QAing UCSC Genes: Difference between revisions
| Line 4: | Line 4: | ||
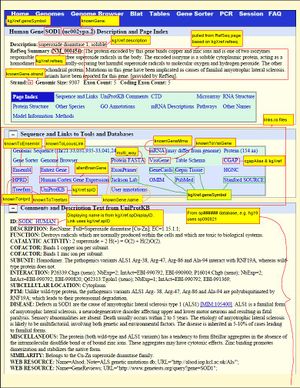
The sources annotated in this image are our best deductions. | The sources annotated in this image are our best deductions. | ||
[[Image:HgGeneAnnoImage.JPG | [[Image:HgGeneAnnoImage.JPG|link=Genomewiki:Hg19uc002yoa.2.pdf|thumb|caption]] | ||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 16:41, 30 November 2009
hgGene Page Source Information:
File:Hg19uc002ypa.2.pdf
Click on the image to see an example of the hgGene page annotated with the sources of the different components.
The sources annotated in this image are our best deductions.
Gene Sorter Column Sources:
|
Name |
Description |
Source |
|
# |
Item Number in Displayed List/Select Gene |
n/a |
|
Name |
Gene Name/Select Gene |
kgXref.geneSymbol |
|
UCSC ID |
UCSC Transcript ID |
knownGene.name |
|
UniProtKB |
UniProtKB Protein Display ID |
kgXref.spDisplayID or kgXref.spID_organism |
|
UniProtKB Acc |
UniProtKB Protein Accession |
kgXref.spID |
|
RefSeq |
NCBI RefSeq Gene Accession |
kgXref.refseq |
|
Entrez Gene |
NCBI Entrez Gene/LocusLink ID |
knownToLocusLink |
|
GenBank |
GenBank mRNA Accession |
kgXref.refseq or kgXref.mRNA |
|
Ensembl |
Ensembl Transcript ID |
knownToEnsembl |
|
GNF Atlas 2 ID |
ID of Associated GNF Atlas 2 Expression Data |
knownToGnfAtlas2 |
|
Gene Category |
High Level Gene Category - Coding, Antisense, etc. |
kgTxInfo.category |
|
CDS Score |
Coding potential score from txCdsPredict |
kgTxInfo.cdsScore |
|
VisiGene |
UCSC VisiGene In Situ Image Browser |
knownToVisiGene |
|
Allen Brain |
Allen Brain Atlas In Situ Images of Adult Mouse Brains |
knownToAllenBrain & allenBrainUrl |
|
U133 ID |
ID of Associated Affymetrix U133 Expression Data |
knownToU133 |
|
GNF Atlas 2 |
GNF Expression Atlas 2 Data from U133A and GNF1H Chips |
gnfAtlas2 |
|
Max GNF Atlas 2 |
Maximum Expression Value of GNF Expression Atlas 2 |
calculated? |
|
GNF Atlas 2 Delta |
Normalized Difference in GNF Expression Atlas 2 from Selected Gene |
gnfAtlas2Distance |
|
BLASTP |
NCBI BLASTP Bit Score |
knownBlastTab.bitScore |
|
BLASTP |
NCBI BLASTP E-Value |
knownBlastTab.evalue |
|
%ID |
NCBI BLASTP Percent Identity |
knownBlastTab.identity |
|
5' UTR Fold |
5' UTR Fold Energy (Estimated kcal/mol) |
foldUtr5.energy |
|
3' UTR Fold |
3' UTR Fold Energy (Estimated kcal/mol) |
foldUtr3.energy |
|
Exon Count |
Number of Exons (Including Non-Coding) |
knownGene.exonCount |
|
Intron Size |
Size of biggest (or optionally smallest) intron |
knownGene exonStarts - exonEnds |
|
Genome Position |
Genome Position/Link to Genome Browser |
(knownGene.txStart + txEnd)/2 |
|
Mouse |
Mouse Ortholog (Best Blastp Hit to UCSC Known Genes) |
mmBlastTab |
|
Rat |
Rat Ortholog (Best Blastp Hit to UCSC Known Genes) |
rnBlastTab |
|
Zebrafish |
Danio rerio Ortholog (Best Blastp Hit to Ensembl) |
drBlastTab |
|
Drosophila |
D. melanogaster Ortholog (Best Blastp Hit to FlyBase Proteins) |
dmBlastTab |
|
C. elegans |
C. elegans Ortholog (Best Blastp Hit to WormPep) |
ceBlastTab |
|
Yeast |
Saccharomyces cerevisiae Ortholog (Best Blastp Hit to RefSeq) |
scBlastTab |
|
Pfam Domains |
Protein Family Domain Structure |
knownToPfam à pfamDesc |
|
Superfamily |
Protein Superfamily Assignments |
ucscScop & scopDesc |
|
PDB |
Protein Data Bank |
kgProtMap2 & sp###### database |
|
Gene Ontology |
Gene Ontology (GO) Terms Associated with Gene |
kgProtMap2 & sp###### database |
|
M. Vidal P2P |
Human Protein-Protein Interaction Network from Marc Vidal |
humanVidalP2P |
|
E. Wanker P2P |
Human Protein-Protein Interaction Network from Erich Wanker |
humanWankerP2P |
|
HPRD P2P |
Human Protein-Protein Interaction Network from the Human Reference Protein Database |
humanHprdP2P |
|
Description |
Short Description Line/Link to Details Page |
kgXref.description |
Table Descriptions
Attempt to describe the uses of the tables used in or related to UCSC Genes.
UCSC Gene and GS Table Descriptions
- allenBrainGene - "Human Cortex Gene Expression" link in Seq&Links to Tls&Dbs
- allenBrainUrl - w/ knownToAllenBrain creats GS column, "Allen Brain"
- bioCycMapDesc - BioCyc description name in Biochem & Signaling...
- bioCycPathway - BioCyc pathway name in Biochem & Signaling...
- ccdsKgMap - determines the CCDS in the "Other names for this Gene" section
- ceBlastTab - other species C. elegans
- cgapAlias - links cgapID with kgXref.geneSymbol to pull info for gene.
- cgapBiocDesc - BioCarta description in Biochem & Signaling Pathways
- cgapBiocPathway - BioCarta pathway name in Biochem & Signaling Pathways
- dmBlastTab - other species D. melanogaster - leave as open issue for now
- drBlastTab - other species zebrafish
- foldUtr3 - mRNA Secondary Structure....section
- foldUtr5 - mRNA Secondary Structure....section
- gnfAtlas2 - own track, QA'd with that track, GS, micrary exp data sxn
- gnfAtlas2Distance - GS sort by "Expression (GNF Atlas2)" & GS clmn "GNF Atlas 2 Delta"
- humanHprdP2P - Gene Sorter column "HPRD P2P" & "sort by"
- humanVidalP2P - Gene Sorter column "M. Vidal Protein-to-Protein" & GS sort by
- humanWankerP2P - Gene Sorter column "E. Wanker Protein-to-Protein" & "sort by"
- keggMapDesc - KEGG pathway description in Biochem & Signaling Pathways
- keggPathway - KEGG pathway name in Biochem & Signaling Pathways
- kg4ToKg5 - allows searching of an old ID from previous gene set in new gene set or users can check the kg3ToKg4 table directly to find corresponding gene IDs.
- kgAlias - pops "Alternate Gene Symbols" in Other Names... section
- kgColor - colors the gene in browser
- kgProtAlias - intermediate table?
- kgProtMap2 - Scop Domains in Protein Domain & Structure Info & Protein Data Bank in GS needs this to work, also involved with proteome browser (not releasing with proteome browser with hg19; being phased out)
- kgSpAlias - duplicate of kgAlias w/ extra field, spID, that is blank in all records
- kgTxInfo - provides "Gene Model Information"
- kgXref - provides the "other names for the gene"
- knownAlt - separate track "Alt Events"; see tracks/altEvents/hg19/methods
- knownBlastTab - Gene Sorter (GS "ID%"=identity, GS "BLASTP E-Value"=eValue, GS "BLASTP Bits"=bitScore)
- knownCanonical - best transcript from each clusterId - don't display splice variants
- knownGene - primary table
- knownGeneMrna - "mRNA" link in Seq & lnks to Tools &Db section
- knownGenePep - "protein" link in Seq & lnks to Tools & Db section
- knownIsoforms - groups transcripts into clusterId
- knownToAllenBrain - w/ allenBrainUrl creates GS "Allen Brain" column/link
- knownToCdsSnp - Not pushing; Coding SNP column in gene sorter
- knownToEnsembl - used in link to Ensembl
- knownToGnf1h - dropped & didn't see changes on hgGene or Gene Sorter, gnfAtlas1?
- knownToGnfAtlas2 - "Microarray..." sxn & Microarray link, GS "GNF Atlas 2 ID"
- knownToHprd - creates the "HPRD" link in the Seq&lnks to Tls&Dbs section
- knownToLocusLink - used in link to Entrez Gene, see issues below
- knownToPfam - gives Pfam Domains section of Prot Dom & Stre info & GS
- knownToRefSeq - used in link to RefSeq (Other Names)
- knownToSuper - contains scop domain info with gene name & start/end
- knownToTreefam - used for link to Treefam website in Seq&lnks to Tls&Dbs
- knownToU133 - Gene Sorter column "U133 ID"
- knownToVisiGene - used in link to VisiGene
- mmBlastTab - other species mouse
- pfamDesc - gives Pfam description in pfam domains section (step in GS)
- rnBlastTab - other species rat
- scBlastTab - other species S. cerevisiae
- scopDesc - prints acc and description in "SCOP Domains" of Prot Dmn * Strtr Info
- spMrna - intermediate table? Doesn't seem to directly affect hgGene or GS
- ucscScop - from ucscID gets scop domainName
Tables Related to UCSC Genes are Separate tracks
- affyU133
- allenBrainAli
- exoniphy - created by Adam Siepel of Cornell for each assembly (2nd choice is to lift from previous assembly)
- gnfAtlas2
- nibbImageProbes
- omimGene
- omimGeneMap
- omimMorbidMap
- omimToKnownCanonical
- vgAllProbes
No longer UCSC Genes Tables
- knownToCdsSnp - dropping on all assemblies. Found too many issues; Populated Cds Snp column in Gene Sorter.
- knownToGnf1h - part of GNF Atlas 1, which is not on hg19
Proteome Browser Tables (no longer releasing)
- pbAnomLimit
- pbResAvgStd
- pepCCntDist
- pepExonCntDist
- pepHydroDist
- pepIPCntDist
- pepMolWtDist
- pepPi
- pepPiDist
- pepResDist
- pepMwAa