Gene Set Summary Statistics: Difference between revisions
From genomewiki
Jump to navigationJump to search
| Line 72: | Line 72: | ||
<TR><TH>db</TH><TH COLSPAN=5>gene name (CDS extent size: thickEnd-thickStart)</TH></TR> | <TR><TH>db</TH><TH COLSPAN=5>gene name (CDS extent size: thickEnd-thickStart)</TH></TR> | ||
<TR><TH>hg17</TH> | <TR><TH>hg17</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr7:145251477-147555734 NM_014141] (2298740)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX:30897002-33117383 NM_000109] (2217347)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr11: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chr11:82846625-85015962 CR749820] (2138880)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX:30897002-32989330 NM_004006] (2089394)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg17&position=chrX:30898403-32989184 X14298] (2089394)</TD> | ||
</TR> | </TR> | ||
<TR><TH>hg18</TH> | <TR><TH>hg18</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr7: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr7:145444386-147749019 uc003weu.1] (2298740)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chrX:31047266-33267647 uc004ddb.1] (2217347)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr11: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr11:82843701-85015962 uc001pak.1] (2138880)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chrX:31047266-33139594 uc004dda.1] (2089394)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr8: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg18&position=chr8:2782789-4839736 uc003wqd.1] (2055833)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm8</TH> | <TR><TH>mm8</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chrX:79201622-81457760 NM_007868] (2253366)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr6:44989695-47230955 NM_001004357] (2238304)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2:40418782-42475607 NM_053011] (2055883)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr2:140086871-142081491 AK134694] (1988713)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr8: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm8&position=chr8:15895480-17535258 NM_053171] (1639258)</TD> | ||
</TR> | </TR> | ||
<TR><TH>mm9</TH> | <TR><TH>mm9</TH> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chrX: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chrX:80194242-82450380 uc009tri.1] (2253366)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr6: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr6:45010087-47251368 uc009bst.1] (2238325)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr16: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr16:39984484-42176405 uc007zfr.1] (2189582)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:40452293-42509118 uc008jon.1] (2055883)</TD> | ||
<TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2: | <TD>[http://genome.ucsc.edu/cgi-bin/hgTracks?db=mm9&position=chr2:140221166-142215786 uc008mpv.1] (1988713)</TD> | ||
</TR> | </TR> | ||
</TABLE> | </TABLE> | ||
Revision as of 17:36, 19 September 2007
gene sets measured
- hg17 - knownGenes version 2
- hg18 - knownGenes version 3
- mm8 - knownGenes version 2
- mm9 - knownGenes version 3
The min, max and mean measurements are per gene
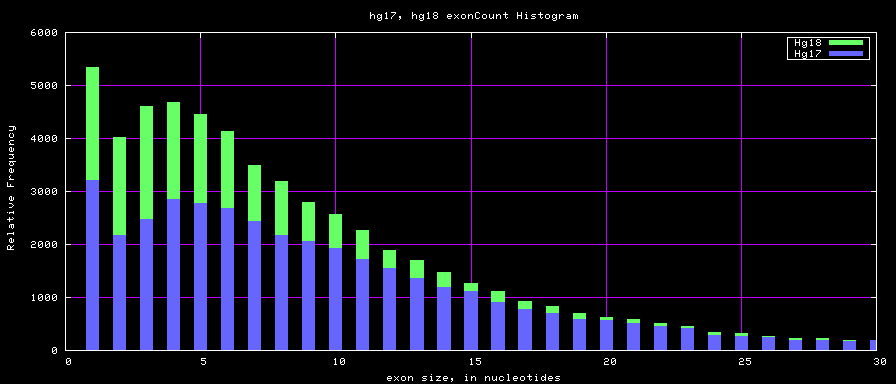
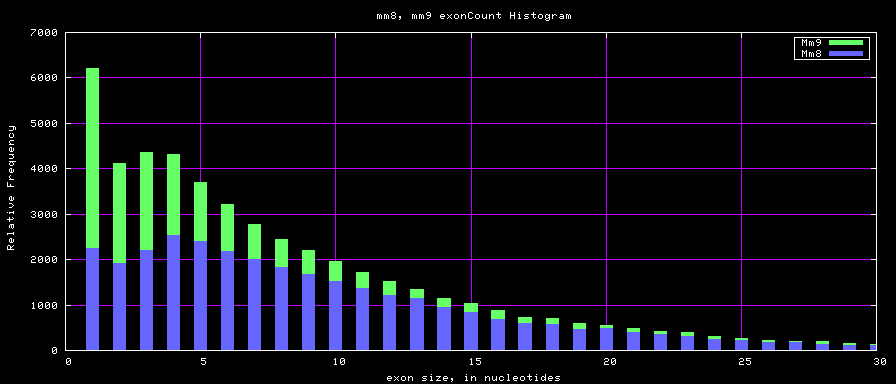
summary of gene and exon counts
| db | gene count | total exon count | min exon count | max exon count | mean exon count |
|---|---|---|---|---|---|
| hg17 | 39368 | 405720 | 1 | 149 | 10 |
| hg18 | 56722 | 519308 | 1 | 2899 | 9 |
| mm8 | 31863 | 314628 | 1 | 313 | 10 |
| mm9 | 49220 | 417114 | 1 | 610 | 8 |
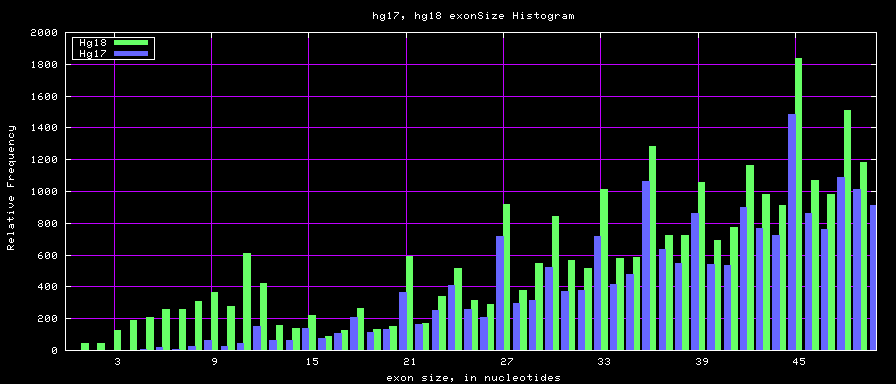
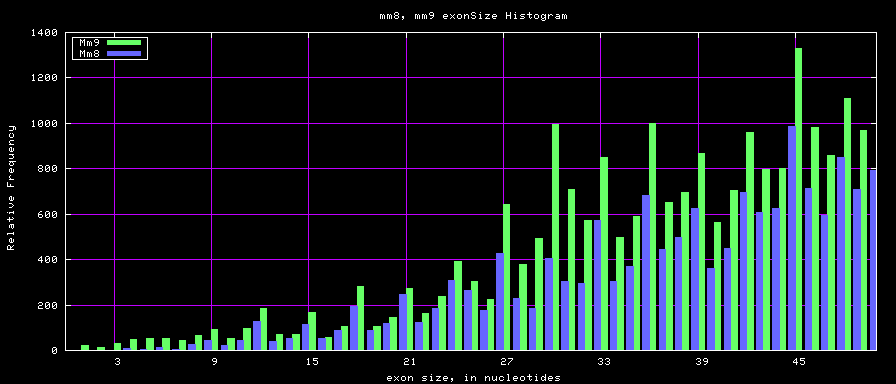
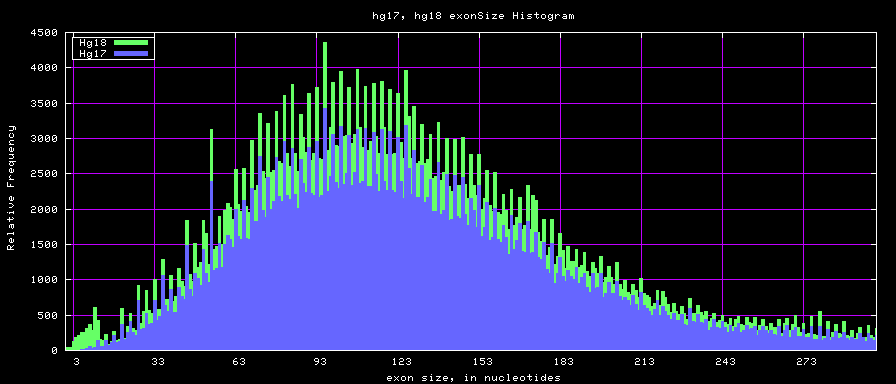
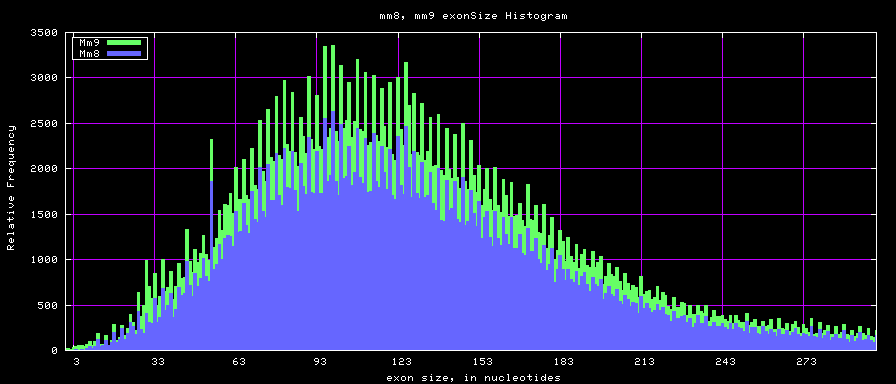
summary of exon size statistics
| db | sum exon sizes | min exon size | max exon size | mean exon size |
|---|---|---|---|---|
| hg17 | 106839720 | 1 | 18172 | 263 |
| hg18 | 146371091 | 1 | 36861 | 282 |
| mm8 | 83159087 | 4 | 17497 | 264 |
| mm9 | 117671086 | 1 | 29698 | 282 |
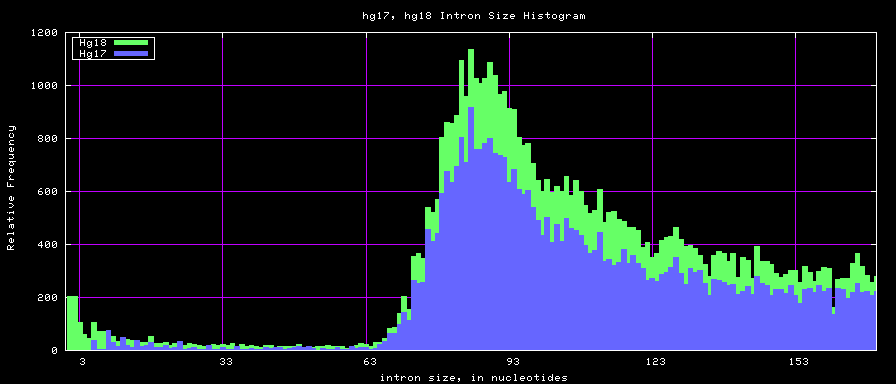
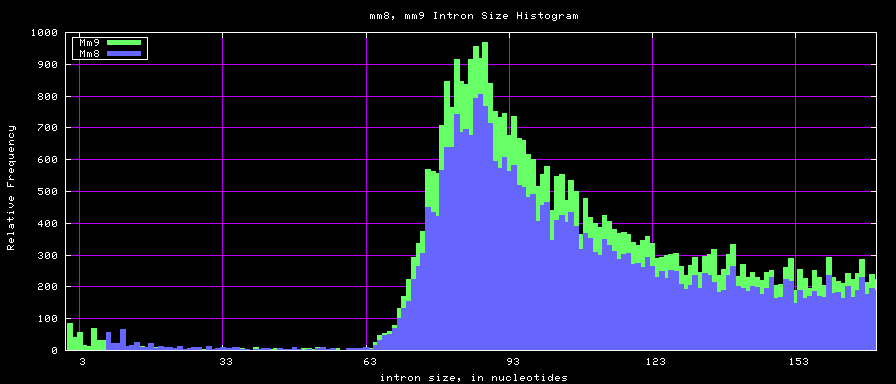
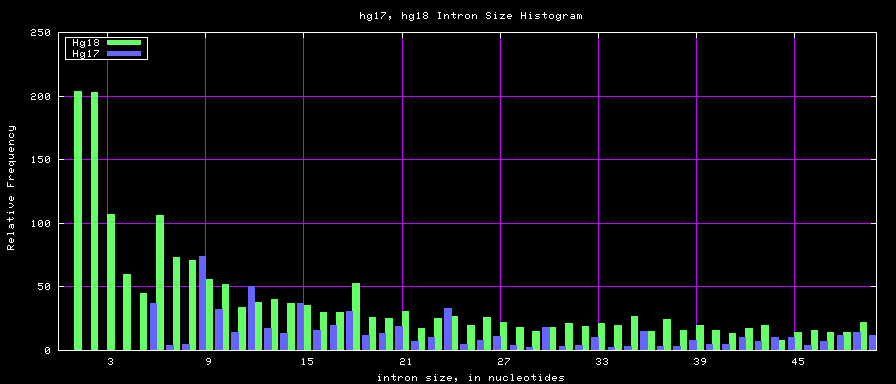
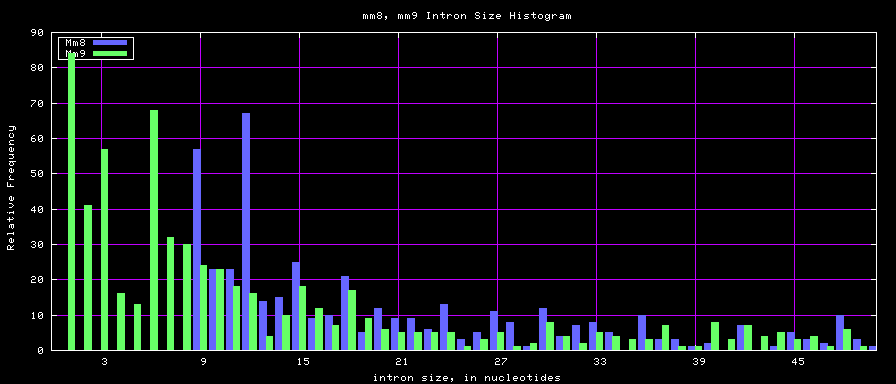
summary of intron size statistics
| db | sum intron sizes | min intron size | max intron size | mean intron size |
|---|---|---|---|---|
| hg17 | 2223224397 | 6 | 1096450 | 6069 |
| hg18 | 2784923600 | 1 | 1047320 | 6023 |
| mm8 | 1476081990 | 9 | 1347550 | 5220 |
| mm9 | 2055504784 | 1 | 1253430 | 5589 |
Top five exon count genes
| db | gene name (exon count) | ||||
|---|---|---|---|---|---|
| hg17 | NM_004543 (149) | AF535142 (146) | AF535142 (146) | NM_033071 (146) | AF495910 (146) |
| hg18 | uc001yrq.1 (2899) | uc002zvw.1 (322) | uc002umr.1 (313) | uc002stk.1 (217) | uc002umt.1 (194) |
| mm8 | NM_011652 (313) | NM_028004 (192) | NM_007738 (118) | NM_134448 (99) | DQ067088 (99) |
| mm9 | uc007pgj.1 (610) | uc008kfn.1 (313) | uc008kfo.1 (192) | uc008jqv.1 (157) | uc009rrh.1 (118) |
Top five largest CDS extent genes
| db | gene name (CDS extent size: thickEnd-thickStart) | ||||
|---|---|---|---|---|---|
| hg17 | NM_014141 (2298740) | NM_000109 (2217347) | CR749820 (2138880) | NM_004006 (2089394) | X14298 (2089394) |
| hg18 | uc003weu.1 (2298740) | uc004ddb.1 (2217347) | uc001pak.1 (2138880) | uc004dda.1 (2089394) | uc003wqd.1 (2055833) |
| mm8 | NM_007868 (2253366) | NM_001004357 (2238304) | NM_053011 (2055883) | AK134694 (1988713) | NM_053171 (1639258) |
| mm9 | uc009tri.1 (2253366) | uc009bst.1 (2238325) | uc007zfr.1 (2189582) | uc008jon.1 (2055883) | uc008mpv.1 (1988713) |
Top five smallest transcript genes
| db | gene name (transcript size: txEnd-txStart) | ||||
|---|---|---|---|---|---|
| hg17 | AF241539 (168) | AF277175 (176) | AY459291 (240) | AY605064 (243) | AF503918 (258) |
| hg18 | uc004buj.1 (20) | uc001dcm.1 (22) | uc001seo.1 (22) | uc001sqn.1 (22) | uc002wpa.1 (22) |
| mm8 | AJ319753 (217) | BC107019 (231) | BC016221 (286) | NM_130876 (303) | NM_130873 (304) |
| mm9 | uc007bma.1 (22) | uc007gmr.1 (22) | uc007khz.1 (22) | uc007pay.1 (22) | uc007qpn.1 (22) |
Custom Track of Small Exons and Introns
Custom track: Hg18 small exons and introns on the UCSC Genes track
These are exons of size less than 22 bases, and introns of size less than 12 bases. The score column contains the size and thus you can filter smaller subsets via the score column in the table browser.
These small exons and introns are used to maintain frame coding boundaries as found in mRNAs compared to the reference genome coordinates.
Histogram graphs
Methods
- From the table browser, request three different bed files for the knownGenes track:
- whole gene
- exons only
- introns only
- From those bed files, stats can be extracted
- gene count from: 'wc -l wholeGene.bed'
- exon count stats from:
STATS=`ave -col=10 wholeGene.bed -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
COUNT=`echo $STATS | cut -d' ' -f8 | awk '{printf "%d", $1}'`
- for exon or intron size stats:
STATS=`awk '{print $3-$2}' {introns,exons}.bed \
| ave -col=1 stdin -tableOut | grep -v "^#"`
MIN=`echo $STATS | cut -d' ' -f1`
MAX=`echo $STATS | cut -d' ' -f5 | awk '{printf "%d", $1}'`
MEAN=`echo $STATS | cut -d' ' -f6 | awk '{printf "%d", $1+0.5}'`
SUM_SIZE=`awk '{sum += $3-$2} END{printf "%d", sum}' {introns,exons}.bed`
- top five exon count genes
sort -k10nr wholeGene.bed | head -5
- top five CDS size genes
awk '{cdsSize=$8-$7
if (cdsSize > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,cdsSize}
}' wholeGene.bed | sort -k5nr | head -5
- top five smallest transcript genes
awk '{size=$3-$2
if (size > 0) {printf "%s\t%s\t%s\t%s\t%d\n", $1,$2,$3,$4,size}
}' wholeGene.bed | sort -k5n | head -5