CDH23 SNPs: Difference between revisions
Tomemerald (talk | contribs) |
Tomemerald (talk | contribs) |
||
| Line 21: | Line 21: | ||
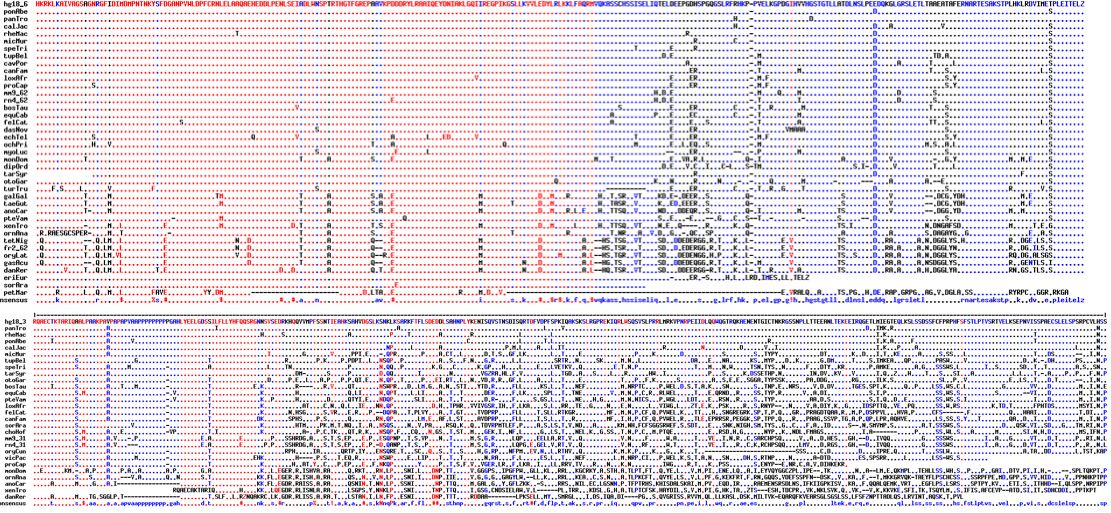
Orthologs of CDH23 are available from 42 vertebrates in the exon containing L1122V. The following exon is quite short, therefore difficult to obtain by blast and transcripts are uncommon so deep in a gene. An unusual GC-AG phase 0 intron separates these exons and may help identify ancient orthologs. Residue 1122 lies in a linker region between two adjacent cadherin domains but to model this region it will [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=18395225 prove necessary] to model consecutive globular domains as well. | Orthologs of CDH23 are available from 42 vertebrates in the exon containing L1122V. The following exon is quite short, therefore difficult to obtain by blast and transcripts are uncommon so deep in a gene. An unusual GC-AG phase 0 intron separates these exons and may help identify ancient orthologs. Residue 1122 lies in a linker region between two adjacent cadherin domains but to model this region it will [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=18395225 prove necessary] to model consecutive globular domains as well. | ||
Observe that while leucine is sometimes found at this position in other species, that | Observe that while leucine is sometimes found at this position in other species, that occurrence is concentrated strictly in early diverging vertebrates. In all 33 species of tetrapods (where sound is conducted primarily through air), the value here is exclusively valine. Note in particular the four other species of great apes have valine with no indication of heterozygosity. | ||
From this perspective, L1122V may reflect retention of the ancestral value in one allele, rather than result from de novo back mutation from a L1122L homozygote. In other words, L1122 could be viewed as a mutation apparently fixed for better or worse in all other human populatons -- at a position conserved over billions of years of branch length in phylogenetically related species. | From this perspective, L1122V may reflect retention of the ancestral value in one allele, rather than result from de novo back mutation from a L1122L homozygote. In other words, L1122 could be viewed as a mutation apparently fixed for better or worse in all other human populatons -- at a position conserved over billions of years of branch length in phylogenetically related species. | ||
Dozens of disease alleles are known, for example D124G, P240L, E247K, R301Q, A366T, N452S, L480Q, R582Q, H755Y. These often directly disrupt a Ca+2 binding motif (thus in EC1 at 124: DVNDNAPTF --> GVNDNAPTF) so demonstration of residue phylogenetic conservation would support that criterion in the analysis of L1122V. | |||
[http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=18395225 Posy et al] observe cadherin Ca2+ binding domains like DxNDN are located in a linker region so cannot be clearly associated with one of the folded beta-sheet domains, observing further that the domain definition at SMART v.34 omits the N-terminal region critical to EC domains (A*, A and part of B strands) whereas Pfam35 drops the first two. These tools also err in defining calx-beta domains important to vlgr1 binding to usherin. | |||
To see if the domain swap dimer model is applicable to link EC1 of PCDH15 to EC1 of CDH25 -- it seems not because the conserved tryptophan is not present -- it is preferable to associate the Ca2+ binding inter-domain residues with the domain from which they originate but not include residues more naturally part of the following domain. Note the first 3 EC regions of human CDH23 lack any cysteine so a disulfide linkage can be ruled out. | |||
<1......................exon 26.................0> <0..exon 27.......1> <2...............exon 28........................0> | <1......................exon 26.................0> <0..exon 27.......1> <2...............exon 28........................0> | ||
Revision as of 03:41, 24 August 2009
CDH23 SNPs
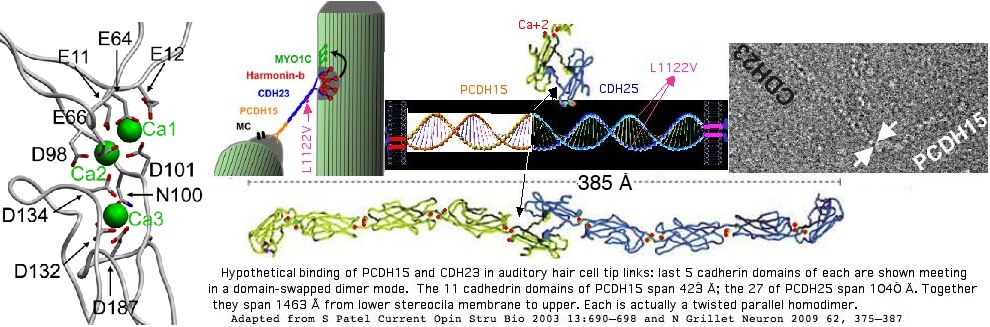
CDH23 (cadherin 23) on 10q22.1 is one of the better understood genes of the Usher disease complex. These genes generally encode structural proteins utilized in both hearing and visual systems -- and so at the mutational level by effects on both. Stop codons within CDH23 cause both deafness and blindness (USH1D) whereas missense alleles can affect hearing only (DFNB12). Both conditions are autosomal recessive. However one bad copy of CDH23 in conjunction with one bad allele of PCDH15, protocadherin 15 on 10q21.1 (17 million bp over, not tandem) can perhaps give rise to digenic disease USH1H. That could have a simple physical explanation in defective heteroligomeric binding of the two terminal domains where the respective cSNPs occur.
Many Usher genes function both transiently during development of cochlea and retina and permenantly in adult structures. These functions may localize to multiple sites within each organ, for example ribbon synapses and stereocilia. CDH23, like many of these proteins, has different binding partner issues in cytoplasmic (USH1C harmonin, MYO7A myosin, USH1G sans) versus extracellular and transmembrane domains. Other unrelated cell types elsewhere in the body may use these gene products (or particular splice variants) though mutant alleles manifest most sensitively in hearing and vision (where mouse serves erratically as human disease model). The role of CDH23 in hair tip links has recently been disentangled from its transient but critical role in hair cell development.
However some coding variants of CDH23 are simply near-normal (or even adaptive) polymorphic variants not giving rise to problems during the carrier's lifespan, though subtle subclinical effects on age related (or noise-induced) hearing loss or night vision acuity might still occur. In the past, such variations would be occasionally be detected within geneologies of affected indiviuals but not track with their disease; today, coding SNPs are far more likely to emerge -- and in far greater numbers -- simply in the course of genomic screening. That trend will only accerate with the advent of rapid screening platforms such as Nimblegen that can affordably screen the entire human proteome.
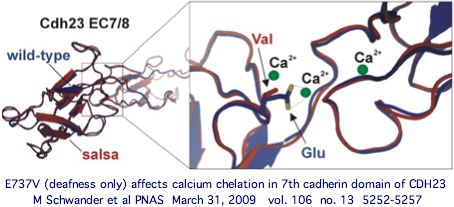
Note these myriad new cSNPs needing interpretation will come with accurate population frequencies further stratified by ethnic group distribution. That can be viewed as 'close-up' comparative genomics that complements the longer view of reduced alphabet afforded currently by CDH23 orthologs in 50-odd vertebrate genome phylogenetic tree. These considerations, along with accurate 3D models of both the cadherin module affected and protein binding partner, greatly help in interpreting disease implications of particular observed SNPs (for example E737V), yet uncertainty will remain in many instances.
Here a newly observed cSNP in a Kalahari Bushmen, heterozygous L1122V in exon 26, lies fall just before the boundary of the 11th of 27 cadherin ectodomains of the 3354 residue, 67 exon protein. This would appear unremarkable except for the observation that valine is the ancestral mammalian value here and it is conserved over a vast phylogenetic time scale.
It does not suffice to consider the structual impact of L1122V in isolation (say by modeling the two adjacent cadherin domains and the intervening region). Even though CDH23 is highly extended (meaning other cadherin domains are irrelevent to L1122V), it forms a parallel dimer in hair tip links implying a side-by-side uncharacterized interaction with a second copy of the relevent L1122V-containing domains. Leucine, valine and isoleucine are hydrophobic residues often important to tight packing of globular fold interiors (where they are often not interchangeable) but also can occur similarly occluded in dimer surfaces patches.
The current view of the placement of this residue in auditory hair cell stereocilia tip links involves the relationship between PCDH15 and CDH25 as shown below. Twisted homodimers of each form before they meet at their special first cadherin domain to form the two domain-swapped dimers because both proteins are anchored by transmembrane regions and cytoplasmic interactions. If PCDH15 continues the twist of CDH25 (say clockwise viewed N to C, then the net result is neither homodimer can unravel. However if a substitution weakens the initial twisting, the connection might not stably form.
Comparative genomics
Orthologs of CDH23 are available from 42 vertebrates in the exon containing L1122V. The following exon is quite short, therefore difficult to obtain by blast and transcripts are uncommon so deep in a gene. An unusual GC-AG phase 0 intron separates these exons and may help identify ancient orthologs. Residue 1122 lies in a linker region between two adjacent cadherin domains but to model this region it will prove necessary to model consecutive globular domains as well.
Observe that while leucine is sometimes found at this position in other species, that occurrence is concentrated strictly in early diverging vertebrates. In all 33 species of tetrapods (where sound is conducted primarily through air), the value here is exclusively valine. Note in particular the four other species of great apes have valine with no indication of heterozygosity.
From this perspective, L1122V may reflect retention of the ancestral value in one allele, rather than result from de novo back mutation from a L1122L homozygote. In other words, L1122 could be viewed as a mutation apparently fixed for better or worse in all other human populatons -- at a position conserved over billions of years of branch length in phylogenetically related species.
Dozens of disease alleles are known, for example D124G, P240L, E247K, R301Q, A366T, N452S, L480Q, R582Q, H755Y. These often directly disrupt a Ca+2 binding motif (thus in EC1 at 124: DVNDNAPTF --> GVNDNAPTF) so demonstration of residue phylogenetic conservation would support that criterion in the analysis of L1122V.
Posy et al observe cadherin Ca2+ binding domains like DxNDN are located in a linker region so cannot be clearly associated with one of the folded beta-sheet domains, observing further that the domain definition at SMART v.34 omits the N-terminal region critical to EC domains (A*, A and part of B strands) whereas Pfam35 drops the first two. These tools also err in defining calx-beta domains important to vlgr1 binding to usherin.
To see if the domain swap dimer model is applicable to link EC1 of PCDH15 to EC1 of CDH25 -- it seems not because the conserved tryptophan is not present -- it is preferable to associate the Ca2+ binding inter-domain residues with the domain from which they originate but not include residues more naturally part of the following domain. Note the first 3 EC regions of human CDH23 lack any cysteine so a disulfide linkage can be ruled out.
<1......................exon 26.................0> <0..exon 27.......1> <2...............exon 28........................0>
<-----------EC10-------------><---interdomain----- -><----------------- -------------------------EC11-------------------->
....................Ca+2........................^. ....Ca+2............ ..................................................
CDH23_homSap DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSILQ LKATDADEGEFGRVWYRILH GNHGNNFRIHVSNGLLMRGPRPLDRERNSSHVLIVEAYNHDLGPMRSSVR
CDH23_panTro DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_gorGor dNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILR
CDH23_ponAbe DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASIPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_nomLeu DNGPVGKRHTGTATVFITVLDVNDNRPIFLQSSYEASIPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_rheMac DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_calJac DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILQ
CDH23_tarSyr DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_micMur DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILR
CDH23_musMus DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_ratNor DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_cavPor DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_speTri DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_oryCun DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LRATDADEGEFGRVWYRILH
CDH23_ochPri DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIVEGHSIVQ LRATDADEGEFGRVWYRILH
CDH23_bosTau DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVSEDIPEGHSIVQ LKATDADEGEFGRVWYRIVH
CDH23_canFam DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKAADADEGEFGRVWYRILH
CDH23_felCat DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_pteVam DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LMATDADEGEFGRVWYRILH
CDH23_turTru DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LTATDADEGEFGRVWYRILH
CDH23_susScr DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ
CDH23_equCab DNGPVGKRRTGTATVFITVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILQ
CDH23_eriEur dNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_loxAfr DNGPVGKRRTGTTTVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGLVWYRILH
CDH23_proCap DNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASIPEDIPEGHSIVQ LKATDADEGEFGRVWYRILR
CDH23_echTel dNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSIVQ LKATDADEGEFGRVWYRILH
CDH23_choHof dNGPVGKRRTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGRSIVQ
CDH23_monDom DNGPVGKRRTGTATIYVTVLDVNDNRPIFLQSSYEASVPEDIPEGSSIVQ LMATDADEGDNGRVWYRILH
CDH23_macEug DNGPVGKRRTGTATVYVTVLDVNDNRPIFLHSSYEASISEDIPEGSSIVQ LMATDADEGDNGRVWYRILH
CDH23_ornAna DNGPSGKRRTGTATVYVTVLDVNDNRPIFLQSSYEASVPEDIPEASSIVQ LKATDADEGEYGRVWYRIIS
CDH23_galGal DNGPTGNRRTGTATVYVTVLDVNDNRPIFLQSSYEASVPEDIPAASSIVQ VKATDADEGVNGRVWYRIVK
CDH23_taeGut DNGPSGNRRTGTATVYVTVLDVNDNRPIFLQSSYEVSVPEDIPAASSIVQ VKATDADEGINGRVWYRIVK
CDH23_anoCar DNGPTGKRRTGTATVHVTVLDVNDNRPYFLQSSYEATVPEDIPDYSSIVQ VKATDADEGINGRVWYRIVK
CDH23_xenTro DNGPAGNRKTGTATVSVTVLDINDNKPIFLKSSYEASVPENVPFSSSIVQ LEATDADEGDNGLVWYRILS
CDH23_oryLat DNGPAGSRRTGTATVFVEVLDVNDNRPIFLQNSYETSVLETVPQGTSILQ VQATDADQGENGRVLYRILS
CDH23_takRub DNGPAGSRRTGTATVFVEVQDVNDNRPIFLQNSYETGILESVPQGTSILQ VQATDADQGENGRVLYRILT
CDH23_danRer DNGPAGGRRTGTATVYVEVLDVNDNRPIFLQNSYETSVLENIPRGTSILQ VQATDADQGENGKVLYRILS
CDH23_gasAcu DNGPAGSRRTGTATVFVEVQDVNDNRPIFLQNSYETSILESVPQRTSILK VQATDADQGENGKVLYRILT
CDH23_tetNig DNGPAGSRRTGTATVFVEVQDVNDNRPIFLQNSYETSVLESVPQGTSILQ VQATDADQGENGSVLYRILT
CDH23_ictPun DNGPAGDRKTGTATVYVEVLDVNDNRPIFLQNSYETTVLENVPRGSSVLQ
CDH23_calMil DNGPAGSRRTGTATVYIRVLDVNDNRPIFLQNTYEASVPENITMSTSILQ VSATDADTGQNGRLTYQILQ
CDH23_petMar DHGPAGSRRTGTTTLDVLVLDVNDNRPLFLEGSYZVSVPDNVTRGAIFLQ
................................................^.
CDH23_homSap DNGPVGKRHTGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSILQ
CDH23_panTro ................................................V.
CDH23_gorGor ................................................V.
CDH23_rheMac ................................................V.
CDH23_calJac ................................................V.
CDH23_pteVam ................................................V.
CDH23_ponAbe .....................................I..........V.
CDH23_nomLeu ................I....................I..........V.
CDH23_tarSyr ........R.......................................V.
CDH23_micMur ........R.......................................V.
CDH23_musMus ........R.......................................V.
CDH23_ratNor ........R.......................................V.
CDH23_cavPor ........R.......................................V.
CDH23_speTri ........R.......................................V.
CDH23_oryCun ........R.......................................V.
CDH23_canFam ........R.......................................V.
CDH23_felCat ........R.......................................V.
CDH23_turTru ........R.......................................V.
CDH23_susScr ........R.......................................V.
CDH23_echTel ........R.......................................V.
CDH23_eriEur ........R.......................................V.
CDH23_proCap ........R............................I..........V.
CDH23_equCab ........R.......I...............................V.
CDH23_loxAfr ........R...T...................................V.
CDH23_choHof ........R....................................R..V.
CDH23_bosTau ........R.............................S.........V.
CDH23_ochPri ..........................................V.....V.
CDH23_monDom ........R.....IY.............................S..V.
CDH23_macEug ........R......Y..............H......IS......S..V.
CDH23_ornAna ....S...R......Y............................AS..V.
CDH23_galGal ....T.N.R......Y...........................AAS..V.
CDH23_taeGut ....S.N.R......Y...................V.......AAS..V.
CDH23_anoCar ....T...R......H...........Y........T......DYS..V.
CDH23_xenTro ....A.N.K......S.....I...K....K.........NV.FSS..V.
CDH23_oryLat ....A.S.R........E.............N...T..L.TV.Q.T....
CDH23_takRub ....A.S.R........E.Q...........N...TGIL.SV.Q.T....
CDH23_tetNig ....A.S.R........E.Q...........N...T..L.SV.Q.T....
CDH23_gasAcu ....A.S.R........E.Q...........N...T.IL.SV.QRT...K
CDH23_danRer ....A.G.R......Y.E.............N...T..L.N..R.T....
CDH23_ictPun ....A.D.K......Y.E.............N...TT.L.NV.R.S.V..
CDH23_calMil ....A.S.R......YIR.............NT.......N.TMST....
CDH23_petMar .H..A.S.R...T.LD.L.........L..EG..ZV...DNVTR.AIF..
Consensus .n..a...r...a.v..t.l.......i..qs..#as!p#.!p.g.siv.
................................................^.
Comparative anatomy
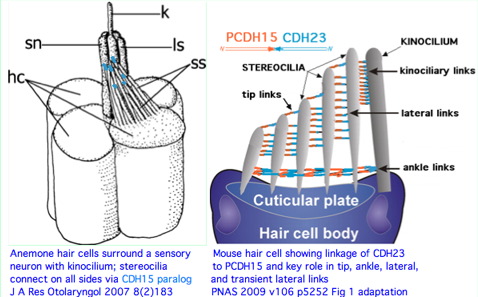
The remarkable auditory hair cell linker provided by CDH23 and PCDH15 is not a vertebrate innovation. Instead it must date back to the pre-bilateran ancestor because contemporary cnidarians such as Nematostella have a very similar overall structure that incorporates a strong homolog to CDH23. This cannot be plausibly attributed to convergent evolution given the extent of structural agreement of kinocilium, sterocilia, lateral and tip links. Note in mouse that the link is polarized with PCDH15 attached to the shorter sterocilium and CDH23 to the longer.
The Nematostella protein most resembling CDH23 has 6,074 residues, three transmembrane helices and 44 contiguous cadherin ectodomains with 4x-periodicity. Thus the correspondence at the protein level is imperfect. However antibodies show it is distributed on stereocilia of anemone hair bundles and required for tentacle sensitivity to vibration (prey detection). It provides both lateral and tip linkages. It can be predicted to form a coiled parallel homodimer like mammalian CDH23.
Nematostella also has long but weak matches to PCDH15, namely XM_001638202 and EU289217. However upon back-blast to human, these do not quite pull up PCDH15 as best match but rather its closest paralog FAT4 (unsurprising given its 34 cadherin, 6 EGF and 2 lamG domains), perhaps because of lineage-specific expansion or because the blast score is inflated. Possibly CDH23 had not yet undergone duplication and divergence to protocadherins and it alone may play a double homodimer linker role in anemone stereocilia.
Nematocyst discharge is sensitive to calcium levels and streptomycin (like vertebrate mechanotransduction channels) but is insensitive to the MET channel blocker amiloride. The channel itself has not been identified in anemone either.
Prey capture can result in signficant trauma to anemone tenacle hair bundles but this can be repaired using a protein again with similarities to a vertebrate stereocilia repair protein ARL5B which acts on the extracellular face of the plasma membrane along stereocilia in the vicinity of tip links. Human and cnidarian protein XM_001629283 are 77% identical:
homSap ARL5B MGLIFAKLWSLFCNQEHKVIIVGLDNAGKTTILYQFLMNEVVHTSPTIGSNVEEIVVKNTFLMWDIGGQESLRSSWNTYYSNTEFIILV
MGL+FAK +S F N+EHKVIIVGLDNAGKTTILYQFLMNEVVHTSPTIGSNVEEIV KNHF+MWDIGGQESLRS+WNTYY+NTEF+ILV
nemVec repair MGLLFAKFFSWFSNEEHKVIIVGLDNAGKTTILYQFLMNEVVHTSPTIGSNVEEIVWKNIHFIMWDIGGQESLRSAWNTYYTNTEFLIL
homSap ARL5B HVDSIDRERLAITKEELYRMLAHEDLRKAAVLIFANKQDMKGCMTAAEISKYLTLSSIKDHPWHIQSCCALTGEGLCQGLEWMTSRI
+DS DRERLAI+K ELY+MLA+E+L++AA+ LI ANKQD+KG M+ AEIS+L L+ IKDH WHIQ+CCALTGEGL QGLEW+T+++
nemVec repair VIDSTDRERLAISKAELYQMLANEELKQAALLILANKQDIKGSMSVAEISEQLNLACIKDHGWHIQACCALTGEGLYQGLEWITTQL
Note 'stereocilia' is an anatomical misnomer. These are instead actin-based membrane protrusions. It is the kinocilium that is a true cilium in both anemone and developing vertebrate hair cells. If parallels to ciliary photoreceptors are sought, these should be with the kinocilium rather than stereocilia. Since no known counterpart to the PCDH15-CDH23 linker occurs in vision, the commonality (Usher syndrome) may reside primarily in ribbon synapses of auditory and photoreceptive neurons.
Pseudogene and paralog issues
No potential exists here for mis-determining orthologous exons, even in remote species such as lamprey with poor assemblies. Exceedingly long genes such as this are not well-represented as retrogenes (which begin 3' and truncate early). Position 1122 is too remote from the 3' terminus. Relevent pseudogenes are not observed by Blat of human, macaque, and dog. Indeed, this exon gives a unique match at this level of sensitivity, even though cadherins are very widespread in the proteome.
The top matches to CDH23 within the human genome are shown as provided by GeneSorter at UCSC. Observe the established binding partner PCDH15 is by no means the best match; the e-value is high but only because of many weak alignments of tandem cadherin domains.
PCDH15 has 11 cadherin domains and a transmembrane region C-terminally. It's problematic to call it a paralog of CDH23 even though all cadherin domains must ultimately coalesce. PCDH15 tandem domains, which cannot be put in 1:1 correspondence with the 27 cadherin domains of CDH23, might have had quite a different history of sources in its domain histories. The evolution of these gene families might be better worked out using interdomain spacer regions and intron position and phasing. Particular regions can be very conserved in themselves while not display much conservation between spacers.
Here the spacer region of CDH23 containing L1122 has best match within the human proteome to PCDHB14 (protocadherin beta 14), far down on the list of overall best matches. The interdomain region is shown in blue below. Note the best matches internally to other spacers are quite weak and neither L nor V is conserved in them.
PCDH15 S-TLTLAIKVLDIDDNSPVFTNSTYTVLVEENLPAGTTILQIEAKDVDLG---ANVSYRIRS
+ | |+ + |||++|| |+| |+| | |++| | +|||++| | | | | |||
CDH23 TGTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSILQILKATDADEGEFGRVWYRILH
+GT V + VLD+NDN P F QS YE VPED P G I + A D D G +G++ Y H
PCDHB14 SGTTLVLIKVLDINDNAPEFPQSLYEVQVPEDRPLGSWIATIISAKDLDAGNYGKISYTFFH
IFLQSSYEASVPEDIPEGHSILQLK 1122
TFQNLPFVAEVLEGIPAGVSIYQVV 928 CDH23 internal spacer
IFSQPLYNISLYENVTVGTSVLTVL 497 CDH23 internal spacer
TFFPAVYNVSVSEDVPREFRVVWLN 1034 CDH23 internal spacer
TFHNQPYSVRIPENTPVGTPIFIVN 169 CDH23 internal spacer
TWKDAPYYINLVEMTPPDSDVTTVV 809 CDH23 internal spacer
CDH23 0 chr10:72826710-73245710 cadherin-like 23
FAT4 0 chr4:126457017-126633537 FAT tumor suppressor homolog 4
DCHS1 0 chr11:6599134-6633650 dachsous 1
FAT3 0 chr11:91724910-92269283 FAT tumor suppressor homolog 3
FAT1 0 chr4:187745931-187881981 FAT tumor suppressor 1
FAT2 0 chr5:150863846-150928698 FAT tumor suppressor 2
DCHS2 0 chr4:155375138-155531899 dachsous 2 isoform 1
CELSR2 1e-115 chr1:109594164-109619901 cadherin EGF LAG seven-pass G-type receptor 2
CELSR1 5e-113 chr22:45135395-45311731 cadherin EGF LAG seven-pass G-type receptor 1
CELSR3 5e-109 chr3:48637835-48684985 cadherin EGF LAG seven-pass G-type receptor 3
PCDH24 3e-87 chr5:175908971-175955375 protocadherin LKC
...
PCDHB14 5e-53 chr5 140,584,653 protocadherin beta 14
Structural significance to normal function
Blastp at PDB of the region around residue L1122 establishes that the best fit to an already determined structure is the 39% identity match to mouse cadherin CDH8, 2A62. Within the 25 residue interdomain region, the percent identity is somewhat higher at nearly 50%. While not ideal, this should still allow accurate modeling of the adjacent cadherin domains and the critical spacer region, although the structural effects of the L1122V may be fairly subtle.
CDH23 1 GTATVFVTVLDVNDNRPIFLQSSYEASVPEDIPEGHSILQLKATDADEGEFGRVWYRILHGNHGNNFRI
GT T+ VT+ DVNDN P F QS Y SVPED+ G +I ++KA D D GE + Y I+ G+ F I
CDH8 174 GTTTLTVTLTDVNDNPPKFAQSLYHFSVPEDVVLGTAIGRVKANDQDIGENAQSSYDIIDGDGTALFEI
Since CDH23 is known to form a helical homodimer in tip links -- and such binding patches can involve hydrophobic residues that otherwise would be buried -- the quaternary structure here is the main unknown. Crystallographic adjacency in the unit cell does not always reflect oligomeric solution structure. Consequently, it may not be possible to fully evaluate L1122V despite the favorable match at PDB.
There is no reason to think L1122V would directly affect the calcium binding motifs (LDRE, D.ND, D.D) of either adjacent cadherin domains in the manner of the E737V salsa mouse mutation in exon 22 or D124G, R1060W, E1595K and D2202N, none of which have syndromic effects on the retina but demonstrably weaken calcium dependent binding to PCDH15 even though they do not lie in the amino-terminal cadherin binding domains.
An effect of L1122V on the MET (mechanotransduction) channel is also implausibly indirect because this is at the lower (PCDH15) end of the link tip, though that is disputed for larger sound displacement effects.
Similarly an up-link intracellular effect of extracellular L1122V on CDH23 binding to harmonin would be a stretch. That binding is now thought mediated by an autonomously folding region proximal to the harmonin PDZ motif with a short internal peptide of CDH23 that extends from 3180-3211, KPDDDRYLRAAIQEYDNIAKLGQIIREGPIK, over 2,000 residues away.
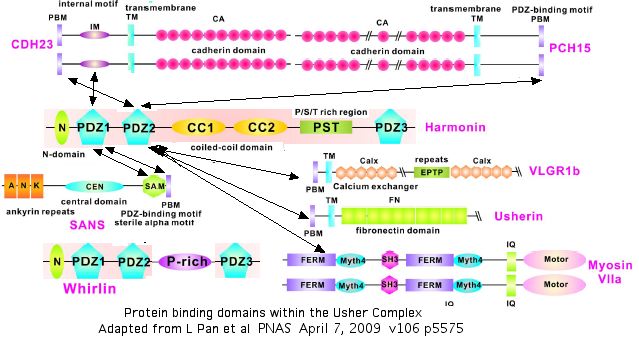
The diagram below summarizes what is currently known about homotypic and heterotypic binding of proteins within the Usher network. Some of these remain unclear, like those of whirlin USH2D which also localizes at stereocilia tips and has an N-terminal domain like that of harmonin but yet does not bind CDH23. These interactions must be understood before SNPs such as V1122L can be modelled in their quaternary context and assessed with any confidence.
Comparison of CDH15 domains to PCDH15
Given that CDH23 and PCDH15 binding constitutes the critical part of hair links joining a lower stereocilia to the next higher, it's worth investigating the evolutionary relationship betweeen the 27 and 11 cadherin ectodomains (ie the extent to which these proteins are paralogs and so naturally dimerize). It emerges that the cadherin domains have little residual sequence identity to each other within or across proteins, even though each individually has very considerable phylogenetic conservation.
Further, the cadherin domain of CDH23 best corresponding to a cadherin domain of PCDH15 (as linearly ordered in its primary sequence) are out of order with respect to the primary sequence of CDH23: 25, 16, 15, 13, 12, 16, 14, 2, 9, 18, 12. This suggests if a deep relationship ever existed between these two proteins, different rates of cadherin domain evolution has obscured it.
However when the 27 spacer domains separating cadherin units are concatenated, SMART detects an internal repeat within CDH23 of the first 11 to the last 11 spacers. Furthermore, these repeats give weak but full length alignments to the 11 concatenated spacer units of PCDH15. (Use of spacers avoids confounding issues of cadherin domain cross-matching.) This suggests the latter is ancestral length and that CDH23 experienced an internal duplication that lengthened its ectodomain. Spacers is surely a misnomer as the degree of phylogenetic conservation suggests each is under considerable selection along with its associated cadherin domains.
When the cytoplasmic domains are aligned (using uncorrected genomic alignments from the 46 species UCSC data set), it emerges that CDH23 (top) is far better conserved to far better phylogenetic depth than PCDH15 (bottom). This conservation does not extend into early deuterostomes, protostomes or cnidarians but rather emerged abruptly in lamprey (ie synchronoously with ciliary opsins and the retina).
Presumably both are specifically anchored to the internal Usher protein network at one or more domains, with the terminal PDZ binding motif ITEL* and STSL* respectively) and internal motif IM KPDDDRYLRAAIQEYDNIAKLGQIIREGPIK serving to anchor CDH23 but not PCDH15 bidentately to harmonin N-domain. However these domains can explain only a small fraction of observed CDH23 cytoplasmic conservation. Note runs of compositionally simple sequence cannot form ordered secondary or tertiary structure.
Intriguingly, whirlin (USH2D) has a similar domain structure to harmonin, in particular the N-domains align. Although this does not bind the cytoplasmic internal motif of CDH23, it could plausibly bind a comparable region in PCDH15, in effect quasi-paralog binding to quasi-paralog.
>CDH23_homSap domains: signal, 27 spacers, 27 cadhedrin domains (last weak), unknown extracellular, single pass transmembrane, unknown cytoplasmic: MGRHVATSCHVAWLLVLISGCWG QVNRLPFFTNHFFDTYLLISEDTPVGSSVTQ LLAQDMDNDPLVFGVSGEEASRFFAVEPDTGVVWLRQPLDRETKSEFTVEFSVSDHQGVITRKVNIQVGDVNDNAP TFHNQPYSVRIPENTPVGTPIFI VNATDPDLGAGGSVLYSFQPPSQFFAIDSARGIVTVIRELDYETTQAYQLTVNATDQDKTRPLSTLANLAIIITDVQDMDP IFINLPYSTNIYEHSPPGTTVRI ITAIDQDKGRPRGIGYTIVSGNTNSIFALDYISGVLTLNGLLDRENPLYSHGFILTVKGTELNDDRTPSDATVTTTFNILVIDINDNAP EFNSSEYSVAITELAQVGFALPLF IQVVDKDENLGLNSMFEVYLVGNNSHHFIISPTSVQGKADIRIRVAIPLDYETVDRYDFDLFANESVPDHVGYAKVKITLINENDNRP IFSQPLYNISLYENVTVGTSVLT VLATDNDAGTFGEVSYFFSDDPDRFSLDKDTGLIMLIARLDYELIQRFTLTIIARDGGGEETTGRVRINVLDVNDNVP TFQKDAYVGALRENEPSVTQLVR LRATDEDSPPNNQITYSIVSASAFGSYFDISLYEGYGVISVSRPLDYEQISNGLIYLTVMAMDAGNPPLNSTVPVTIEVFDENDNPP TFSKPAYFVSVVENIMAGATVLF LNATDLDRSREYGQESIIYSLEGSTQFRINARSGEITTTSLLDRETKSEYILIVRAVDGGVGHNQKTGIATVNITLLDINDNHP TWKDAPYYINLVEMTPPDSDVTT VVAVDPDLGENGTLVYSIQPPNKFYSLNSTTGKIRTTHAMLDRENPDPHEAELMRKIVVSVTDCGRPPLKATSSATVFVNLLDLNDNDP TFQNLPFVAEVLEGIPAGVSIYQ VVAIDLDEGLNGLVSYRMPVGMPRMDFLINSSSGVVVTTTELDRERIAEYQLRVVASDAGTPTKSSTSTLTIHVLDVNDETP TFFPAVYNVSVSEDVPREFRVVW LNCTDNDVGLNAELSYFITGGNVDGKFSVGYRDAVVRTVVGLDRETTAAYMLILEAIDNGPVGKRHTGTATVFVTVLDVNDNRP IFLQSSYEASVPEDIPEGHSILQ LKATDADEGEFGRVWYRILHGNHGNNFRIHVSNGLLMRGPRPLDRERNSSHVLIVEAYNHDLGPMRSSVRVIVYVEDINDEAP VFTQQQYSRLGLRETAGIGTSVIV VQATDRDSGDGGLVNYRILSGAEGKFEIDESTGLIITVNYLDYETKTSYMMNVSATDQAPPFNQGFCSVYITLLNELDEAV QFSNASYEAAILENLALGTEIVR VQAYSIDNLNQITYRFNAYTSTQAKALFKIDAITGVITVQGLVDREKGDFYTLTVVADDGGPKVDSTVKVYITVLDENDNSP RFDFTSDSAVSIPEDCPVGQRVAT VKAWDPDAGSNGQVVFSLASGNIAGAFEIVTTNDSIGEVFVARPLDREELDHYILQVVASDRGTPPRKKDHILQVTILDINDNPP VIESPFGYNVSVNENVGGGTAVVQ VRATDRDIGINSVLSYYITEGNKDMAFRMDRISGEIATRPAPPDRERQSFYHLVATVEDEGTPTLSATTHVYVTIVDENDNAP MFQQPHYEVLLDEGPDTLNTSLIT IQALDLDEGPNGTVTYAIVAGNIVNTFRIDRHMGVITAAKELDYEISHGRYTLIVTATDQCPILSHRLTSTTTVLVNVNDINDNVP TFPRDYEGPFEVTEGQPGPRVWT FLAHDRDSGPNGQVEYSIMDGDPLGEFVISPVEGVLRVRKDVELDRETIAFYNLTICARDRGMPPLSSTMLVGIRVLDINDNDP VLLNLPMNITISENSPVSSFVAH VLASDADSGCNARLTFNITAGNRERAFFINATTGIVTVNRPLDRERIPEYKLTISVKDNPENPRIARRDYDLLLIFLSDENDNHP LFTKSTYQAEVMENSPAGTPLTVLNGP ILALDADQDIYAVVTYQLLGAQSGLFDINSSTGVVTVRSGVIIDREAFSPPILELLLLAEDIGLLNSTAHLLITILDDNDNRP TFSPATLTVHLLENCPPGFSVLQ VTATDEDSGLNGELVYRIEAGAQDRFLIHLVTGVIRVGNATIDREEQESYRLTVVATDRGTVPLSGTAIVTILIDDINDSRP EFLNPIQTVSVLESAEPGTVIAN ITAIDHDLNPKLEYHIVGIVAKDDTDRLVPNQEDAFAVNINTGSVMVKSPMNRELVATYEVTLSVIDNASDLPERSVSVPNAKLTVNVLDVNDNTP QFKPFGITYYMERILEGATPGTTLIA VAAVDPDKGLNGLVTYTLLDLVPPGYVQLEDSSAGKVIANRTVDYEEVHWLNFTVRASDNGSPPRAAEIPVYLEIVDINDNNP IFDQPSYQEAVFEDVPVGTIILT VTATDADSGNFALIEYSLGDGESKFAINPTTGDIYVLSSLDREKKDHYILTALAKDNPGDVASNRRENSVQVVIQVLDVNDCRP QFSKPQFSTSVYENEPAGTSVIT MMATDQDEGPNGELTYSLEGPGVEAFHVDMDSGLVTTQRPLQSYEKFSLTVVATDGGEPPLWGTTMLLVEVIDVNDNRP VFVRPPNGTILHIREEIPLRSNVYE VYATDKDEGLNGAVRYSFLKTAGNRDWEFFIIDPISGLIQTAQRLDRESQAVYSLILVASDLGQPVPYETMQPLQVALEDIDDNEP LFVRPPKGSPQYQLLTVPEHSPRGTLVGNV TGAVDADEGPNAIVYYFIAAGNEEKNFHLQPDGCLLVLRDLDREREAIFSFIVKASSNRSWTPPRGPSPTLDLVADLTLQEVRVVLEDINDQPP RFTKAEYTAGVATDAKVGSELIQ VLALDADIGNNSLVFYSILAIHYFRALANDSEDVGQVFTMGSMDGILRTFDLFMAYSPGYFVVDIVARDLAGHNDTAIIGIYILRDDQRV KIVINEIPDRVRGFEEEFIHLLSNITGAIVNTDNVQFHVDKKGRVNFAQTELLIHVVNRDTNRILDVDRVIQMIDENKEQLRNLFRNYNVLDVQPAISVRLPDDMSALQM AIIVLAILLFLAAMLFVLMNWYY RTVHKRKLKAIVAGSAGNRGFIDIMDMPNTNKYSFDGANPVWLDPFCRNLELAAQAEHEDDLPENLSEIADLWNSPTRTHGTFGREPAAVKPDDDRYLRAAIQEYDNIAKLGQIIREGPIKGSLLKVVLEDYLRLKKLFAQRMVQKASSCHSSISELIQTELDEEPGDHSPGQGSLRFRHKPPVELKGPDGIHVVHGSTGTLLATDLNSLPEEDQKGLGRSLETLTAAEATAFERNARTESAKSTPLHKLRDVIMETPLEITEL >PCDH15_homSap domains: signal, 11 spacers, 11 cadhedrin domains, unknown extracellular, single pass transmembrane, unknown cytoplasmic: MFRQFYLWTCLASGIILGSLFEICLG QYDDDCKLARGGPPATIVAIDEESRNGTILVDNMLIK GTAGGPDPTIELSLKDNVDYWVLMDPVKQMLFLNSTGRVLDRDPPMNIHSIVVQVQCINKKVGTIIYHEVRIVVRDRNDNSP TFKHESYYATVNELTPVGTTIFTGFSGD NGATDIDDGPNGQIEYVIQYNPDDPTSNDTFEIPLMLTGNIVLRKRLNYEDKTRYFVIIQANDRAQNLNERRTTTTTLTVDVLDGDDLGP MFLPCVLVPNTRDCRPLTYQAAIPELRTPEELNPIIVTPP IQAIDQDRNIQPPSDRPGILYSILVGTPEDYPRFFHMHPRTAELSLLEPVNRDFHQKFDLVIKAEQDNGHPLPAFAGLHIEILDENNQSP YFTMPSYQGYILESAPVGATISDSLNLTSPLR IVALDKDIEDTKDPELHLFLNDYTSVFTVTQTGITRYLTLLQPVDREEQQTYTFSITAFDGVQESEPVIVNIQVMDANDNTP TFPEISYDVYVYTDMRPGDSVIQ LTAVDADEGSNGEITYEILVGAQGDFIINKTTGLITIAPGVEMIVGRTYALTVQAADNAPPAERRNSICTVYIEVLPPNNQSP PRFPQLMYSLEISEAMRVGAVLLN LQATDREGDSITYAIENGDPQRVFNLSETTGILTLGKALDRESTDRYILIITASDGRPDGTSTATVNIVVTDVNDNAP VFDPYLPRNLSVVEEEANAFVGQ VKATDPDAGINGQVHYSLGNFNNLFRITSNGSIYTAVKLNREVRDYYELVVVATDGAVHPRHSTLTLAIKVLDIDDNSP VFTNSTYTVLVEENLPAGTTILQ IEAKDVDLGANVSYRIRSPEVKHFFALHPFTGELSLLRSLDYEAFPDQEASITFLVEAFDIYGTMPPGIATVTVIVKDMNDYPP VFSKRIYKGMVAPDAVKGTPITT VYAEDADPPGLPASRVRYRVDDVQFPYPASIFEVEEDSGRVITRVNLNEEPTTIFKLVVVAFDDGEPVMSSSATVKILVLHPGEIPR FTQEEYRPPPVSELATKGTM VGVISAAAINQSIVYSIVSGNEEDTFGINNITGVIYVNGPLDYETRTSYVLRVQADSLEVVLANLRVPSKSNTAKVYIEIQDENNHPP VFQKKFYIGGVSEDARMFTSVLR VKATDKDTGNYSVMAYRLIIPPIKEGKEGFVVETYTGLIKTAMLFHNMRRSYFKFQVIATDDYGKGLSGKADVLVSVVNQLDMQV IVSNVPPTLVEKKIEDLTEILDRYVQEQIPGAKVVVESIGARRHGDAFSLEDYTKCDLTVYAIDPQTNRAIDRNELFKFLDGKLLDINKDFQPYYGEGGRILEIRTPEAVTSIKKRGESLGYTE GALLALAFIIILCCIPAILVVLV SYRQFKVRQAECTKTARIQAALPAAKPAVPAPAPVAAPPPPPPPPPGAHLYEELGDSSILFLLYHFQQSRGNNSVSEDRKHQQVVMPFSSNTIEAHKSAHVDGSLKSNKLKSARKFTFLSDEDDLSAHNPLYKENISQVSTNSDISQRTDFVDPFSPKIQAKSKSLRGPREKIQRLWSQSVSLPRRLMRKVPNRPEIIDLQQWQGTRQKAENENTGICTNKRGSSNPLLTTEEANLTEKEEIRQGETLMIEGTEQLKSLSSDSSFCFPRPHFSFSTLPTVSRTVELKSEPNVISSPAECSLELSPSRPCVLHSSLSRRETPICMLPIETERNIFENFAHPPNISPSACPLPPPPPISPPSPPPAPAPLAPPPDISPFSLFCPPPSPPSIPLPLPPPTFFPLSVSTSGPPTPPLLPPFPTPLPPPPPSIPCPPPPSASFLSTECVCITGVKCTTNLMPAEKIKSSMTQLSTTTVCKTDPQREPKGILRHVKNLAELEKSVANMYSQIEKNYLRTNVSELQTMCPSEVTNMEITSEQNKGSLNNIVEGTEKQSHSQSTSL Numbered cadhedrin domains of CDH23 >CDH23.CA.01 LLAQDMDNDPLVFGVSGEEASRFFAVEPDTGVVWLRQPLDRETKSEFTVEFSVSDHQGVITRKVNIQVGDVNDNAP >CDH23.CA.02 VNATDPDLGAGGSVLYSFQPPSQFFAIDSARGIVTVIRELDYETTQAYQLTVNATDQDKTRPLSTLANLAIIITDVQDMDP >CDH23.CA.03 ITAIDQDKGRPRGIGYTIVSGNTNSIFALDYISGVLTLNGLLDRENPLYSHGFILTVKGTELNDDRTPSDATVTTTFNILVIDINDNAP >CDH23.CA.04 IQVVDKDENLGLNSMFEVYLVGNNSHHFIISPTSVQGKADIRIRVAIPLDYETVDRYDFDLFANESVPDHVGYAKVKITLINENDNRP >CDH23.CA.05 VLATDNDAGTFGEVSYFFSDDPDRFSLDKDTGLIMLIARLDYELIQRFTLTIIARDGGGEETTGRVRINVLDVNDNVP >CDH23.CA.06 LRATDEDSPPNNQITYSIVSASAFGSYFDISLYEGYGVISVSRPLDYEQISNGLIYLTVMAMDAGNPPLNSTVPVTIEVFDENDNPP >CDH23.CA.07 LNATDLDRSREYGQESIIYSLEGSTQFRINARSGEITTTSLLDRETKSEYILIVRAVDGGVGHNQKTGIATVNITLLDINDNHP >CDH23.CA.08 VVAVDPDLGENGTLVYSIQPPNKFYSLNSTTGKIRTTHAMLDRENPDPHEAELMRKIVVSVTDCGRPPLKATSSATVFVNLLDLNDNDP >CDH23.CA.09 VVAIDLDEGLNGLVSYRMPVGMPRMDFLINSSSGVVVTTTELDRERIAEYQLRVVASDAGTPTKSSTSTLTIHVLDVNDETP >CDH23.CA.10 LNCTDNDVGLNAELSYFITGGNVDGKFSVGYRDAVVRTVVGLDRETTAAYMLILEAIDNGPVGKRHTGTATVFVTVLDVNDNRP >CDH23.CA.11 LKATDADEGEFGRVWYRILHGNHGNNFRIHVSNGLLMRGPRPLDRERNSSHVLIVEAYNHDLGPMRSSVRVIVYVEDINDEAP >CDH23.CA.12 VQATDRDSGDGGLVNYRILSGAEGKFEIDESTGLIITVNYLDYETKTSYMMNVSATDQAPPFNQGFCSVYITLLNELDEAV >CDH23.CA.13 VQAYSIDNLNQITYRFNAYTSTQAKALFKIDAITGVITVQGLVDREKGDFYTLTVVADDGGPKVDSTVKVYITVLDENDNSP >CDH23.CA.14 VKAWDPDAGSNGQVVFSLASGNIAGAFEIVTTNDSIGEVFVARPLDREELDHYILQVVASDRGTPPRKKDHILQVTILDINDNPP >CDH23.CA.15 VRATDRDIGINSVLSYYITEGNKDMAFRMDRISGEIATRPAPPDRERQSFYHLVATVEDEGTPTLSATTHVYVTIVDENDNAP >CDH23.CA.16 IQALDLDEGPNGTVTYAIVAGNIVNTFRIDRHMGVITAAKELDYEISHGRYTLIVTATDQCPILSHRLTSTTTVLVNVNDINDNVP >CDH23.CA.17 FLAHDRDSGPNGQVEYSIMDGDPLGEFVISPVEGVLRVRKDVELDRETIAFYNLTICARDRGMPPLSSTMLVGIRVLDINDNDP >CDH23.CA.18 VLASDADSGCNARLTFNITAGNRERAFFINATTGIVTVNRPLDRERIPEYKLTISVKDNPENPRIARRDYDLLLIFLSDENDNHP >CDH23.CA.19 ILALDADQDIYAVVTYQLLGAQSGLFDINSSTGVVTVRSGVIIDREAFSPPILELLLLAEDIGLLNSTAHLLITILDDNDNRP >CDH23.CA.20 VTATDEDSGLNGELVYRIEAGAQDRFLIHLVTGVIRVGNATIDREEQESYRLTVVATDRGTVPLSGTAIVTILIDDINDSRP >CDH23.CA.21 ITAIDHDLNPKLEYHIVGIVAKDDTDRLVPNQEDAFAVNINTGSVMVKSPMNRELVATYEVTLSVIDNASDLPERSVSVPNAKLTVNVLDVNDNTP >CDH23.CA.22 VAAVDPDKGLNGLVTYTLLDLVPPGYVQLEDSSAGKVIANRTVDYEEVHWLNFTVRASDNGSPPRAAEIPVYLEIVDINDNNP >CDH23.CA.23 VTATDADSGNFALIEYSLGDGESKFAINPTTGDIYVLSSLDREKKDHYILTALAKDNPGDVASNRRENSVQVVIQVLDVNDCRP >CDH23.CA.24 MMATDQDEGPNGELTYSLEGPGVEAFHVDMDSGLVTTQRPLQSYEKFSLTVVATDGGEPPLWGTTMLLVEVIDVNDNRP >CDH23.CA.25 VYATDKDEGLNGAVRYSFLKTAGNRDWEFFIIDPISGLIQTAQRLDRESQAVYSLILVASDLGQPVPYETMQPLQVALEDIDDNEP >CDH23.CA.26 TGAVDADEGPNAIVYYFIAAGNEEKNFHLQPDGCLLVLRDLDREREAIFSFIVKASSNRSWTPPRGPSPTLDLVADLTLQEVRVVLEDINDQPP >CDH23.CA.27 weak VLALDADIGNNSLVFYSILAIHYFRALANDSEDVGQVFTMGSMDGILRTFDLFMAYSPGYFVVDIVARDLAGHNDTAIIGIYILRDDQRV Numbered cadhedrin domains of PCDH15 with individual best blastp to a CDH23 cadhedrin domain >PCDH15.CA.01 CDH23.CA.25 GTAGGPDPTIELSLKDNVDYWVLMDPVKQMLFLNSTGRVLDRDPPMNIHSIVVQVQCINKKVGTIIYHEVRIVVRDRNDNSP >PCDH15.CA.02 CDH23.CA.16 NGATDIDDGPNGQIEYVIQYNPDDPTSNDTFEIPLMLTGNIVLRKRLNYEDKTRYFVIIQANDRAQNLNERRTTTTTLTVDVLDGDDLGP >PCDH15.CA.03 CDH23.CA.15 IQAIDQDRNIQPPSDRPGILYSILVGTPEDYPRFFHMHPRTAELSLLEPVNRDFHQKFDLVIKAEQDNGHPLPAFAGLHIEILDENNQSP >PCDH15.CA.04 CDH23.CA.13 IVALDKDIEDTKDPELHLFLNDYTSVFTVTQTGITRYLTLLQPVDREEQQTYTFSITAFDGVQESEPVIVNIQVMDANDNTP >PCDH15.CA.05 CDH23.CA.12 LTAVDADEGSNGEITYEILVGAQGDFIINKTTGLITIAPGVEMIVGRTYALTVQAADNAPPAERRNSICTVYIEVLPPNNQSP >PCDH15.CA.06 CDH23.CA.16 LQATDREGDSITYAIENGDPQRVFNLSETTGILTLGKALDRESTDRYILIITASDGRPDGTSTATVNIVVTDVNDNAP >PCDH15.CA.07 CDH23.CA.14 VKATDPDAGINGQVHYSLGNFNNLFRITSNGSIYTAVKLNREVRDYYELVVVATDGAVHPRHSTLTLAIKVLDIDDNSP >PCDH15.CA.08 CDH23.CA.02 IEAKDVDLGANVSYRIRSPEVKHFFALHPFTGELSLLRSLDYEAFPDQEASITFLVEAFDIYGTMPPGIATVTVIVKDMNDYPP >PCDH15.CA.09 CDH23.CA.09 VYAEDADPPGLPASRVRYRVDDVQFPYPASIFEVEEDSGRVITRVNLNEEPTTIFKLVVVAFDDGEPVMSSSATVKILVLHPGEIPR >PCDH15.CA.10 CDH23.CA.18 VGVISAAAINQSIVYSIVSGNEEDTFGINNITGVIYVNGPLDYETRTSYVLRVQADSLEVVLANLRVPSKSNTAKVYIEIQDENNHPP >PCDH15.CA.11 CDH23.CA.12 VKATDKDTGNYSVMAYRLIIPPIKEGKEGFVVETYTGLIKTAMLFHNMRRSYFKFQVIATDDYGKGLSGKADVLVSVVNQLDMQV Difference alignment of human CDH23 cadherin domains CDH23.CA.01 LLAQDMDN----------DPLVFGVSGEEASRFFAVEP--DTGVVWLRQ----PL------DRETKSEFTVEFSVSDH--QGVITRKVNIQVGDVNDNAP CDH23.CA.02 VN.T.P.LGAG------GSV.YSFQPPS-----QFFAIDSAR.I.TVIRELDYET----TQAYQLTVNA.DQ---DKTRPLST-LANLA.IIT..Q.MD. CDH23.CA.03 IT.I.Q.KGRP------RGIGYTI...NTN.---IFALDYIS..L-TLNGLLDRENPLYSHGFILT-VKGT.LNDDRTPSDATV.TTF..L.I.I..... CDH23.CA.04 IQVV.K.ENLG------LNSMFEVYLVGNN.HH.IIS.TSVQ.KADI.IRVAI..------.Y..VDRYDFDLFANESVPDH.GYA..K.TLINE...R. CDH23.CA.05 V..T.N.AGTF------GEVSY.FSDDPDR-----FSLDK...LIM.IARLDYE.----IQRFTLTIIARDG---G---GEET-.GR.R.N.L.....V. CDH23.CA.06 .R.T.E.SPPN------NQITYSI..ASAFGSY.DISLYEGY..ISVSRPLDYEQ-----ISNGLIYLTVMAMDAGNPP--LNS.VP.T.E.F.E...P. CDH23.CA.07 .N.T.L.RSRE------YGQESIIY.L.GST---QFRINARS.EITTTSLLDRET----KSEYILIVRAVDG--GVG.NQKTG-IAT...TLL.I...H. CDH23.CA.08 VV.V.P.LGEN------GTLVYSIQPPN--K---FYSLNST..KIRTTHAMLDRENPDPHEAELMRKIVVSVTDCGRPPLKATSSAT.FVNLL.L...D. CDH23.CA.09 VV.I.L.EGLN------GLVSYRMPV.MPRM---DFLINSSS..--VVTTTELDR-----ERIAEYQLRV.ASDAGTPT--KSS.STLT.H.L....ET. CDH23.CA.10 .NCT.N.VGLN------AELSY.ITG.NVDG---KFSVGYRDA..RTVVGLDRET----TAAYMLIL.AIDN--GPVGKRHTG-.AT.FVT.L.....R. CDH23.CA.11 .K.T.A.EGEF------GRVWYRILH.NHGN---NFRIHVSN.LLMRGPRP-LDR-----ERNSSHVLIVEAYNHDLGP--MRSSVR.IVY.E.I..E.. CDH23.CA.12 VQ.T.R.SGDG------GLVNYRIL..A.GK----F.IDES..LIITVNYLDYET----KTSYMMNVSA.DQ---APPFN..F-CSVYITLLNELDEAV CDH23.CA.13 VQ.YSI..LNQ------ITYRFNAYTSTQ.KAL.KID--AI...ITVQGLV----------...KGDFY.LTVVAD.GGPKVDS.V..Y.T.L.E...S. CDH23.CA.14 VK.W.P.AGSN------GQVVFSLA..NI.GA.EI.TTNDSI.E.FVARPLDREE-----L.HYI--LQV.ASDRGTPP--RKKDHILQVTIL.I...P. CDH23.CA.15 VR.T.R.IGIN------SVLSYYITE.NKDM---.FRMDRIS.EIAT.PAP-PDR-----ERQSFYHLVATVEDEGTPT--LSA.TH.YVTIV.E..... CDH23.CA.16 IQ.L.L.EGPN------GTVTYAI.A.NIVN---TFRIDRHM..I-TAAKELDYE---ISHG.Y.L-IV.ATDQCPILSHRLTS.TT.LVN.N.I...V. CDH23.CA.17 F..H.R.SGPN------GQVEYSIMD.DPLG---EFVISPVE..LRV.KDVELDR-----ETIAFYNLTICARDRGMPP--LSS.ML.G.R.L.I...D. CDH23.CA.18 V..S.A.SGCN------ARLTFNITA.NRER---.FFINAT..I.TVNRPLDRER----IPEYKLTISVKDNPENPRIARRDY-DL-LL.FLS.E...H. CDH23.CA.19 I..L.A.QDIY------AVVTYQLL-.AQSG---LFDINSS....TV.SGVIIDR-----EAFSPPILELLLLAEDIGL--LNS.AHLL.TIL.D...R. CDH23.CA.20 VT.T.E.SGLN------GELVYRIEA.-AQD---RFLIHLV...IRVGNAT-IDR-----EEQ.SYRLTV.ATDRGTVP--LSG.AI.T.LID.I..SR. CDH23.CA.21 IT.I.H.LNPKLEYHIVGIVAKDDTDRLVPNQED.FAVNIN..S.MVKSPMNRE.----VATY.VTLSVIDNA.DLPERSVS.PNA.LTVN.L.....T. CDH23.CA.22 VA.V.P.KGLN------GLVTYTLLDLVPPG--YVQLEDSSA.K.IANRTVDYEE-----V-HWL-NFTVRASDNGSPP--RAAEIP.YLEIV.I...N. CDH23.CA.23 VT.T.A.SGNF------ALIEYSL--.DGE.---KFAINPT..DIYVLSSLDREK----KDHYILTALAKDNPGDVASNRREN-SVQ.V...L....CR. CDH23.CA.24 MM.T.Q.EGPN------GELTYSLEGPGV----E.FHVDM.S.L.TTQRPLQSYE-----KFS----LTV.ATDGGEPP--LWG.TMLLVE.I.....R. CDH23.CA.25 VY.T.K.EGLN------GAVRYSFLKTAGNRDWEFFIIDPIS.LIQTA.RLDRES----QAVYSLILVASDL---GQPVPYET-MQPLQVALE.ID..E. CDH23.CA.26 TG.V.A.EGPN------AIVYY.IAA.N.EKN.HLQPDGCLLVLRD.DREREAIFSFIVKASSNRSWTPPRGP.PTLDLVADLTLQE.RVVLE.I..QP. CDH23.CA.27 V..L.A.IGNN------SLVFYSILAIHYFRALANDSEDVGQVFT-MGSMDGILRTFDLFMAYSPGYFVVDIVARDLAGHNDTAIIGIY.LRD.QRV Consensus !.......G.n g.v.Y.i..g........f........!.......... ............................v.!.......... Difference alignment of human CDH23 cadherin domains PCDH15.CA.01 GTAGGPDPTIELSLKDN---VDYWVLMDPVKQMLFLNSTGRVLDRDPPMNIHSIV-----VQVQCINKKVGTIIYHEVRIVVRDRNDNSP PCDH15.CA.02 NGATDIDD..NGQ..YVIQY.PDDPTSNDTFEIPLMLTGNIVLRKR.NYEDKTRYFV.I----QANDRAQ.LNERRTTTTTLTVD.L.GD.LG. PCDH15.CA.07 VKATDPDA.INGQVHY..G------NFNN.FRITS--NGSIY.AVK.N.EVRDYYELV.----VATDGAVH---PRHSTLTLA.K.L.ID.... PCDH15.CA.05 LTAVDADE.SNGE.TYEILV-----GAQGDFIIN.T-TG.ITIAPGVEMIVGRTYALT.----QAADNAPPA-ERRNSICT.Y.E.LPP.NQ.. PCDH15.CA.06 LQATDREGDS.TYAIEN----G.PQRVFNLSET-TGILTL.KA...ESTDRYIL--------IITASDGRPDGTSTAT.N...T.V...A. PCDH15.CA.10 VGVIS.AAINQS.VY.IVS----GNEEDTFGINNI-TGVIYVNGP..YETRTSYVLR.QADSLEV.LANLRVPSKSNTAK.Y.EIQ.E.NHP. PCDH15.CA.08 IEAKDVDLGANVSYRIRS----PEVKHFFALHPF-TGEL.LL.S..YEAFPDQEASI---TFLVEAFDIYGTMPPGIAT.TVI.K.M..YP. PCDH15.CA.03 IQAIDQDRNIQPPSDR.G.LY.ILVGTPEDYPRFFHMHPR--TAEL.LLEPVN..FHQKFDLVI-------KAEQDNGHPLPAFAGLH.EIL.E.NQ.. PCDH15.CA.04 IVALDKDIEDTKDPELHLFL------NDYTS.FTVTQTGITRYLTLLQPV..EEQQTYTFSI-------TAFDGVQESEPVI--.N.Q.M.A...T. PCDH15.CA.09 VYAEDAD-PP.L.ASRVRYRVD.VQFPYPASIFEVEED--SGRVI.RVN.NEE.TTIFKLV.-------.AFDDGEPVMSSSAT.K.L.LHPGEIPR PCDH15.CA.11 VKATDKD-.GNY--SVMAYR.IIPPIKEGKEGFVVETY--TG.IK.AMLFHNMRRSYFKFQ.-------IATDDYGK.LSGKAD.LVS.VNQL.MQV Consensus .........d......!.Y.i...........ff..... TG.i.l...l#r#....%.l.! ....aa..........at.....l..##... >CDH23_spacers color shows repeat detected by SMART QVNRLPFFTNHFFDTYLLISEDTPVGSSVTQ TFHNQPYSVRIPENTPVGTPIFI IFINLPYSTNIYEHSPPGTTVRI EFNSSEYSVAITELAQVGFALPLF IFSQPLYNISLYENVTVGTSVLT TFQKDAYVGALRENEPSVTQLVR TFSKPAYFVSVVENIMAGATVLF TWKDAPYYINLVEMTPPDSDVTT TFQNLPFVAEVLEGIPAGVSIYQ TFFPAVYNVSVSEDVPREFRVVW IFLQSSYEASVPEDIPEGHSILQ VFTQQQYSRLGLRETAGIGTSVIV QFSNASYEAAILENLALGTEIVR RFDFTSDSAVSIPEDCPVGQRVAT VIESPFGYNVSVNENVGGGTAVVQ MFQQPHYEVLLDEGPDTLNTSLIT TFPRDYEGPFEVTEGQPGPRVWT VLLNLPMNITISENSPVSSFVAH LFTKSTYQAEVMENSPAGTPLTVLNGP TFSPATLTVHLLENCPPGFSVLQ EFLNPIQTVSVLESAEPGTVIAN QFKPFGITYYMERILEGATPGTTLIA IFDQPSYQEAVFEDVPVGTIILT QFSKPQFSTSVYENEPAGTSVIT VFVRPPNGTILHIREEIPLRSNVYE LFVRPPKGSPQYQLLTVPEHSPRGTLVGNV RFTKAEYTAGVATDAKVGSELIQ >PCDH15_spacers color shows 26% identity alignment to CDH23 spacer repeat QYDDDCKLARGGPPATIVAIDEESRNGTILVDNMLIK TFKHESYYATVNELTPVGTTIFTGFSGD MFLPCVLVPNTRDCRPLTYQAAIPELRTPEELNPIIVTPP YFTMPSYQGYILESAPVGATISDSLNLTSPLR TFPEISYDVYVYTDMRPGDSVIQ PRFPQLMYSLEISEAMRVGAVLLN VFDPYLPRNLSVVEEEANAFVGQ VFTNSTYTVLVEENLPAGTTILQ VFSKRIYKGMVAPDAVKGTPITT FTQEEYRPPPVSELATKGTM VFQKKFYIGGVSEDARMFTSVLR
CDH23 allele assessment by PolyPhen
Here L1122V will test out as benign because it is a common conservative substitution and comparative genomics support in fish. PolyPhen and SIFT have comparative genomics limited to sequences at SwissProt and do not incorporate phylogenetic relations; consequently they miss the L-->V transition at the level of the tetrapod clade. Consequently such tools do not utilize a significant part of the available information and are not informative here.