADLPAM >LOXHD1_schMan_PLAT:14 IFVKTGNMPASSTDANVYVQLFGEYGDSGEILLKQTVSNQKPFQNNSIDHFKIPSILKLGNLARCRIWHDNKGSSPNWYCEWLEVKEVLIPGEKNLACNWKFAFNKWLSVSDDNKQLLRDAPCS >LOXHD1_schMan_PLAT:15 VYEVVIETGNLKDSGTTCDAWIILEGKHGRSPKLELVNQVGNPILQINQMNTFQLPSFPLGDLETIRLGIQERNINKQTNPNDIQSQKWFCEKVSIKDPVSKRTYIFTIHQWLS >LOXHD1_schMan_PLAT:16 VMYKVSIYTGTKGCANTDANIFITMFSTTPGLNSGRIALKRENNNLFDRKQLDEFYVESIDLESIQRIIIEHDNTGVSPDWYLDKVLITNQTNNQIHLFQCYQWISKKKGDCRLWKELLVS >LOXHD1_monBre_PLAT:01 Monosiga brevicollis (monosiga) GHASLEIFTSKRSNPVSKSEKYITVVGTKGRSDAIQLAAPGVHFRAGNKDVFQVNLTGIGKPTKVILENTGTKRTDGWCVSKVVLVKKTDKGTRRYRFSGPVWLSKHHDEMKLKR >LOXHD1_monBre_PLAT:02 Monosiga brevicollis (monosiga) NGYRIDCYTGDVANAGTDAVATIQLFGSKGQSPMVELRRSDGQAFQRARVATFTLDDLANLGKLKKLVISHNGHGMASGWFLDKIIVTSLSSNKATVFPCDAWLDRKNGRSKELVAR >LOXHD1_monBre_PLAT:03 Monosiga brevicollis (monosiga) TTFTIRVMTGDRRGAGTDANVQCTLFGEDGESGPHTLNTSRNDFRRGHTDVFAVSSRKIGTLKRLRIWHDNGGAGPAWFLDAVEVVDEASGQTYRFECNRWLAKDEDDGQISRELTCN >LOXHD1_monBre_PLAT:04 Monosiga brevicollis (monosiga) KSYKLTIFTGDKRNAGTSANVFCKLVGERGASDNVILENSSKNFQRDRTDIFTVEASDLGSLRHIVLGHDNHGMGAGWYVERFSLEVPSEGKLYNVDVKQWFATDMSDGAIERTFRLD >LOXHD1_monBre_PLAT:05 Monosiga brevicollis (monosiga) LDWKCTIYTSDVANAGTDANVFMQVYGKKGKTDVVPLKNKSDTFERGQTDELRVQLINVGSLRKLRVWHDNKGMASGWHLDRIVLSRDGEEYIFPCAEWLAVSEGDKEIVRELPAT >LOXHD1_monBre_PLAT:06 Monosiga brevicollis (monosiga) VEYTVRVATGHARFAGTNADVFVMLTGELGDSGKRALLRSQTNRNKFERGKEDVFTVAAVDLGKLTSVTVGHNNAGTSAGWFLDKIVVLDPRRGEEEEFPCHRWLAVDADDGQIERELVPK >LOXHD1_monBre_PLAT:07 Monosiga brevicollis (monosiga) TTYIVKIKTGDVRHAGTDANVFVQLFGKTGESTQLKLRNSETYSDAFERNKMDIFKFELLDLGDLSRILVGHDNKGMGAAWFLDYVEVEVPSIRTRWKFPCSRWFSKSQDDGLTER >LOXHD1_monBre_PLAT:08 Monosiga brevicollis (monosiga) APYLFRFYTSDVAFAGTDANVSVVLYGDEGKTEELVVNNQSDNFERGKADDFKLACKPVGRPSKIRLSAHGGGMSADWHLEKIEVHELGQARIYTFEHNDWLRKGTKAKPFMVEL >LOXHD1_monBre_PLAT:09 Monosiga brevicollis (monosiga) RTYRVKVHTGDQKGAGTDANVYVNLHGSLGDSGDRHLKNSLTHTNKFQRKTVDEFDIDAVTLGDINKVKVWHDNAGLGAAWYLEKIEVVDTADDKTYIFPCAQWFAKSMGDGQIAREL >LOXHD1_monBre_PLAT:10 Monosiga brevicollis (monosiga) FKYRISVHTSDVKHAGTDANVDIVLYGEKGDTGKIRLAKSETHRDMWERGNCDVFTVSAIELGDLKRVDIMHDGKGVGSGWHLNKVVVDAPQAGKTWTFMCDAWLDKATDDGTMAK >LOXHD1_monBre_PLAT:11 Monosiga brevicollis (monosiga) VPYEIIIKTSDVRNAGTDANVFIDLYGRDQEERDLTAHHEFKDAVKAHFERNLEDRFNVELPDVGSIYKIRLGHDGKGMSSSWHVASVVVINQRTHERFEFPCDAWLSKDKDDKKLVREFAVG >LOXHD1_monBre_PLAT:12 Monosiga brevicollis (monosiga) AIYKIHVFTGDIKHAGTDANIYVQIFGDTGDSGEIKLEKSETYRDKFERGHEDIFTHRCLDLGPLRKIKVRSDGKGLMGGDWYLDRVEVHQENDLSEPPVRFVCQDWFKRGKQEGDTLEREITAQ >LOXHD1_monBre_PLAT:13 Monosiga brevicollis (monosiga) LEYKVTVYTGTDTQAGTTANVWLQLFGEKDAPLTPAPPSPSKLSRSGSLFGRRRRASSDAQSVSSSLSAGASGPRETSTGRLQLNNAPADLQSGAKTTFTVTGLDVGELVGLEIGHDDERDKWYLEQVVVEVPKSATHAARRYEFKAGVWLAARADGSTSGSAK >LOXHD1_monBre_PLAT:14 Monosiga brevicollis (monosiga) LEYTLKVYTASADGAGCTAVPQVQLFGDKHTTEALPLRAGGDILPASVVETQHRVPDLGALLKVRLMVPRGSSWTVEKVEFGRAGQTPITFVGSDGSAVTLGAERLS >LOXHD1_monBre_PLAT:15 Monosiga brevicollis (monosiga) TSYRVYVTTADERGTGTDANVSIILYGAMGDSGEHSLTKSETFDDPFERGNTDVFTLEVPDLGELQRARIWHDGKGMFSSWKLDKIVVVVEATQSRYELPCGQWLSKNKGDKQLTRDLAVASKR >LOXHD1_monBre_PLAT:16 Monosiga brevicollis (monosiga) GTYKLEVATDSRAGGGCKGPVRIMLLDAEKNQLPLTLEPPGGEFAPGSVEHLVFDNVMLLGPLTELRIRRKPSGASSRRGAEDDDEDDNEASGASSQSGSAVSPWHLEHIIVKHLQSGQSFVFKGPSKGLSRSRAKLSVHTEA >LOXHD1_monBre_PLAT:17 Monosiga brevicollis (monosiga) AQYEVAVTTGTERGAGTDSNVFVTLFGKNGDSGERALAKSKTFRNMFESGNTDVFDVECQDLGELTKIEVKSDLKGFGAAWQLDKIKVTRTGSQNSWQFKCDQWFDKKQGAEHTFSVA
Reference PLAT:07 domains
>PLAT:07_homSap Homo sapiens (human) 2 LVHYEVEIWTGDVGGAGTSARVYMQIYGEKGKTEVLFLSSRSKVFERASKDTFQ 0 0 LEAADVGEVYKLRLGHTGEGFGPSWFVDTVWLRHLVVREVDLTPEEEARKKKEKDKLRQLLKKERLKAKLQRKKKKRKGSDEEDEGEEEESSSSEESSSEEEEMEEEEEEEE FGPGMQEVIEQHKFEAHRWLARGKEDNELVVELVPAGKPGPE 1 >PLAT:07_monDom Monodelphis domestica (opossum) 2 LIHYEVEIVTGDMGNASTSARVYMQIYGEEGKTEVLFLKSRSKVFQRGNTDTFQ 0 0 LQEAEVTPEEEARKKKEMDKLRQLMRKERLKAKEEQKRKRKLAKGSDVEDEEESSSEESSEESEEETEIEEEDEE FQEVVDVYRFHAGRWLATDEEDKDLDMELAPSGRSGPQR 1 >PLAT:07_ornAna Ornithorhynchus anatinus (platypus) 2 LIHYEVEIVTGDMGYAGTNARVYMQIYGELGKTEVLHLTSRTNVFEQGATDTFQ 0 0 LEATDVGEIYKVRLGHSGEGFGSGWFIESLVLKRLVLKEVEPNPEEEKRKAKERERAREQRRKERLKAKQQRKKKKKMKKSSDDEDSEAEDSEEEEGSSEEESSSSSSEEEVEEEFGPGIKEVIDVYKFEAHRWLARDEDDKELIVELEPANRPGPE 1 >PLAT:07_galGal Gallus gallus (chicken) 2 LINYEVSVVTGDVRAAGTNAKVFMQIYGETGKTELIILENRSNNFERGATDIFE 0 0 VEAADVGKIYKIRIGHDGKGIGDGWFLESVTLKRLATKMDETDKKKKKKKKKSEEEEEEEETKVEEVMDVYTFVAHRWLAKDEGDKELVVELVPDGESELE 1 >PLAT:07_taeGut Taeniopygia guttata (finch) 2 LINYEVSVVTGDVRAAGTNAKVFMQIYGETGKTELIILENRSNNFERGATDIFK 0 0 REAADVGKIYKIRIGHDGTGIGDGWFLESVTLKRLATKTEGSDKKKKKKKKSEEEETKEEEGMDVYTFVAHRWLAKDEGDKELVVELVPDGESDLE 1 >PLAT:07_anoCar Anolis carolinensis (lizard) 2 LINYEVCVATGDVRNAGTNANVFMQIYGELGKTELLILKNRSNNYERGASETFR 0 0 VEAVDVGKVYKIRIGHDGKGFGDGWFLDSVVVKKLPTKVPKKKKKKKKKKTPEEEEAEEGPGIMEVYNFTPCRWLASDEEDKELVVELVPDEGSELE 1 >PLAT:07_oryLat Oryzias latipes (medaka) 2 VINYEIAVVTGDVRAGGTNASVFCQIYGEEGKTEVLNLKSRSNNFERGTTEIFK 0 0 IEALDVGKIYKIRIYHDGSGIGDGWFLETVDIKRLTMALVQVEVKKEEAPKKDKKKDKKKKKKEEEEVEIIEEMQEVVETFTFTCNRWLARDEEDGEIVVELLTEENEDLE 1 >PLAT:07_gasAcu Gasterosteus aculeatus (stickleback) 2 VINYEVTVVTGDVTFAGTNARVIIQIYGDKGKTEVIILESRSNNYERNTTEIFK 0 0 IEAKDVGKIFKIRIGHDGSGIGSGWFLETVDVKRLILALVPKEKKKEDKKKKKKKKEDVDEEGGEEMQEVVLTYSFPCSRWLAGGEEDGELVVELLPDDAKELE 1 >PLAT:07_takRub Takifugu rubripes (fugu) 2 VINYEVTVVTGDVTFAGTNAKVFVQIYGDKGKTEVIMLESRSNNYERNAMEIFK 0 0 IEAKDVGKIFKIRIGHDGLGIGSGWFLEKVYVKHLIMALVPRENKKDDKKKKKKKKKDKEDEEEVGGEEMQEVVVTYHFPCSRWLASGEDDDDLVVELLPEDAEELE 1 >PLAT:07_petMar Petromyzon marinus (lamprey) 2 GINYQVSVHTGNVRSAGTNANVFIQLYGDMGKTEVHNLRNRSDNFERASNDIFK 0 0 VEAMDVGKVVKLRVGHDNSGMGSGWFLDSIVIRRLRQSSPHRPQPVDAEEDEDEEDDEEAEDEDSEEVQTYTFPCKRWLARDEDDGEIVRELLPQDCAEME 1 >PLAT:07_sacKow Saccoglossus kowalevskii (acornworm) Bilateria|Deuterostomia|Hemichordata 2 GIPYEITVITGDKSGAGTDAQVFIRMYGLKGKTDEFILNNRTDNFERGMTDKFK 0 0 IEAADVAMLTKIRIGHDNSGRSAGWYLERVIIERFPPKRKMKRKRSGTPRRRGEYDEEDYDDIPETNVVNFVCNRWFAKDEEDHQIVRELLPTDEEALKGH 1 >PLAT:07_strPur Strongylocentrotus purpuratus (sea_urchin) 2 GIPYEITVITGDKSGAGTDANVFLTMYGDDGAKTEEFSLRNRTDNFEKNMTDKFK 0 >PLAT:07_triAdh Trichoplax adhaerens (placozoan) Eukaryota|Metazoa|Placozoa 2 KIPYQISVHTGDIRHAGTDSNVFAVIYGENGKTEELKLRNKSDNFERGQVDVFK 0 0 VECEDVGKLRKLRIGHDSAGMGSAWFLDK 00 VYVRRLPPKSGKKSKETDEREEETAKKDADEPEKLDENNYLFVANRWLSKEEGDRQTVIEISPVGVDGALA 1 >PLAT:07_monBre Monosiga brevicollis (choanoflagellate) Eukaryota|Choanoflagellida 2 PMEEDVSAPYLFRFYTSDVAFAGTDANVSVVLYGDEGKTEELVVNNQSDNFERGKADDFK 0 0 LACKPVGRPSKIRLSAHGGGMSADWHLEKIEVHELGQARIYTFEHNDWLRKGTKAKPFMVELPLRRIETVDDNGREVVEELALDANKRTYR 1