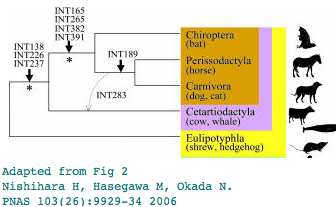
Pegasoferae?
Can rare genomic events establish Pegasoferae?
Pegasoferae is a novel proposal using rare genomic events involving retroposon insertions to establish the phylogenetic ordering within Laurasiatheres, grouping bats, perissodactyls and carnivores to the exclusion of the other hoofed mammalian group, artiodactyls. Bats have been placed in many previous locations, notably in the Euarchonta wing (outgroup to primates). While that particular idea is clearly refuted by many lines of evidence, the proper placement of bats remains under discussion.
Rare genomic events may be more useful for this than maximal likelihood because the orders of Laurasiatheres may have diverged relatively rapidly. Retroposon events are so numerous per million years however that they may be able to resolve branching at these tight nodes. However they suffer from homoplasy in that separate insertion events from a given parental element can look very similar and because deletions over time (no selection for their retention) can cause their disappearance and so confusion with lineages that never had the insertion.
Qualifying retroposons need to be situated between two well-conserved flanking markers because orthology is otherwise difficult to decisively establish in intergenic regions. These markers ideally are no more than 1500bp apart to allow tiling of traces for species without assemblies (eg vicugna, pig, dolphin, macrobat in Laurasiatheres) and spanning PCR runs. Higher sampling density greatly enhances the ability to correctly infer the sequence of events.
Short coding indels in coding exons can also be phylogenetically informative. Here if the exon is otherwise quite conserved, the risk of homoplasy (recurrent events at the same or indistinguishably similar position) is fairly low. These events are inherantly rare first because conserved regions of a protein may not admit indels structurally (ie are inactivated) and second because the window of relevancy for a given tree topology issue may only be a small fraction of elapsed evolutionary time (eg 1 million year stem on a 85 myr branch).
Coding indels can exhibit the usual problems of lineage-sorting: two co-existing alleles at the time of speciation that resolve differently in descendent lineages. Insertions, while a third as common as deletions and so less likely to have arise multiple times, are more subject to subsequent confusing reversion; deletions are less likely to revert to ancestral length for lack of genetic mechanism. It goes without saying that indels from repetitive regions or in dna of anomalous composition are wholly unsuitable for taxonomic purposes.
Methods for re-analysis and supplementation of L1MA9 data
Nishihari et al located 4 retroposon insertion events that support Pegasoferae. In the ensuing 2-3 years, it has become possible to reinvestigate these events with additional species utilizing bioinformatic methods on the 10 available Laurasiathere genomes (not all of which will completely cover a given event).
This can be done fairly rapidly (though only a subset of the putatively informative L1MA9 insertions are analyzed here). First, the primers used by the Okada group are blatted into the UCSC genome browser of an appropriate species (dog). The reverse strand primer must be first reverse-complemented with the UCSC utilities tool for a single line of Blat output to span the region of interest.
In every case studied here, the L1MA9 elements lie within small introns of coding genes. That means typically 30-40 amino acids from each flanking exon are available to create reliable anchors for future blast searches at NCBI. Without these anchors, L1MA9 elements do not necessarily retrieve orthologous elements in other species via blast, for the reason that the active L1MA9 parent element may have spun off subsequent fragments that are closer in sequence.
Next, dna spanning the exons and their internal intron are collected from Laurasiatheres with UCSC genome browsers, notably dog, cat, horse and cow. This dna is then marked up for all retroposons using the RepeatMasker track. Here it is important to track orientation of the repeat element (letting the reading frame of the exons define the positive strand convention), the fragment coordinates relative to an idealized repeat of the given class in the underlying RepeatMasker library, and the percent identity to this element.
These three steps allow in-depth comparison of putatively orthologous retroposons, noting however in the case of L1MA9 elements that fragment coordinates are very similar in many thousands of these insertions due to the replication mechanism favoring distal fragments of similar lengths. Thus homoplasic events cannot truly be ruled out even upon this closer examination.
The comparative genomics is extended now to additional species using tblastn of the WGS contig division of GenBank. This target database stores 2x assemblies of species such as microbat, hedgehog, and shrew. The spanning contig GenBank entries are then uBlasted against the growing database of fiducial sequences to locate the exons and intervening sequence. The latter is most conveniently retrieved by screen-scraping of 6-frame translation, dropping out the alternate lines containing the amino acids.
The latter two species exons can sometimes be retrieved faster from the 28-species alignment track at UCSC -- genomic alignment provides more reliable orthologs -- some lineages could have idiosyncratic segmental duplications that could cause cross-matching. If the coding regions are evolving too fast, the whole enterprise becomes questionable. Another pitfall is paralogs. Here GeneSorter at the UCSC site can be consulted, at least for human, to rapidly examine the situation.
Next, the est_other and nr divisions of GenBank are probed to retrieve any data that might be available from non-genomic projects. Transcripts per se don't seem useful here (lack the intron of interest) yet they provide precise probes later of the trace archives for the given species.
Finally, the trace achives are consulted for residual species such as macrobat, vicugna, sheep, pig, and dolphin. Note too that contigs and genomes often leave out singleton traces so these species can be completed here as necessary. The approach here is to tile in from both directions extending the reliable exon matches (if any). Since the L1MA9 elements needed to occur in small introns given the Nishihari et al discovery approach, usually 3 traces suffice to tile the intron successfully.
Trace reads are inherently error-prone (hence the need for 6x coverage to obtain a reliable genomic assembly). However here it is unlikely that a whole L1MA9 element would be missing unless the species in question had two distinct alleles. In evaluating rare genomic events, it is important to check all high-quality covering traces to rule out such polymorphisms because these would seriously undercut the whole exercise.
After obtaining data from all available Laurasiatheres, the sequences are marked up with different colors for their various constituent elements as illustrated below. Note the fasta header can carry the exonic sequence as well as quantitative information about the repeat elements. By extracting header lines only, a simple flatfile database emerges suitable for making simple summary comparative diagrams/
Analysis of L1MA9 retroposon INT189
The phlogenetic distribution of the L1MA9 retroposon INT189 has been taken as evidence for bats being the immediate outgroup of horse + dog. That interpretation can be revisited using newly available genomes. Yet only two sequences representing perissodactyl and carnivore are at GenBank as cat assembly has a gap in the critical region. But other new data in 3 bats and 4 cetartiodactyls and 2 shrew/hedgehog confirm the lack of L1MA9 near the distal exon.
The trouble is a second L1MA9 element lies upstream of the MER58A middle marker. This is lacking in both carnivores. Evidently it was deleted in stem carnivore -- otherwise it would be providing evidence for carnivores being outgroup to cows + bats + horse. In short this single intron is providing 'support' for two contradictory topologies.
The sizes of many bat genomes have been experimentally determined: the 30-genus average of 2.6 gbp is about 500,000,000 bp less than human. Since bats in essence have the same 20,000 coding genes as other mammals, that discrepancy has to arise from less intronic and intergenic dna. Possibly bats had fewer active retroposing elements. Far more likely, bats they have an average number and the discrepancy arises from a faster rate of deletions than insertions.
Thus for taxonomically informative (ancestral laurasiathere) retroposons, many millions of deletion events have occured. Since the L1MA9 elements here are only 100bp or so, it would come as no surprise if a high percentage of the older relevent ones have experienced partial (or full) deletions making them unrecognizable with RepeatMasker.
Thus presence of a retroposon in a given orthologous position bat can be informative but absence is not so informative. INT189 is an absence. That one event isn't insufficient anyway to establish branching order. So bat/horse/carnivore tree topology remains unresolved. If horse is the outgroup to carnivore + bat -- and cow outgroup to all of these -- then hoofed animals are parsimoniously ancestral (rather than arising twice by convergent evolution) and bat and carnivore lost hooves (a bit unreasonable as dog and bats retain the ancestral 5 digits).
Summary of the phylogenetic distribution of the L1MA9 retroposon INT189: >PGM2_canFam Canis familiaris (dog) abseny -MER58 182-265 23% -L1MA9 6069-6302 27% >PGM2_felCat Felis catus (cat) genomic del absent -MER58 no data >PGM2_equCab Equus caballus (horse) -L1MA9 6172-6264 26% -MER58A 1-145 23% -L1MA9 6050-6302 23% >PGM2_myoLuc Myotis lucifugus (microbat) -L1MA9 6174-6264 20% -MER58A 38-157 26% >PGM2_pteVam Pteropus vampyrus (macrobat) -L1MA9 6161-6291 25% -MER58A 35-145 29% >PGM2_pipAbr Pipistrellus abramus (microbat) -L1MA9 6180-6301 252% -MER58A 38-157 24% >PGM2_bosTau Bos taurus (cow) -L1MA9 6155-6263 28% -MER58A 7-157 21% >PGM2_turTru Tursiops truncatus (dolphin) -L1MA9 6155-6265 29% -MER58A 37-148 21% >PGM2_susScr Sus scrofa (pig) cdna + tiled -L1MA9 6159-6264 27% -MER58 212-271 28% >PGM2_vicVic Vicugna vicugna (vicugna) tiled -L1MA9 6162-6310 24% -MER58A 35-157 20% >PGM2_ateAlb Atelerix albiventris (hedgehog) ... ... >PGM2_sorAra Sorex araneus (shrew) ... ... >PGM2_canFam Canis familiaris (dog) -MER58 182-265 23% -L1MA9 6069-6302 27% VISAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICHDQGTIKKLFENLRNY GTCATCAGCGCCGAGTTGGCTAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAGGCCATTTATGTTGAGTACGTTTCTATTAACTCTG TTTAATTGAAATAATACTTTTTAAAAGTTTTATTATGTTTTTATGTGTGACACTAATATTCTAACCCTCTTACTTTGGGTGAGGGTTCTTCTGAAAACTA AAGGATCACTTTTTCTTTTAATGCTTAACTATTCAATACTAATTATCACTTATGACTGTGTTAATCCTTAACAAATGAGAACATCAGTTGCAGAAATAGC TAATTGAGGAGGGTGATTCCCTGATGTCAGAAAGGACAAAGGTTTTCGTGAAACATCTATTACGTGTTTAGAgccactagtcaagtctgcctttgtagtg caaaagcagctgatggcaagacgtacaggaatgggtgtggtgtggctgcaatgaaaTGAAACTTTCACCTCCCAAGATAGGCCGAAGGCCAGGCAGCAGT TTGGCAATACCTGGGGTCAATAGTTATACCTCTTTTTTATGCTAAATTATTCCTTTGAAGCTAGTCATTGTTATCGTTTCATTTAGCTTAAAATATACTG ATTGCTACATGTTCTGTATACACCACGTGAGATTATTTGTTCCTCATTTTGCATATTTGTACTTTTtttattgagatgtaattgacattaatgtcaggta taataacataatgattcgatatttatatattattacaaagtgatcaccatagtaagtcgagttaacatccacaccacatataatcacaaatattcattct tgtgatgatagcttttatgatctgtggtcttagcaactttcaaatatacagtacaatactagtagatacagtcaccaagttatatatATATAATTTTATT TCTTTTGATAGATATGGCTACCATATTACCAAAGCTTCCTATTTTATCTGCCATGATCAAGGCACCATTAAAAAATTGTTTGAAAACCTTAGAAACTAC >PGM2_felCat Felis catus (cat) genomic del incomplete coverage -MER58 ASFLATKNLsLSQQLKAIYGE YGYRITKASYFICHDQGTIKQLFENLRNY GCTAGCTTTCTAGCAACCAAGAATTTGTTTGTCTCAGCAGCTAAAGGCCATCTACGGCGAGTAAGTGTCTTCTAACCTGGTAAAGAAGTAATAG TGTTAAATATTTTCTTATGGTTCTACGTGTGAGATATTAATATTCTTTCTAATGCTCTTTGGTTGTGAATTCTATTTCTTTTTCTTTTTTTAATGTTTAT TTATTTTTGAGAGAGAGAGAGAGAGAGATGGAGTATGAGCAGGGGAGGGGCAGAGAGAGAGGGAGATACAGAATCCAAAGCAGGCTCCAGGCTCTGAGCT GTCAGCACAGAGCTCCACACGGGGCTTAAACTCACAAACCATGAGATCATGACCTGAGCTGAAGTCAGACACTCAACCGTTTGAGCCACCCACGTGCCCC ATGAATTCTATTTCTTATGAAACTAAATAATCATCTTTTCTTTTGATACTTAACCATGTAATGGTAATTATCATTCACGATTGCACGAATCCTTAACAAA TGAGGGCATCAGTTGCAGAAATAGCTAATTGAAGAATGTGATTTTAAGTGTGTGATGTCAAAAAAGATTAAAGGTGTTCATGAAATCTCTATTAAGTTTT TAGAGCAATGACCCAGGTCTGCCTTTATAAAGTGCAAAAGCAGCCCGTGGCAACACGTTGCAGTAAGACTCTTACTTACAAATACAGGCTAAAGGCCAGG CAGCAGTTTGGCAATCCCCAGGGTTAATTGTTGTACCTCTTTTTTATGCTAAATTATTCCTTTGAAGGTACTCATGGCTATTTGTTTCATTTGGTTTAAA ATATACTGGTTGACAAATGTACACTGTGTGGAATTATGTGTTCCTCATTTTGCATATTTGTATTTCCTTAACTGAGATATAACTGACATTAGTTTCAGGT ATGCGATACAGTGTTTCAATATCTGTATATATTACAAAATGATCATCACAGTACATCTAGTAACAGTCGCACCACACTTAATACAAAAGT TCCaTATGGCTACCGTATTACCAAAGCTTCATATTTTATTTGCCATGATCAAGGCACCATTAAACAATTATTTGAAAACCTTAGAAACTAT >PGM2_equCab Equus caballus (horse) -L1MA9 6172-6264 24% -MER58A 1-145 23% -L1MA9 6172-6264 26%-L1MA9 6050-6302 23% VISAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICYDQDTIKKLFENLRNY GTCATAAGCGCAGAGTTGGCTAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAGGCCATCTATGTTGAGTAAGTTTCTATTAACTCTC TTTAACTGAGGTAATTTTTTTTATTAGtttcaaatgtacaacataatgattcaatgtatgtatatattttgaaatgatcgccacaataagtctggctaac ctgtatcaccgacatagGGCTCTTTTTAAATGTTTTATGTTCTTTTGCATGAAACAGTAATATTCTTTTGAATGCTCTTACTTTAGCTATGAATTGTTCC TTATGAAAACTAAGTAAGAGATCACTTTTTCCTTTCGATACTTAACCACTTAGTAGTATTACCCTTTGTGATTGCATTAATCCTTAACAAATGAGAACAT TAGTCACGGAAATGGTGAAGTGAAGAATGTAATTTTCAGTGTCTGAGGTCAAAAAAGATTAAATGTGTTCATGAAACATCTATTTAGTCTTTAACTTCat tgctcagctctgcctttgtagtgcagaaacagccggggacaatacataatgtaatgggtgtggggtggctgtgttccagtagatcttttacttaaaaata caggccgaaggccaggcagcagtttggcaatccctgGGGGAGATTATTGTACCTTTTTTTAATGTTAAATTATCCCTTTGAAGTTAGTCATGGTTATTTC ATTTAGTTTAGAATATAATGGTTAATACATAGTGTATGTACACCATGTGGAATTATTTTTTCCCATTTTGCATTTCTTCTtttgttgagatataattaac atagaacattatattagcttcaggtgtacagtgtaattatttgataattgtatatattgcagattgatcaccaccataagactagttaacatccatcacc acacatagttataaatttttttcttgtgatgagaacttttaaggtctattctcttagcaaccttcaaatatacaatacagtattattaattctagtcacc gtgctgtgtattatatcctcatgacccattTTATTATTTTGTTTCGAAAGGTATGGCTACCATATTACCAAAGCTTCATATTTTATCTGCTATGATCAAG ACACCATTAAAAAATTGTTTGAAAACCTTAGAAACTAC >PGM2_myoLuc Myotis lucifugus (microbat) -L1MA9 6174-6264 20% -MER58A 38-157 26% VISAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICHDQGTIKKLFENLRNY AAPE01636299 GTCATAAGCGCAGAGCTGGCTAGCTTTCTTGCAACCAAAAATTTGTCTCTGTCTCAGCAGCTAAAGGCCATCTACGTTGAGTAAGTTTCTATTGATTATTG AATTGAAGTAATATAGTTTGATTAGTTTCATGTGTACAATGTAATGATTCAATATGTGTATATATTGGGACATGGTTGCCACAATAAGTCGTTAACATAC ATTACCACATGTGGCAATGTATTTTAAGTGTATTATGTTCTTGCGTATGAGATGCTAATGTTCTTTCCAAAGCTCTGACTTTAGTTATGAATTCTATTTC TTAAGAAAACGAAACGAGATTATCTTTTCCTTTTGATACTTACCATTTGTGATAGCACTAATCTTTACTAAATGAGAACATGACACAGAATGTGATTTTA AGTGTCTGATGCCAAAAAAGATTAAATGTGTTCATGAAACGTCTATTTAGTCTTTATAGCAGTTTCTCAACTCTTGCCTTTCTGATGCAAAAGGAGCCAG ACACAGTACATAATGCAATGGGCGTGGTATGGCTGTTCCAGTATAATTTTACTTACAAGTATAGGCTGAAGGCAAGGTAGCAGCTTGGTGAGCCCTCGGG TAAATTGTTGCACCTCCTTTTAATGCTAAATGATTGCTTTGAAGCTAGTCATGGTCATTTGTCTCATTACGTATTTGAGAATGTGCTGGTTGGTGCCCGT TCTGTATATGCTATGCATAATTATTTGTTCCTCATTTTGCATGTATTTGTATTTGTTTTGATAGGTATGGCTACCATATTACCAAAGCTTCATATTTTAT CTGCCATGATCAAGGAACCATTAAGAAATTATTTGAGAACCTTAGAAACTAT >PGM2_pteVam Pteropus vampyrus (macrobat) -L1MA9 6161-6291 25% -MER58A 35-145 29% VISAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICHDQGTIKKLFENLRNY GTCATAAGCGCGGAGTTGGCTAGCTTTTTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAGGCCATCTATGTTGAGTAAGTTTCTATTGACTCTA CATAACTGAAATAATATTTTTTATTAGTTTCAGGTGTACAGCACAGTGATTCGGTATATGTATATATTATGACATGATTGCTATAAGTCTATTGCATGCA TCAGTCTATTACTACATGCATCACCACACGTAGTAATATTTTTAAATGTATTATGTACTTGTGCACAAGATACTAATATTCTTTCCAATGCTCTTACTTT AGTTATGAATTCTATTTCTTATAAAAACCAAATAAGAAATTACCTTTTCCCTTTGATACTTAGCCATTTAATAGTAATTACCATTTGTGATGACAGTAAC CTTTACCAGATGAGACATTAGCCACAGAAACAGCTAAAGAATATGATTTTAAGTGTCCGATGTCAAAAGATTAAATGTGTTTATGAAACATCCTATTTAG TCTTTTTATAGCATTATTCAGCTGTGCCTTTGTAGTACAAAAGCAGCCAGACCCGATGCATATGTAATGGGTGCAGCGTGGCTACATTTCTGTAAAATTT TTACTTACAAATATAGGCTGAAGGCCAGGCAACAGTTTGGTGATCCCCTGAGTAAATTGTTATACTTCTTTCTTAATGCTGAACTATTCCTTTGAAGCTA GTCATGGTCATTTGTTTCATTAAGCGTTTTAGAATGTACTGGTTGATACATGTTCTGTGTACACTATGCAGAATGATTTGTTCCTTATTTTGCATGTGTT TGTATTTATTTTGATAGGTATGGCTACCATATTACCAAAGCTTCATATTTCATCTGCCATGATCAAGGCACCATCAAAAAATTATTTGAAAACCTTAGAAACTAT >PGM2_pipAbr Pipistrellus abramus (microbat) -L1MA9 6180-6301 25% -MER58A 38-157 24% AB258957 AIYVE YGYHITKASYFICHDQGTIKKLFENLRNY GGCCATCTATGTCGAGTAAGTTTCTATTGATTATTGAATTAAAGTAATATAATTTGATTAGATTCATGCGTACAGTGTAATGATTCAATACATGTATATA ATGGGACATGGTTGCCACAATAAGTCGTTAACATACATCACCACCTGTGGCAATATATTTTAGGTGTATTATGTTCTTTAGTATGAGACACTAGTACTAA TATTCTTTCCAAGGCTCTGACTTTAGTTATGAATTCTATTTCTTAAGAAAATGAAACGAGATTATCTTTTCCTTTGGATACTTACCATTTGTGATTGCAC TAATCTTGATTAAACGAGAACATTACACAGAATGTGATTTTAAGTGTCTGATGCCAAAAAAGATTACATGTGTTCATGAAACATCTATTTAGTCTTTATA GCAATTTCTCAACTCTTGCCTTTCTGGTGCAAAAGCAGCCTGACACAATACATAATGTAATCGGCGAGGGATGGCTGGTCCAATAAAACTGTACTTACCA ATGTAGGCTGAAGGCAAGGTAGCAGCGTGGTGTTCCCTCAGAATTATTTGTTCCTCATTTTGCACGTATTATTTGTTTTGATAGGTATGGCTACCATATT ACCAAAGCTTCATATTTTATCTGCCATGATCAAGGCACCATTAAGAAATTATTTGAAAACCTAAGAAACTACGATGGGAAGAATAATTAT >PGM2_bosTau Bos taurus (cow) -L1MA9 6155-6263 28% -MER58A 7-157 21% VITAELASFLATKNLSLSQQLKAIYVE YGYHITRASYFICHDQETIKQLFENLRNY AB258958 [L1_Carn7] GTCATAACTGCAGAGTTGGCCAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAAGCCATCTATGTTGAGTAAGTTTCTATTGACTATT TAATTGAAGTAATTTTTTTTTATCAGttcaggtatacaacacagtgattcagtgtatgtctatattgtgaaatgatcacagtggatacaattaacatgca tccccacacaggaatattttttaatgtTTTACTCTCTTCTTGTGCACCCGATACTCATATTCTTTCTGATGCTCTTGCTTTAGTTATGAATTCTATTTCG TATGAAAACTAAATAAGAGATCACCTTTTCCTTTTGCTACTTAAGCAGTTAATAGTAATTACCATTCATGATGACGTTAATCCTTAATAAATGAGAACGT TAGCTGCAGAAATGGCTAAGGGAAGAATGTGATTTTTTAAATGTCCAGTGTTGAAAAAGACTAAATGTGTTCATTAAACATCTATTTAgtctttgtagca attacttatttctgcctttctagtgcaaaagcaaccagacacaaggtaatgggcatgacgtggctgtattccaatgataaaacttttacttacaaacaga gactgagggccACACAGCAGGGCAGTGATTCCTGGTGTAGATTGTTGGACCTCTTTATTTAATGCTGAATTACTCCTTTGAAATTAGTCATGGTTGTTTG TTTTAGAATATACTGTTTGATAGATACATGTTCAGTGTACACTGTGCCCAATTATTTGTCCCTCATTTGCATGTAACCATGTTTGTATTGATAGGTATGG CTACCATATCACCAGAGCTTCGTATTTTATCTGCCATGATCAAGAAACTATTAAACAATTATTTGAAAACCTTAGAAACTAT >PGM2_turTru Tursiops truncatus (dolphin) -L1MA9 6155-6265 29% -MER58A 37-148 21% FISAEVGSFLAQNCLVSAAKAIYV YGYHITKASYFICHDQGTIKKLFENLRNY TTCATAAGTGCAGAGGTTGGCAGCTTTCTAGCACAGAATTGTCTTGTTTCAGCAGCTAAAGCCATCTATGTTGAGTAAGTTCTTCTATGACTGTTAAATG AGTAATGTTTTTTTTCATTTCAGTTGTGCAACACAATGATTCAATGTATATCTATTATTGTGAAATGATTGCAACAAATACAGTTTACATGTATCCCCAC ATGTAGTAATATTTTTTAATGTTTTACTCCGTTCTTATGCATGAGATACTAATATTCTTTCTGATGTCCTTACTTTGGCTATGAATTCTATTGCCTATAA AAACTAAATAAGGGATCACCTTTTCCTTTCGATATTTAACTACTTAATAGTAGTTACCCCTTCATGATGACATTGATTCTTAACAAATGAGAACATTAGT TGCAGAAATGGCTAAGGGAAGAATGTGATTTTTAAGTGTCCAATGTCAAAAAAGACACATGTGTTCACAAAACATGTTTAGCCTTTAAAGCAATTATTCA CCAGTGTCTTTGTAGTGCAAAAGCAGCCAGACACAATACATAAGGTAATGGGCATGGCATGGCTACGTTCCAATAGAGAAACTTTTACTTAGAAATACAG GCTGAGGGCCACAGAGCAGTTCAGCGATCCCTGGGGTAGATTGTTGGACCTCTTTTATAAAATTGGACCTCTTTTTTTTTTTTTTTTTTTTTGGCGGGGG GTACGTGGACCTCTCACTGTTGTGGCCTCTCCCGTTGCAGAGCACAGGCTCCAGACGCGCAGGCTCAGTGGCCATGGCTCGCGGGCCCAGCCGCTCCACG GCATGTGGGATCTTCCCAGACCGGGGCACGAACCCGTGTCCCCTGCGTCGGCAGGCGGACTCTCAACCACTGCGCCACCAGGGAAGCCCTGAACCTCTTT TTTAATGCTGAATTATTCCTTTGAAATTAGTCGCGGTTATTTGTTTTAGAATATACTGGTTGATACATGTTCAGTGTACACTGTGCAGAATTATTTGTTC CTCGTTTTGCATGTAATTGTGTTTGTATTGATAG GTATGGCTACCATATCACCAAGGCTTCGTATTTTATCTGCCACGATCAAGGCACTATTAAAAAATTATTTGAAAACCTTAGAAACTAC >PGM2_susScr Sus scrofa (pig) cdna + tiled -L1MA9 6159-6264 27% MER58 212-271 28% VISAELASFLATKNLSLSQQLNAIYVE YGYHVTKGTYFICHDQGNVKKLFENLRNY GTCATAAGCGCAGAGTTGGCCAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAATGCCATCTATGTTGA GTAAGTTTCTATTGACTGCATTTAATTGAAGTAATTTTTTTAATCAGTTTCGGGTGTACGACATAATGATTCAGTGTATATGTATTGTGAAATGATCCCAA TGAGTACAGCTAACATGCATCCCACACGTAATAATATTTTTTTTTCTTTCTTTTTCTTTTTTTAGGGCTACTCCTGTGGCATATGGAAGTTCCCAGGCTA AGGGTCGAATAGGATCCATAGCCGCTAGCCTAAGCCACAGCCACAGCAGCACGGAATTCGAGCCACATCTTTGACCTCCGCTACAGCTCATGGCAATGCC AGATCCTTAACCCACTGAGCAAAGCCAGGGATCAAACCCAACATCTCATGGATCCTAGTCGGGTTTGTTAACCCTTGAGCTGCAAAGGGAACTCCCATAA TAATCCTTTTAAATGTTTTACTCTGTTCTGATGCATGAGACTAATATTCTTTCTGATACTCTCATTTTAGCTATAAAGTTGATTTCTTATGAAAACTCAG TAAGAGATCACTCTTTCCTTTTGATATTTAACCCCTTAATAGTAATTACCATTCATGATGACATTAATCCATAACAGATGAGAACAGTAGTTGCAGAAATGGGTAAT GGAAGAATGTGATTTCAACTAAATGTCCAATATCAAAAAAGACTAAGTGTGTTCATGAAACATCTATTTACTATTTATAGCAGTTATTCAGCTCTGCCTT TGTAGTGGTAAAGTGGTCAGACACAATACTTAAGGTAAAAGTTTCCAGTTATGAAACTTTTACTTACAAATATGGGCTGAGACTGGGCAATAGTTCAGTG ATTCCTTGGGGTAGATTCTTGGACCTCTTTTTTTAAATGTTGGACCTCTTTTTTAATGCTAAGTTATTCCTTTGAAATTAGTCTTGCTTATTTGTGTCAT TTGTATTGAAGTATACTGGTGAATTACATGTTCTGTGTATGCTGTGTGGAATTATTTGTTCCTCATTTTGCATGTAATTGTATTTGTATTGATAGG TATGGCTACCATGTTACCAAAGGTACATATTTTATCTGCCATGATCAAGGCAATGTTAAAAAATTATTTGAAAACCTTAGAAACTACGATGGGAAGAATAATTAT >PGM2_vicVic Vicugna vicugna (vicugna) tiled -L1MA9 6162-6310 24% -MER58A 35-157 20% VITAELASFLATKNLSLSQQLKAIYVE YGYHITKASYFICHDQGTVKKLFENLRNY GTCATAACTGCAGAGTTGGCTAGCTTTCTAGCAACCAAGAATTTGTCTTTGTCTCAGCAGCTAAAGGCCATTTACGTTGAGTAAGTTTCTATTAATGCTG TTTAATTGGAGTAAGCTTTTTATCCATTTCAGATGTACCACATTATGACTCAGTATACGTCTACATTGTGAAATGATCACAATTAGTAAAGTTAACGTGT ATCATCACACATAGTAATATTTTATAATGCTTTACTCTGTTCTTGTGCATGGGACACTAATGTTCTTTCTGATGCTCTTTCTTTAGTTATGAATTCTGTT TCTTATGAGAACTAGATAAGAGATCATCTTTTCCTTTTGATACCTAATCACTTAATAGTAATTACCATTCATGATGACATTAATCCTTACAAATGAGAAA ATTAGTTGCAGAAATGGCTAATGGAAGAATGCGATTTTAAGTGTCTAATGTCAAAAAAGACTAAATGTGTTCATGAAACATCTGTTTAGTCTTTATAGCA ATTACTCAACTCTACCTTTGTAGTGCAGAAGCAGCCAGACTCAACACATAAGGTAATGATGTGGCTGTTCCACTAATAAAACTTTTACTCAAAAACACTG GCTGAGGGCCAGGCAACAGTTCAGCAATCCCTGGGGTAGATAGTTGGACCTCTTTTTTTTAATTCTAAATTATTCCTTTGAAACTCATCATGGTTATTTG TGTCATTTATTTTAGAGTATACTGGTTGATGACATGTTCAGTGTACACTGTGCAGAATTCTTTGTTCCTTGTTTGCATGTAATTGTATTTGTATTGATAG GTATGGCTACCATATTACCAAAGCTTCATATTTCATCTGTCACGATCAAGGCACTGTTAAAAAATTATTTGAAAACCTTAGAAACTAC >PGM2_ateAlb Atelerix albiventris (African hedgehog) No repetitive sequences [VISAELASFLATKNLSLSQQLKAIYVE] YGYHITKASYFICHDQVTIKKLFENLRNY AB258952 TTAATGTGTTTGTTAAACATCTATTTATTCTTTACAGCTATCACTCAACTCTGACTTTGTAATACAAAATAGCCACACTTAGTCCATGAGGTCATGGACC TGATGTGACTGCCCCAATAAAACTTATACCTACAGATATAATCAAAATAAGATAAAATGGATGCTATCAATACTTAAGAATATTGGCTAAGTAAAAACAA AGAACTAGTTTAGAAACCTACAGGGGGTTATTGTTCTTCCTTTTTTTCATGCTATATTATTCCTTTGAAGCCAGTCATAGTTATTAGTCTCATTAACTTT ATAATATACTGGTTATATATGTTCTGTGTATACTAGGTAAAGTTATTTCTACCTAATTTTGCATACGTTTTATTTGTTTGCTAGGTATGGCTACCATATA ACCAAAGCTTCATATTTTATCTGCCATGATCAAGTCACCATTAAAAAATTATTTGAAAACCTTAGAAATTAT >PGM2_sorAra Sorex araneus (shrew) +SOR1_SINE +SOR1_SINE VISAELASFLATRNLSLSQQLKAIYVE YGYHITKASYFICHDQSIIKKLFENLRNY AALT01183695 AALT01470682 GTCATTAGCGCGGAGCTGGCCAGCTTTCTCGCCACCAGGAACCTGAGTTTGTCCCAGCAGCTAAAGGCCATCTATGTGGAGTAAGTTCCCTACTGACTGT GCTTAATCAAAATAACCCGTATTTTTGGATCCATTTTTAACGGTTTATTATGCTCTTGTGTGTGTGATACTGATAGTCTCTCTAATGTCCTCACTTCAGT TATAAATCCTATTTCTTAAAAACATGAAGTTAAGGGGCTGAACCGATAGCACAGCGATAGCAAGGTTTGCCTTGCATGTGACCGATCTGGGTTCGATTCC CAGCATCCCATTTGGTCCCCTGAGCACTGCCAGGAGTAATTCCTGAGTGCATGAGTCAGGAGTAATCCCTGTGCATCGCTGGGTGTCACCAAAAAAAAAA AAACCATGAAGTTAAAAAATCACCTTTTGGGGGGGGTCGGAGAGATAGTGCAGCAGTGGGTAGGGAGCTTGAGTCATTCATGGGTCACCCAGCTTCAATC CCTGGCACGCCCTGTGGCCTCCCAAGTCCCGCCAGGAGTGATCCCTGAGCTCAGAACCATAAGCAAGCCCTGAGCACCATTGGTGTGGCCCCAGAATAAA TAAATTAGAGATAGAAATCACTTTTTCATGCTTAACTACTTAATAATACTTATGATTGCCATACTCCCTAATGAATGAGATCTAATCGCAGAACTAGTTA TTAGTTAAAAGTGTGAATTTAAATGTGTAGTGTCAAAAAAATG ACCAAGATAACCAGCTTATAACTTAGACTTATAAATGACTGCTTATCAATATATCT AAGGCCAGACAGCAGTTTACTTTAGCAGTTCCTAGGATAGGTTATTGTTCCTCTTTTTTTTTTCCCTTTATTTTTTGCCCCCTAGAGATCTACCTTTTAA AAAAATATTTTTTTAATTGAATCACAATGAGATACACAGTTACAAATTGTTTCTGATTTGATTTCAGTCAGACAATGTTCAAATATCTGTCCCTTTCACA GTGTACATTTCCCACCACCAGTGTCCCCACTTTCCTTCCTTGTTCCTCTTTTTTCATGCTCAGTGATTCCTTTGAAGCGAATTATGGTCATTCGCTTCAC TTGCTTAAAAGCAAATGAATCAGCGGCCGATTGATGTCCTGTGTTCGAAAGACAGAATTCTTTGTTCCTTATTTTGCGTGTATTTGTATTGATAGGTATG GCTACCATATAACCAAAGCTTCGTATTTTATCTGTCACGATCAAAGCATCATTAAAAAGTTGTTTGAAAACCTTAGAAATTAT
Analysis of L1MA9 retroposon INT283 within gene ZUBR1
This potentially informative retroposon insertion denoted INT283 conflicts with Pegasoferae topology. That is not a fatal flaw because a certain fraction of such events resolve anomalously via lineage-sorting after speciation.
Again, this presented a favorable annotation situation because the two flanking exons are well-conserved and the intonic distance is short at about 1500 bp. Using new genomic data, the species density can be brought up 10 Laurasiatheres, tiling traces in unassembled genomes as necessary. The conflict with Pegasoferae holds up even sampling at this greater taxonomic sampling depth: an orthologous L1MA9 is also missing in microbat, shrew, hedgehog but present with correct orientation and correct fragment coordinates in vicugna, pig, and dolphin.
It can always be asked whether this is the "same" L1MA9 in cetartiodactyls or merely a similar one from a separate insertion event of the active parental element. However pushing the putative event back to basal vicugna narrows the window for that. While this situation can be dismissed with an appeal to lineage-sorting (importantly 4 events support Pegasoferae versus this 1 conflict).
Deletion in bat could equally be argued, though located orthologous dna in macrobat now pushes that putative event back into the shorter stem window as well. Absence in bat is not especially informative given their genomes are greatly reduced in size relative to other placental mammals -- much of this loss would occur in intragenic dna which like much intergenic does not often experience selective pressure for retention.
>ZUBR1_canFam Canis familiaris (dog) 2108 bp -L1MA9 6025-6299 25% >ZUBR1_felCat Felis catus (cat) 1631 bp -L1MA9 5958-6299 30% >ZUBR1_equCab Equus caballus (horse) 1667 bp -L1MA9 5959-6299 23% >ZUBR1_myoLuc Myotis lucifugus (microbat) 1227 bp no repeat detectable >ZUBR1_pteVam Pteropus vampyrus (macrobat) 1388 bp no repeat detectable >ZUBR1_bosTau Bos taurus (cow) 1891 bp -L1MA9 5958-6302 24% also -tRNA-Gly -tRNA-Glu SINEs >ZUBR1_turTru Tursiops truncatus (dolphin) 1688 bp -L1MA9 5936-6312 21% >ZUBR1_susScr Sus scrofa (pig) 1022bp ---- bp -L1MA9 5958-6182 22% not fully tileable >ZUBR1_vicVic Vicugna vicugna (vicugna) 1525 bp -L1MA9 5937-6312 22% >ZUBR1_eriEur Erinaceus europaeus (hedgehog) 1256 bp no repeat detectable >ZUBR1_sorAra Sorex araneus (shrew) 1098 bp no repeat detectable Markup of exons and intronic retroposons of INT283 within ZUBR1: blue: coding exons magenta: L1MA9 INT391 >ZUBR1_canFam Canis familiaris (dog) 2108 bp -L1MA9 6025-6299 25% KKYLSQKNVVEKLNANVMHGK HVIVLECTCHIMSYLADVTNALSQSNGQGPSH AAGAAATACCTATCACAGAAGAACGTGGTTGAAAAACTGAATGCCAATGTGATGCATGGAAAGGTAAGCGAAATGCACCTTGACAGCAGTCAGGAAGTGA TGAGTCTTCTTTGTGGTCATGTAGAAGACTCTCCTTTTGACTGGTTCCTGGTCCTAGCCAGCCTGCCCACAATTTCAAAGCCCTGGGGGTGTGTAACATG AACTACCTGCTCTCAGGCTTCTACTAGGTTATGAGGAATGATGAGGGGAGGGAAGGGGGAATACAAGACGATAGACAAAACATCCAACTGAATCCTTAAA ACCTAGTAAATTCTTGCTATTTTTAGATTTTGTTATGTTGTGGAAAATTGTTCCCTGCTCCTGCATTGGAACTTTGTGCTCTGAGTATGTGTTTTAGGGG GTTACCTACTGCCTTCTCAGCTCTAGTCCACTTGCTAGTATTGTTTACTGCTGAGAAAAAGGGTTTTTAAACTGACAGAAATCTTGCATTCAGGCATTTT TCCTACTTGCCCTACCGTGAACTGGACATAAGTGGCTAAAAAGAGCTAACTGGCCTCTGAGACCTAGCACAGCACCGTTCATGTCTCCTTTCCTTTCTTT CATGTTTCTTCTCTCAGCTCTGAACTGTCAGAGTGCAAAGGGAAGGAACTTAGAAATCTGAGGCCTTGACGCCAGGTGCTGGGATTTTAGGCAAACCTAA GCTACTGGTAGTGGTTTGCTGGAAGACTTTTTGCTTGGTTTCTGAGGCTGTTCCTTTCTTATTGCATTAATTTAAAACCTGGTAATGTAAGTGTCTTTCA CCAGTGGTTAAAAAAATACTGATGGTAGGAAAAAAGCCAATGAGAGCCCATTCATACTTTTAATCTAGTCttttttgttttcttttgttttttattttta ttttttttttattttttatttttttaaagattttatttattcttgagagacacacacacacacacacagagatacagagatacaggcagagggagaagca ggctccatgtagggagcctgatgtgggacttgatccccagactccaggatcaagccctgggccaaaggcaggtactaaaccgctgagccacccaggtgtc cctcttttgttttttAAattgaggtatccttaacatgacactagattattttcaggtggacagcataatcattagatgtttgtgtaggctgtgaaatgat cacagtaaggctagttaacatgtcttcatacatagttccaagatgtttttttctaataatgagggcttttaagatctattctcttagtgactttcaaata tgcaatacagtatgttttttttttttttcagtatgttgttaactatagtccttatactgtctgtgtattatatccctgtgacttatttattttataactg gaaggttgtaggtttttttttggtaaagatttttatttatttattcatgagacagagagaggcagagtcataggtaaagggagaagcaggctccctgcaa ggagcctgatgcgggactcgattccaggaccctgggatcacgacccgagccaaaggcagatgctcaacccctgagtcacccaggcatcccaaggttgtag cttttgaacccacttcatctgttctccactcaccccctccttctccctgcctctggcaactaccattctgttctctgTATCAGTCATCTTTTCTTGAATG ATTTTTGCCTCCAGTTTTACCTGCTTTCCTGATTTCAGGTTTGCCTCATTTTATAGCATTTTGTGGAGAATGGTTTTTGTGCTTAAGAATGAAAGAATCA CATTAATTCAGTACAGCATATTGACTGGCTACATGTGGTTACAGTGCTCAGAGCTGAGGAAATAGATAATGTAAACATGTTTTTTTGTCTGCTTTTAAAC CATTTCTCTGATCTTCAGCTTTATTGGGAAAAGTTTGAGAAAACACAGTATATAATAATTAGGTATGGAGAAGCTAAGGTCTCTTTGGTATGATGGTGGT CTTCTCTTCTAGCACGTGATAGTCCTGGAGTGCACGTGCCATATTATGTCTTACTTGGCTGATGTCACCAATGCCCTGAGCCAGAGTAATGGTCAAGGCC CAAGTCAC >ZUBR1_felCat Felis catus (cat) 1631bp -L1MA9 5958-6299 30% KKYLSQKNVVEKLNANVMHGK HVIVLECTCHIMSYLADVTNALSQSNGQGPSH AAGAAATACCTGTCACAGAAGAATGTGGTTGAAAAACTGAATGCCAATGTAATGCATGGAAAGGTAAGCAAAATGCAGAGAGACAGCAGTCGGTGAGTGA TGATCCTTACCTTTGTGGTCACATAGAAGGCTCTGCTTCTTACTGGCTTGTGTCCCTGGCCAGCCTGCCTGCAATTTCAAAGCCCTGGGAGTGTATAAAT CCGAACTGTGTGCCCTTGGGCCTTTGCTGAGTGATGAGGAATGATAGGAGGGAAATAATATGGGAAACAGTATCCAACTGAATCCTTCATAAAACCTGGT GGATTCTTGCTATTTTTAGCTATTGGTGTGCTGTGGAACTGGAGCTGTGTGCTCTGAGTAGGTGTTCTGTGCGGTTACAGACTCTGCCTTCTCAGCTCTA GTCCAGTTGCTAGAATAGTTTAATTTACCTGAAGATTCAGGGCATCTCTGAGAAGGGTTTTGAACTGACAGAAAGTCCACGTTCAGTTGTTCGCCCGCCC CACCACGGACCTGCCGTAGGTCACTGAGAGCGGGTAACCGGCCTCTGAGACCCAGCAGCATAGCGCTGTTTATTTCTCAGTAATGAGATGATTTAAATTT CAGTAATTTCAGCCTTTCTTGCTCGTCTTTCTCCTCTGAGTCTGCACTGTCGCTGCAGAGTGAAGAGCTCAGAAGTCTGAGGCCTTGATGCCAGGCTGCT GGGTCCTCAGGCAGGCATGAGCTGCTGGCAGCTGTTCGCTGGAAAGCTTGTGGCTTCATTTCCGAAGCTGTTCCTCTCTTACTGCTTTCGCTTAAAAGTC TTTGGCTAATGGTTAGAAAAAATTGCTAATGGTAGGGAGAAAGCCAGCGAGAGCCTATTCATACTTTGAATCTAGTCTGTTTTTCCTTtcttcgtttaaa ttgaggtatacttgacaggtaacactgtatccgtttcaggtgcacagcacagggattcactgtttgggcgtgttgtggagtgatccccacaataaggcta gttgctgtctgtctccatatgtagttctaaaacgtgttttcctcgtaaggagtttagagatctgctcacttagcaactatcaacacgtcatacattgtta gctgtagtcgccatgctgtgctttatttacacccctgtgacttatttattttataaccggaagtttgtgtcttttgaacctcattcatccattctccacc cactgctcccctgactctggcaaccaccagtctcttctctgTATCATTTTCTTCTCTTTTTCTATTTTCCTGACGACAGTTTTACTTCATTTCATAATTT GTAAAGGGTTGTCTTTGTGCTTAAGGATGAGTGAATCCCGTTAATTCAGTACCGTGTATTGACTGGCTACACGTGGCCATAGTGCTCAGAGTGAGGAAGT AGATAATTGAAGCATGTGGATTTTCTTGTCTGCTTTTATACCATTTCTCTGATTGCGGGCTTTATTGGGAAAAGTCTGAGAATCGGATATGGAGAACCGA AGTTTCTGTGACGGGGTAGTGGTCTTCTCTTCTAGCATGTGATAGTCCTGGAGTGCACGTGCCACATTATGTCTTACTTGGCTGATGTCACCAATGCCCT GAGCCAGAGTAATGGTCAAGGCCCAAGTCAC >ZUBR1_equCab Equus caballus (horse) 1667bp -L1MA9 5959-6299 23% KKYLSQKNVVEKLNANVMHGK HVMVLECTCHIMSYLADVTNALSQSNGQGPSH AAGAAATACCTGTCACAGAAGAATGTGGTTGAAAAACTGAATGCCAATGTAATGCATGGAAAGGTAAGAGAAGTGAGGAGTGGCGACAGTCAGGGAGTGA GGGTTCTTACCTTTGTGGTCAGGTGGAGGGCTCTCCTTCTCACTGGCTTCTGTCCCTGGCTGGCCTCCGGAGGATAAAGGCCTACGATCCCAAAGGCCTA GAAGTGTGTAATTGCGAACTGCATGCCCTCAGGCCTCTTCTGGGTGGCAAGAAGTGATAGGAAGGAAATATTCTGGTAAGCAAATATCCAGCAGAATCCT TTTTAAAACATAGTGAATTCCTGCTATTTTTAGATACTGGTATGCTATGGAAAATTCTCCCCTGCCCCTGCAGGGGAGCTTTGCTCTCTGAGCAGGTGCT ATATGGGGTTACAGACTACCTTCTCAGCTCCAGTCCAGATGGTAGAATTGTTTACTTTACCTGAAGTTTCAGGTCACTGTGTTGAGAAAAAAGGGTTTGA AACTGACAAGAGATCTTGCATTCAGGCATTTTTTCCCCCTACCCAACTCGTAAACCTGACAAAGGTGGCTAACAGCAGATAACTGGTCTCTGAGACCCAC CACAGCCCCCTGTTTCTCCCTTCTTTCTTGCTAATCTTTATCCTTTCAGCACTCAGCTGTCAGAGTACAGAGTGAAGGAACTTAGCGATCTGAGGCTTTG ATGCCAGACTGTTGGGTTTTTAAGGCAAGCATGAACTGCCTGCAGTAGTTTGCTGGAAAATTTTTCACTTGATCTCTGAAGCTATTCTGCTCTGCTGCAG TAATTTAAAACCCAATAATTTAAATGTCTTTCACTACAGGTTAAAAAAAATGCTAATCGTAGAAAGTATGTCAGTGAGAGCCTATTCATACTTCTAATCT AGTCTTTTTTTAAattgagatataattgacatattagtttcacatgtacaacatgatgattcaatgtttgtatatatcgtaaaatggtcaccacagtaat tctagttaagatccatcatcaaacatagttacaaattttctttttaatgttgaggacttctaagctctactctcttagcagctttcaaatatgcaatata atattagctgtattgaatgtaatgttgtacattccatctcatggcttatttattttataactggaagtttgtaccttttgaccccatttatccatttcat ccactcctcccaccgcctctctctggcaaccactactctattgtctatcaatctagttttttcttGAATGATTTTTGTCTCTATTTTCACTCTATTTTCC TGGGTTGAATTTTTTTACTTCATTTTACAGAGTTTTATGAAAAATTATCTTTGTGCTTAAGAATGAGTGAATCACACTAATTCAGTACAATTTATTGACT GGCTACATGCAGTCATAGTGCTAAGAGCTGAAGAAATAGATAATTCAAATGTTTTTCTTTTCTGCTTTTATACCATCCCTCTGATCTTGAGCTTTATTGA GAAAACAGTTTATGTAGTAATCAAATACAGAGATCTAAAACTTTCTGTGGTATGGTGGTCTTCTTTTGTAGCATGTGATGGTCCTGGAGTGCACATGCCA TATTATGTCTTACTTGGCTGATGTCACCAATGCCCTGAGCCAGAGTAATGGTCAGGGCCCAAGTCAC >ZUBR1_myoLuc Myotis lucifugus (microbat) 1227 bp No repetitive sequences were detected KKYLSQKNVVEKLNANVMHGK HVVVLECTCHIMSYLADVTNALSQSNGQGPSH AAPE01620425 AAGAAGTACCTGTCACAGAAGAATGTGGTTGAAAAACTGAATGCCAATGTAATGCATGGAAAGGTAAGAAAACTGAGGAGTGACAACAGTCAGCGAGTGA TGATTCTCTGTCTGTGTCAGGTGGAAGTCTCTCCTCTCTGTTGGCTTCTGTCCCTGGCCAGCCTACAGAGGAAGAGGCTACAAGTTTCAAAGACCTAGAA AAATGTAATTGCAAACTGCATGCCCTTTAACACCTTTTGGGTGGCAGAATCCTTAAAACATAATGAACTCCAGCTATTTTTAGATACTGCTATGCTGTGG AACACTGTCTCCTGCTCCTACATTGCCCCCATCTGCTCTGAGGAGGTGTTATGTGGGGTTACAGACTATCTCTTCAGCTCCAGTCTAGATACTAGGATGG TTTACTTTGCCTGAGGGTTCAGGTCACCGTGCTGAGAAAATGGTTTGAAACTGACAGAAATCTTACATTCAGGCATTCCCCAAACCCTCAGCCCTCAATT CATGAACCTGGCATAGGAAGCTAATGGCCAACCACCACGACACCATTCCTTTCTCCCTCTTTTCTTGCTAATCTTTATCCTTTCAGCTCTGAACTGTCTG TGCAGAGTGAGGAACTGAGAAATCTGAGGCTTTGATGCTAGACTGTTGAGTTTTTTAGGCAAGCAAGAACTACCTGCAAAGGGTTGCTGGAAATTTTTCA CTTGATCTCTGAAGCTGTTTCTCCTCTACTGCATTGATTTAAAACCAGTAATTTAAGTGTCTTTTACTTATGGTTAAAAGATGCTAATGAGAACCTAGTC ATACTTTTAATCTAATCTTTTCTTGAATATTTGTCTTCATTTTTCTCCCTTTTCCTTATTTCAGTTTTATTTCATTTTATAATATTTTGTAAAGACTCAT CTTTGGGCTTAAGAATAAATGAATACACAGTAATTCAGTGCAATTTATTGACCAGCTACATGTGGTCATAGTGCTAAGAGTTGGGGAAATGGATAATTAA AGTACTTTTTTTCTCTTATACCATTCCTCTGAAATTGAACTTTATTAGGAAAAGCTTGACAAAACATAGTTTACATAATAATGAAATATGGGGGACCAAG GCTTCTATGGTGTGGTGTTCTTTTCTTGTAGCATGTGGTCGTCCTAGAGTGCACATGCCATATTATGTCTTACTTGGCTGATGTCACCAATGCACTGAGC CAGAGTAACGGTCAAGGCCCAAGTCAC >ZUBR1_pteVam Pteropus vampyrus (macrobat) 1388 bp tiled No repetitive sequences were detected KKYLSQKNVVEKLNANVMHGK HVVVLECTCHIMSYLADVTNALSQSNGQGPSH AAGAAATACCTATCACAGAAGAATGTGGTTGAAAAACTGAATGCCAATGTAATGCATGGAAAGGTAAGAAAAGTGAGGCGTGACTACAGTGAGGGAGTGA TGATTCTGTTTGTGGTCAGATGGAAGGCTCTCCTTTCCATTGGCTTCTGTCTCTGGCTGCTGTACGTTTTCAAAGACCTGACAGTGTGTCTTTGCTAACT GCATGCCCTCAGGCGTCTTTTGGGTGGCAAGAGGTGATAGGAAGAAAACATTTTGGCAAACAAACATCCACCAGAATCCTTCTTAAAACATTGTGAATTC CTGCTATTTTTAGATACCAATATGCTATGGAAAACTGCCCTCTGCTCTTGCATTGGAACTGTGCTCTCTTGAGTAGGTATTATATGGGGTTACAGACTAC CTCTTCAGCACCAGTTTGGATACTAGAATTGTTTACCTTGCCTGAAGATTCAGGTCACTGTGCTGAGAAAAGGGATTCGAATCTGACAGAAATCTTGCAT TGAGGCACAGCCCCTTCCTCCACCCCACCCCCACCCAACTCATATACCTGACAGAGATGGCTAAACAGATAACTCCTCTTTGAGACCCGCCACAGCACGG ATTCATTTCTTTCTCTTTTGCTAATCTGTATCCTTTCAACTCTGAGCCGTAAAGGGAGGGCCTCAACTGCCAGGTAGTTCATCAACCCGCCACAGCACGG ATTCATTTCTTTCTCTTTTGCTAATCTGTATCCTTTCAACTCTGAGCCGTCAGAATGCATAGTGAGGAACCAAGACATCTGCGGCTTTAATGCCAGACTG TTGGGCATGAACTACCTGCAGTGGTTTGCTGGCAAGTTTTTCACTTATCTCTGAAGCTATTCCTCGTCTACTGCATTAATTCAAACCCAGTAATTTAAAT GTCTTTCACTAATGGTTAAAAATGCTAATGATAGGAAAAATGCTAATGAGTGCTTATTCATGCTTTTAATCTAGACGCTTCTTGAATGATTTTTGTTTCC ATTTTTACTCTATTTTCCTGGTTTTGATTTTATAATATAAAGAGTTACCTTTGTGCTTAAAAATAAATGAATATACAGTAATTCAGTACAATTTATTGAC CGGCCTTATGTGGTCATAGTGCTGAGAGCTGGAGAAATAATTAGAACATGTTTTTGTGCTTTGTTTCATTTTTGCTTTTATACCATTCCTTTGAAATTGA GCTTTATTGGGAAAAGCTTGAGAAAACAAGTTTACATATTAATGAAATACAAAGAACCGAGGTTTCAGTGGAATGGTGGTCTTCTTTTTTAGCATGTGGT AGTCCTGGAGTGCACATGCCATATTATGTCTTACTTGGCTGATGTCACCAATGCCCTGAGCCAGAGTAACGGTCAAGGTCCAAGTCAC >ZUBR1_bosTau Bos taurus (cow) 1891bp -L1MA9 5958-6302 24% -tRNA-Gly -tRNA-Glu KKYLSQKNVVEKLNANVMHGK HVMVLECTCHIMSYLADVTNALSQSNGQGPSH AAGAAATATCTGTCACAGAAGAATGTGGTTGAAAAACTGAATGCCAATGTAATGCACGGAAAGGTAAGGAGAGTGAGGGGAGCGATGGGTCCTGTCTTGG TGGCCGGGTGGAGGGAAGTCCCTTTCCTGTTGGTTTCTGTCCCTGGCTCCCCTGCAGAGGGGAAGGCCTAGGATCTCAAAGACCTCGAAGCGTGTCATCA CCAAGTGCACGCCCACAGGCCTCTTCCTGGGGGCAAGAAGTGGTCTGGAGGAAATGTTCTGCGAAACAGACATCCAACAGAATCCTTCTGAAAACAGTGA ATTACTGCTGTGTTTAGCTATTGGTATTCTCTGGGAAATTGTCTGCTGCTCCTGCATTGGGATTGTGTGCTCTGAGTGTCAGACACAGGTTCCCGTTTAC TTCCTCAGCGCCAGTCCAGATGCTAGAATCGTTTATGTTATTTGGAGGTTCAGGCCACTGTGCTGAGAAAAAGGGTGGCAACTGGCAGAAATCTTGCATT TGAACATTTTCTCCCTCTACCCAACCTGCGAGCCCGTCATTGTCGCTAATTTAAATGTTTTTCACTAATGGTTAAAAAAATGATTATGGCAGGGAAAAAA GCCAATGAGAGGTTACTCATACTTTAATACAATCTTTTTATCTTTtttcttgaggtataattgacacagatattagtttcaggtttacagcatagtgatc tgacatttgtctgtattgcaaaatgatcacagtctagttaatatccatcaccatacatagttaagttttttttcttgtaatgaggacttttaagacctgc tctcttggcgactttcagatgtgcatacattagtattaactctggtcaccatgcttgacatttcatctccatggcttacttattttataagtggaagttt gtatcttttgactcacatcacccacttcactgaatcaccctcctaccccctgcctctggtaccaccactctgttctctgtatcactctagttttttCTTG AGTGATTTTTGTGTCTATCTCAACTTTATTTTCCTGGTTTACACTGTACTTCATTTTCTAAGGTTTTTAAATATATACACatttatttcgctgtgctggg cccttgcagctgcttgggcgttctctcgtcactgcgagcaggggctgtgctctagtgccgttgggctctcttgttgtggaacctgggccccagggctcga gctcttcagtaactgcagctcccaggctctagagcgcaggctcagtagttgtggcccatgggcttcattgccccgtggcttgtgggatcttcctggatca gggatcaaacccacgtctgttgtgttggcaggtggattctttaccactgaaccaccagggaagcccTATTTTATGATTTTTTAAAGAGTTGTCTTTAAAG GATGTCTTTAAAGTTGTGCTTAAAGGATGAATCATACTGATTCAGTACAGTTTCtttaatttttaaagtttttaaattttttGGTTACCCCATGCAGCAT ATAGACTCTtagttccctgaccagagatcagacctgcatcctttgcattggaagtgctgaatcctaaaccactggcccccagggaAGTCCCCATGCAGTT TATTGGCCAGCCACTTGCTGTTGTAGTGCGAAGAGCTAAGGAAATAGATAATTAAAACATGTTATTTTTCTTCTCTGCTTTTTGTGCTGTTCCTTTGATC TTGAGCTTTATCGGAAAAAGCGTGAGAAAACACAATTTGCATGATAATGGAATGTGGAGAACTGACCTTTCATGGTGTGGTGGTCTTCTTTATAGCACGT GATGGTCCTAGAGTGCACCTGCCATATTATGTCTTACTTGGCTGATGTCACCAATGCCCTGAGCCAGAGTAACGGGCAAGGCCCGAGTCAC >ZUBR1_turTru Tursiops truncatus (dolphin) 1688 bp tiled -L1MA9 5936-6312 21% KKYLSQKNVVEKLNANVMHGK HVVVLECTCHVMSYLADVTNALSQSNGQGPSH AAGAAATACCTGTCACAGAAGAATGTGGTCGAAAAACTGAATGCCAATGTAATGCATGGAAAGGTAAGACAAGTGAGGAGTGACGGCAGTCAGGGAGTGG TGATTCTTATCTTTGTGGCCAGGTAGAAGTCTCCTTCCTGTTGGTTTCTGTCCCTGGCTAGCCTACAGAGGATAAGGCCTATAATCTCAAAGGCCTGGAA GTGTGTAATTGCTAACTGCATGCCCTTAGGCCTCTTCTCGGTGGCAAGAAGTGATAGGAAGGAAATATTCTGGTAAGCAGACATCCAACAGAGTCCTTCT TAAAACAGTGAATTCCTGCTATTTTTAGGTATTGGCATGCTCTGGAAAATTGTCCCCTGCTCCTGCATTGGAACTGTGTGCTCTGAGTGTTATACGGAGT TACGGTCTCCCTCCTCAGCTCCAGTCCAGATGCTAGAATTGTTTACTTTACCTGAAAGTTCAGGTCACTGTGCTGAGAAAAAGGGTGGAAACTGGCAGAA ATCTTGCATTCAGGCATTTTCTTCCCCCACCCAACCCATGAACTTGACATAGTGGCTAAGAGCAGATAACTGGCCTCTGACCCACCACAGCACCATTCAT CTCTCCCTGCTTTCTTGCTGATCTTTATCCTTTCCACACTGAACTGTTAAGAGTGCAGAGTGAAGGAACTTAGAAATCTGAGGCTTTGATGCCAGACTTT TGGGTTTTTTTAGGCAAGCACGAACTATCTGCAGTGATATGCTGGAAAAATTTTCCCTTGATCTCTGAAGCTATTCCTCCCTTACTGCATTAATTAAAAA CCCAGTAATTTAAATATCTTCCACTAATGGTTAAAAAAAATGGTAACAGCAGGAAAAAAGCCAATGAGAGCTTATTCATACTTTTAATATAGCTTTTTTT CTTTTTTCTTGAGGTGTAATTGACATAGAATATTATATTAGTTTCAGATGTACAACATAATGATTTGATATTTGTTTTTATTGCAGAATGATCACAGTAA GTCTAGTTAATACATCACCATACGTAGTTAACAAAATTTGTTTTTTCTTGCAATGAGGACTTTTAAGACCTACTCTCTTCATAACTTTCAGATATGCATA CAATAGTATTTATTAACTCTAGTCACTATGTTGGACATTACATCCCCATGACTTACTTATTTTATAACTGGAAGTTTGTACCTTTTGACCCCCATCACCC ATTTCGCCCACCCTCCTACCCACTGCCTCTGGCACCACCAGTCTGTTCTTTGTATCAGTCTATTTTTTTCTTGAATGAGTTTTGTCTCCATTTTAACTTT TTTTTCTTGATTTCCATCGTACTTCATTTTATAAAATTTTTTAAAGAGTTGTCTTTGTGCTTAAGAAAGAATGAATCACACTGATTCAGTACAGTTTATT GACCAACTACATGTAGTCATAGTGCTACGAGCTGAGGAAATAGATAATTAAAACATGTTGTTTTTCTTCTCTGCTTTTATACTATTCCTTTGATATTGAG CTTTATTGGAAAAAGCTTGAGAAAACACAGTTTACATAATAACCAAATATGGAGAACTGAGGTTTCCGTGGTGTGGTGGTCTTCTCTTACAGCATGTGGT AGTCCTGGAGTGTACCTGCCATGTTATGTCTTACTTGGCTGATGTCACCAATGCCTTGAGCCAGAGTAACGGTCAAGGCCCAAGTCAC >ZUBR1_susScr Sus scrofa (pig) 1022bp not fully tileable -L1MA9 5958-6182 22% KKYLSQKNVVEKLNANVMHGK HVIVLECTCHIMSYLADVTNALSQSNGQGPSH AAGAAATACCTGTCACAGAAGAATGTGGTTGAAAAACTGAATGCTAATGTGATGCATGGAAAGGTAAGAAGAGTGAAGAGTGACAGCAGTCAGGGAGTGA TGATTCTTATCTTTGTGGCCAGGTGGGAGTGTCTTTNCCCATGGCTTCTGTCCCTGGCTAGCCTGCAGAGGATAGGCCTATAACTCAAGGCCTCGAGTGT GTAATGCTAATACTGCCTTAAGCCTCTCGGGGGCAGGATGATAGAAGAATATCTGTAACAACTCAGCATATCTCTGAACAGGATATGGATTTTAAGTGTG TGTCGAAAGCCCCGCTGNATGGGGCCTGAGTAATGGGTAGTCATCCTATCAACGTGCGAAAATGACTGAAATG CGAGGCATGCTAAACTGGACCGGTCTTTAAACCATACATAGTGGAATATCTTTTTTCGTGTGATGGCTGAAGACTTCCTCTCTTAGCAGCATGCACATAT GCACTATAGTCACCCTGCTTGACAGTAGATCCCCATGACTTATGTTATAACTGAAAGTTTGTACCTTTTGACCTTCCCTTCACTCCTTTNGCCCATCCTC CCACCCCTGCCTCTGGCACCACCAATATATTCTCTGTACCAACCTAGTTTTTTCTCGAACAGTTTTTGTTTCCACTTTAATTTTATTTTCCCGAGTTTAA TTGTGTTTCATTTTACAAGATTTTAAAAGTGTTACCTTTTTGCTTAAGAACGAATGAATTTATTGACCAACTACATGTAGTCATAATAGTGTTAAGAGCT GAGGGAATAGATAATTAACATGTTTTTCTTCTTTGCTTTTTATACCATTTCTTTGATCTTGAGCTTTATTGGAAAAAGCTTGAAAAACAGTTTACATAAT AATTAAAATATGGAGGCAGCAGTTTCCACGGTGTGGTGGTCTTCTCTTTTAGCATGTGATAGTCCTGGAGTGCACCTGCCATATTATGTCTTACTTGGCC GATGTCACCAATGCCCTGAGCCAGAGTAATGGTCAAGGCCCAAGTCACC >ZUBR1_vicVic Vicugna vicugna (vicugna) 1525bp not fully tileable -L1MA9 5937-6312 22% KKYLSQKNVVEKLNANVMHGK HVVVLECTCHIMSYLADVTNALSQSNGQGPSH AAGAAATACCTGTCACAAAAGAATGTGGTTGAAAAGCTGAATGCCAATGTAATGCATGGAAAGGTAAGAAAAGTGAGGAGTTGACAACAGTCAGGGAGTG ATGATTTTTTTCTTTGTGGCCGGGTGGAATTCTCTTTCCTGTTGGTTTCTGTCCCTGGGAAACCTACAGAGGATGAGGCCTATAATCTCAGAGACCTAGA AAGTAATTGCTAACTGCATGCCCTTGGGCCTCTTCTCGGTGGCGAGAAGTGATGGGAAGGAAATATTCTGGTAAACGAGCATCCATCAGAATCCTTCTTA AAACAATGAATTACTGCTCTTTTTACATATTGGTATACTCTGGAAAATTGTCCCCTGCTCCTGCGTTGGAACTGTGCTCTGAGTGTTATTTGAGGTTACA GTCTGCCTCCTCAACTCTAGTCCAGATGCTAAAATTGTTTCTTTACCTAAATGATCTGTCTGTGTGATGAGAAAAAGAGTTAACAGTGGCATAAGTCTTG CATTCCCGCCTTTTCTCCCCTGACCCAACTCTGAACCTGACCTAGTGGTTAAGATGCAAATACTGGCCTCTGACTCCCGCATCTACAGTTGGCTCTCCTT CCCTCCTTGCTAATCTTCATCTTAGCCCCTGATCTGTCAAGAGTGATGATGTACGGAGCTAGAATTCGGAGGCTTGAATGCCAGAGATTTTGAATTTTTTCAGGAAGCCGAAAACTATCTGG GTCATCGCACTGTGGTGGATTCTCATACTTTTAATATAGTCTTTTTTCCTTCTTTATTGAGGTATAATTGACACAGAACATTAGTTTCAGGTGTACAACA TAAAGATTTGATATTTGTATATACTGCAGAATGACTACAGTAAGTCTAGTTAATACCGTCACCATACAGTTAAAACTGTTTTTTCTTGTGGTGAGGACTT TTAAGACCTACTCTCTTAGCAACTCTCAAATATACAGTACGATATTATTAACTATAGTCAGCATGTTTGACATTACATCCCCATGACTTACTGATTTTAT AACTGGAAATTTATACCTTTTGATCCCTTTACCCATTTTGCCCACTTCCCGCCCCCTGCCTCTGGCATCACCAGTTCTGTTTTCTCTATCAGTGTAGTTT TTTCTTGAATGATTTTTGTCTCCATTTTAACTTTACTGTCCTGATTTCAGTAGTAGATTTTTAAAGAGTTATCTTTGTGCTTAAGGATAAATGAATCACA TTAGATCAGTACAGTGTATTGACCAACTGCATTTATCATAATGCTGAGAGCTGAGGAGGTAGATAATGAAAGCATGCTGTTTTTCTTCTCTGCTTTTATA CCATTGCTTGATCTTAAACTTTATTGGAAAAAGCTTGAGAAAACACAGCTTCCATACTAATCAAATATGGAAAACTGAAGTTTCCTTGGTGTGGTCTTCT CTTGTAGCATGTGGTAGTCCTCGAGTGCACATGCCATATTATGTCTTACTTGGCTGATGTCACCAATGCCCTGAGCCAGAGTAACGGTCAAGGCCCAAGTCAC >ZUBR1_eriEur Erinaceus europaeus (hedgehog) 1256 bp tiled No repetitive sequences were detected KKYLSQKNVVEKLNAGVMHGK HVTVLECTCHIMSYLADVTNALSQSNGQGPSH AAGAAATACCTGTCACAGAAGAACGTGGTTGAAAAACTGAACGCCGGTGTAATGCATGGGAAGGTAAGGAGAGCAGAGTGGCAGAAGTAGGGGACGGCGA TTCTTGACCCCGTGCCCTGGCAGAAGCCTCTCCTTCCTACTTGCCGCCTTCTGTCCAGAAGATAAGGCTTGAAATCTCCAGGGCCTTCAAGGGCGTCACT GCTAACAGCATGCCCTCAAGCCTGCTCTGGGGGGGCTGCTAATTCCGAAATAGCACATTTCTGCTCATGTTTAAAAAGTGTTTGCGCGTGTGTTTGCATG CGCGCCCTGCCCCGCTCTTGTTTGGGTAGGTGGCCCTCAGCTCTAGTATAGGCCTGGGCTCCTTCACTGTAGCGGAGACGTGCCACTAACTCCAGCAGCT GCGCATCCTGGCAGCCTCTCTCTCTCTCTCTCGCCACATCACAACCCTGACGTAGGTGGCTGAGAGCAGACAGTTGACCTCTGAGACCCACCTCTCTCAC TTTTCCCTCTGTTGCTGACTTGTATCCTTTCAGCTCTGACACTCAGTCAAGACACCTAGAAGCCGAAGACCTTAATGCCTGCAGTGGTTTTCTGGAAAAG TCTTCAGTTCCTCTCTGGAACTGTTTCTCCCTGACTCAAACGCCTTTGACTAGTGGCTAAAAAACACCCCACTAGTTGTAGGACCAATGCGAATGAGAGC GTGCTCGAGGCCCCAGCGGTTCCAGGTTCGATCCCCAAGCCACTGTAAGCCTAGGCTGACTAGTGCGCTGGTCCAGCGGAGCGGAGCGGAGCGCAGAGCA GTGCTCTTACATCAGGCGGGGTCCTCTCTGATGGCAGGCCTCACGTGAGCGGGGGGTTGGCTCTGGCGCAGGGGGTGGAGCGAAGGGTGTGCTCGGTCGC CTAGTTCCCTCTCTGGTACACAGTAGTTTTGAATGATTTTTGTTGCGTTTTACTGTTTTCCCTGGTTTCATTTCTCTTCCTCTTAGGAGATCTTGTAAAG AATCACCTTGGTACTTAGGATGGGTGTGTACTTACTCAGTACATTTACTGAGTGGCTCCATGAGATGAGCGGGGGACATAAGATAGTTCAGTCACACTGC TTTTCTGTCATTCCTGTGACTGGAGCTGCATTGGAAAACACTTGCCTTCTCCTCTTGTAGCACGTGACAGTCCTGGAGTGCACATGCCACATCATGTCGT ACTTGGCTGATGTCACCAATGCCTTGAGCCAGAGCAACGGTCAAGGTCCAAGTCAC >ZUBR1_sorAra Sorex araneus (shrew) 1098bp No repetitive sequences were detected KKYLSQKNVVEKLNANVMHGK HVIVLECTCHIMSYLADVTNALSQSNGQGPSH AALT01499066 AAGAAATACCTGTCACAGAAGAATGTGGTTGAAAAACTGAATGCCAATGTAATGCATGGAAAGGTAAGAGGAATGAGCGGTGATGATGCCATGGGGTGGT GATTCCTACCCCTGTGACCTCTTTCTTAAAGCCTTGTGACCTGGCCACCCTGTCTAAGATCGGGTGTACACACCTCAGAGTTATAGTGCAAATCTCAACC CCCTTTGGACAGTTAGGAAGATATAGGCAGGAAATACTGTGGGAAATACATACCCAGTAGCATCTTTCTGAAGATACATTAAATTTCTGCTATTTTAATA TACTGCTATATGTTGTAACACTGGCATGCACTGCTGCTTTGTAACTGTACTCACTGTGCTGGTAAGAGGGGTCGACGCTGCTAGAAGTCTCACATTTCAA CATTTTCCCAAGACCTGTCTCAAGCCTCTGACATTGGTGGCTAGAGCAGATAACTGGCCTCTAAGGCCTACCACCTTCAGGTCTTTTTTTTTTTTTTTTT TGCTAATCTTTATCCTTTTGGCACTGAGCTGTCAGAATGCAGAGTGAAGGTATTAAGAAATCTGAAGCTTTGATGCCAGACTGTTTATTTTTACCAAGCA TGAGCTACCTGTAGTGATATGCGGCAAAAGTTGTCATTTAGTCTCTGAGGTTAGTCCTTCCCTACTGAACTAATTTAAAACTCAGCAATTTAAAAGTTTC ACGCTTTCATTTACATATAGATAATATGTCTTTTGGGGCCTTGGGCCAAAGGTCAAAAATACTGAAAGAGTAAAAGTTTGCAACTTACAGTGAATGACTA TGCTCTCCATATTATTTCCTGACTTGCTGAATATGGTTATATTGTGGAAAGTTAAGGAAACAGATAATTAAAACCTGTGTTTATCCTTTGTTCTAATAAC ATCCCTTGGCTTTATTAGTAGAAACTTGAGAAACCAGTCAGATAATAAATTATGAGTTGTGGAGAAATGACTCTCCTGTGGTATGGTATTCTTCTCTTGT AGCACGTGATAGTCCTGGAGTGTACCTGCCATATTATGTCTTACCTGGCTGATGTCACCAATGCTCTGAGCCAGAGTAATGGTCAGGGCCCAAGTCAC
Analysis of L1MA9 retroposon INT391
Extended validation of this L1MA9 insertion, which occurs in a short intron of the gene ACSL5, is feasible in May 2008 because of newly available genomes. elements. The basic technique consists of first establishing the comparative genomics of the two outside coding exons. These are needed to reliably probe contig assemblies and trace archives with tblastn and blastn respectively. A complete intron can often be obtained by tiling out to the center from the two ends. It is imperative to avoid paralogous exons in doing so.
In the case of INT391, dog, cat, horse, microbat, macrobat had the retroposon judging by location, fragment coordinates relative to the full length retroposon, and strand orientation relative to the coding exons (minus strand here). Cow, dolphin, pig, vicugna, and shrew did not have it. Since cetartiodacytl L1MA9s might have been interrupted by a later retroposon breaking the L1MA9 into two unrecognizable shorter pieces, it is necessary to remove other repeats and re-run RepeatMasker.
Despite more intensive phylogenetic sampling, INT391 continues to support Pegasoferae as Nishijimi et al originally stated. It should be noted that the MER-class retroposon, while not at issue here, exhibits the type of homoplasy that makes retroposons dicey as tree topology markers. Introns are often susceptible to multiple insertions of similar retroposons as well as to complicated patterns of micro deletions that prevent their recognition even if they aren't fully deleted.
Summary of the phylogenetic distribution of the L1MA9 retroposon INT391: >INT391_ACSL5_Peg_canFam +MER91 85-140 23%9 -L1MA9 6082-6298 24% span 762bp >INT391_ACSL5_Peg_felCat -MER91C 97-140 27% -L1MA9 6082-6301 22% >INT391_ACSL5_Peg_equCab -MER91B 62-128 26% -L1MA9 6082-6302 21% >INT391_ACSL5_Peg_myoLuc no MER -L1MA9 6079-6277 23% >INT391_ACSL5_Peg_pteVam -MER91B 8-62 24% -L1MA9 6060-6302 25% >INT391_ACSL5_Peg_bosTau -MER91C 55 -85 28% No L1MA9 >INT391_ACSL5_Peg_turTru -MER91 261-311 22% No L1MA9 >INT391_ACSL5_Peg_susScr -MER91 257-306 8% No L1MA9 >INT391_ACSL5_Peg_vicVic -MER91 284-337 24% No L1MA9 >INT391_ACSL5_Peg_sorAra no MER91 No L1MA9 Markup of exons and intronic retroposons of INT391 within ACSL5: blue: coding exons magenta: L1MA9 INT391 red: MER91 retroposon >INT391_ACSL5_Peg_canFam +MER91 85-140 23% -L1MA9 6082-6298 24% span 762bp GDPKGAMLTHQNIISNVSSFLKCME YTFKPTPEDVTISYLPLAHMFERIVQ ACAGGTGACCCTAAAGGAGCCATGCTGACCCATCAAAATATTATTTCAAATGTTTCTTCTTTCCTCAAATGTATGGAGGTCAGTGGTCAATTGTCAAGGA GGTCTTCATTAAAATGTAAATCTGTCATAAGATTTTAATCCTGATGTAAGAGGAGTCAGAGACTAACACAAAACAAAACAAAAACAAAACTCATGATAAA GGCCTGAAGAAGGGACAAATAGTGGTGTCTCTTTGTCCAGAGGACTGTGCATTTTCAAGCCTTGGCCTTTTAGAATCACTGCACATCTCTACACTCAGTG AAATTAAGGggcacctctcagagttatacagtgcaccacctgtacaactgggtgtggcagtcctgGGAAGGAGCAGTTTTTTTTAAATTAAAGAAAAAAT Tttgagatacaattaacataacactatattaatttcagatacacaacataatgatttcatatatatgttgcaaaatggttcccacaataaatctaacatc cattatcacacatagctatagtttctttttcttgtgatgagaatttttaagatctgctcacttactaacttgcagatatgcaatacagtattattaacta tagttaACGGGAGTTACTTTTAAGTCTCCTTCGGAAGAGAAAGTTGGCATTAACACAATGTCTCCTCCTTGTTCTAATCTACAGTATACTTTCAAGCCCA CCCCTGAAGATGTGACCATATCCTACCTGCCCTTGGCTCATATGTTTGAGAGGATTGTACAG >INT391_ACSL5_Peg_felCat -MER91C 97-140 27% -L1MA9 6082-6301 22% GDPKGAMLTHENIVANSSAFLKCME CIFKPTTEDVSISYLPLAHMFERIVQ GGTGACCCTAAAGGAGCCATGTTGACCCATGAAAATATTGTTGCAAACAGTTCTGCTTTTCTCAAATGTATGGAGGTCAGTGGTCAATTTAAAAAGAGGT AGTCATTAAAATGTAAATCCATCATAAGATTTTGATCTTGATGTCAGAGGAGGCAGAGACAAAAAACAAAACAAAACCAAAAGCCACGTTAAAGGCCTGA CAATGAATCAGTGTGGACAAATACTGGTGCATCTTTGTCCAGAGGACTGTGCATTTTCCAGCCTTGGTCTCTTAGAATCACTGCATGTATCTACACTCAG TGAAGTTAAGGAGCACCTTAACTTCagtcatacagtgcaaaacctgtgcaactatgtgtggcaatcctgGCAATTTCTTTAAAAGTAAAGAAAAAAAttt gttgagatattattgacgtattaatttcaggtgtacaacgtgattccatatatgtatgtactgcaaaatggtccctgtgataaattccaagtccatcaac acacataattttttttcttgtgatgagaacttttcagatctactcacttaacaactttcaaatctgcaacacagcattattaactgtagttaATAGGAGC TGCTTTTAAATCTCCTTTAGAATAGAAAGTTAGCACTAATCCAATGGTGTCTCTTTCTTGTTCTGGTCTATAGTGTATTTTCAAGCCCACCACTGAGGAT GTGTCCATTTCCTACCTCCCCTTGGCTCATATGTTTGAGAGGATTGTACAG >INT391_ACSL5_Peg_equCab -MER91B 62-128 26% -L1MA9 6082-6302 21% GDPKGAMITHQNITSNTAAFLRSME GTFEINLEDVTISYLPLAHMFERVVQ GGTGACCCCAAAGGAGCCATGATAACCCATCAAAATATTACTTCAAATACTGCTGCTTTTCTTAGATCTATGGAGGTCAGTGATCAATTGAAAAAGAGGA ATTCCTAATTAAATTTCAATTGAAAATTCCTAATTAAAATAGGAATCTGCCATAAGATTTTAATCTTGAAATTAGAGAAGGCATAGAGGAAAAAAATAGG TTTAAGGCCTAAGTATGCACACATATCAGTGCCTCTTTGTCCAGAGGACTGTGCATTTTCACGTCTTGGTCTTTTAGGATCACTGCAGAGCTCTACACTC TGTGCAgttaagggtacctcttacagttgtacagtacatcacctgcacaaccatatgtggcagttctgGGAAGGAGTAGttttttaaaaattaaaaaaat attttattgagatatgattgacatataacattatgctagtttcagatgtacaacataatgatttgaggtttgggtatattgcaaaatgatccccacaata agtctagttaacatccatcaccacgcatagttacaaattttttcttgtgatgaaaacgtttaagatctactctcttagcaaatttctaatatataataca gtattactaactagaattaATAGTAGTTTTTAAATCTCCTTCGAAGAGAAAGTTGGATTAATACAATGTTGTCTCCTCTTTGTTCCCTGATCTGTAGGGT ACTTTTGAGATCAACCTTGAGGATGTGACCATATCCTACCTCCCCTTGGCTCATATGTTTGAAAGGGTTGTACAG >INT391_ACSL5_Peg_myoLuc no MER -L1MA9 6079-6277 23% GDPKGAMLTHQNVVSNASAFLRCVE ESFAPTPEDVSISYLPLAHMFERVVQ AAPE01034117 GGTGACCCCAAAGGAGCCATGCTAACCCATCAAAATGTTGTTTCAAATGCTTCAGCTTTCCTCAGATGCGTGGAGGTTAGTGGTAGCTTGAAAAAGAGGT CTTCGTTAGAATGTGACTCTGTCATAAGATTTTAATCTTGAAGCTAGAGGAGGCAGAGAAGAAAAAAACCAAAACAGGTTAAGGGCCTGAGTGTGGACAA ACACATGTGCATCTTTGTGTGGAGGGCTGTGCATTTTCAAGCCGTGATCTTTGAGGATCCCTGCAGACCTCTACTCCAGCGCAGTCCAGGGCACCTCTCC CAGTTCTTCAGGGCACCCCCTGCATGACTGTATGGGGCACTCATGGAAGGAAATAGTTAAAAAAAAATTTAAATTTTAAATGAGATGTAACGATGCCTaa cattataatagtttcaggtgtgcaacataatgattcaatatttatatgtattgcaaaatgatcctcatagtaagtgtagttaatatccatcactgcacac agttacaaattctttgttcttgtgatcagaacttctaagatcaactctctcagcaactttcgaatatacaatagagtgttattaactatagttaacaAGG GTAGTTCTTAAATCTCTTTGGTAAAGAAGGTTGGCATTAATCCGATTTTGTCTCCTCCCCCTTCCCGATCTGTAGGAAAGCTTTGCACCCACCCCCGAGG ATGTGAGCATATCCTACCTCCCCTTGGCTCATATGTTTGAGAGGGTTGTACAG >INT391_ACSL5_Peg_pteVam -MER91B 8-62 24% -L1MA9 6060-6302 25% GEPKGAVLTHQNVISNAAAFLKLLEVS DSFQVTPKDVTISYLPLAHMFERIVQ ti|1386642117 ti|1371644127 GGTGAGCCCAAAGGGGCCGTGCTAACCCATCAAAATGTCATTTCAAATGCTGCTGCTTTTCTCAAACTTTTGGAGGTCAGTCGATCAAATGAAAAAGAAG TCCTGATCAAAATGTGAATTTGTCATAAGATTTTAATCTTGAAGTCAGAGGAGGCAGAGAGGGGGAAAAAAAACAGGTTAAGGGCCTGAATGTGGGCAAA TATTTGTGCATCTTTGTCTGGAGGACTGTGCATTTTCAAGCCTTGGTCTTTTAGGATCACTGCAGACCTTTGTACTCAGTTAAGGGCACCTCTTAGAGTG ATGCAGTGTACCGCCCGCACAACTGTATGTGGCCCACCTAGAAAGAAGTAGCTTAAATTTTTTAAAAATTTTAATTGAGATATAATTGATATCTAACATT GCCTTAGTTTCAGGTGTACAATGTAATGATTCAATATTTGTATATGTTGCTAAACGATCCTCAAAATAAGTCTAGCTAAGAAAGATCACCACACTTAGAT AAAAACTCTTTTTTTGTGTGTGACAAGAACTTTTAGCAACTTTCATTATTAACTGTCGTTAACAGGGTAGTTCTTAAATCTCCTTTGGAAGAGAAAGTTG GCATTAATCCAATGTCATTTCCTCTTTGTTCTTTATCTATAGGACAGCTTCCAGGTCACTCCCAAGGATGTGACCATATCCTACCTCCCCTTGGCTCATA TGTTTGAGAGGATTGTACAGGTGAGT >INT391_ACSL5_Peg_bosTau -tRNA-GluSine -MER91C 55-85 28% No L1MA9 GDPKGAMLTHANIVSNASGFLKCME GVFEPNPEDVCISYLPLAHMFERIVQ GGTGATCCCAAAGGAGCCATGTTAACCCATGCAAATATTGTTTCCAATGCTTCTGGTTTTCTCAAATGTATGGAGGTCAGTGGTCAATTGAAAACAAGGC CCTCATTAAAATGTAAATCTGTCGTAAGATTTTAATCTTAAAGTGAGAGGAGGCAGAGAGGGAAAAAACTGATTGAAGGCCTGAGTGTGGATGAATACCA GTACATCTTTGTCTGGAGTTTTGCCCTTTTATTTATTTATTAatatatatatatatatatatatatTTTTTAATCTGGACCATTTTTAAAGTTTTTATCG AATGTGTTATAGTATTGGTTCTGTTTTATGTTTTGATTTTTGGGGGGCTACAAGgtacatgggatctcagctccctgaccaggggtagaactcacaccct ctgcattggaaggtgaagtcttaaccactggacctctggggaagtccCATAGAGTTTTGCTGTGTTAGGGTCACTGCAGATCTCCACACTCAATGCAGTT AGAgcagcccttagatttacacagggcacatctgcacagctgtatgcagcagtcctAGAAAGAAGTGTTTAAATCCTCTTTGGAAGAGGAAATTGACATT AACCCATTGTTGTCTCTTTTCCATTTCCTGATCTCTAGGGTGTTTTTGAGCCCAATCCTGAGGACGTGTGTATATCCTACCTCCCCTTGGCTCATATGTT TGAAAGGATTGTACAG >INT391_ACSL5_Peg_turTru -MER91 261-311 22% No L1MA9 GDPKGAMLTHENIVSNAAAFLKCVE HTFEPSSEDVTISYLPLAHMFERVVQ GGTGACCCCAAAGGAGCCATGTTAACCCATGAAAATATCGTTTCAAATGCTGCTGCTTTTCTCAAATGTGTGGAGGTCAGTGGTCAATTGAAAAGGAGGC CCTCGTTAAAATGGGAATCTGTCATAAGATTTTAAAGTTAGAGGAGGCAGAGGGGGAAGAAACAGGTTGAAGGCCTGAGTGTGGACAAATACTGGTGCAT CTTTGTCTAGAGTTTTGCTCTTTTAGGGTCACTGCAGATCTCTGCACTCAGTGCAGTTAGGGCACCCCTTAGGGCACAGTGCACACCTGTACAACTGTAT GCAGCAGTCCTAGAAAGAAGAAGTGTTTAAATCTTCTTTGGAAGAGAAAGTTGGCATTAATCCACTGTTGTCTCCTTTCCATTTCCTGATCTATAGCATA CTTTTGAGCCCAGTTCTGAGGACGTGACCATATCCTACCTCCCCTTGGCTCATATGTTTGAGAGGGTTGTACAG >INT391_ACSL5_Peg_susScr -MER91 257-306 8% No L1MA9 GDPKGAMITHQNIVSNVASFLKRLE YTFQPTPEDVSISYLPLAHMFDRIVQ ti|2023263948 GGTGACCCCAAAGGAGCCATGATAACCCATCAAAATATTGTTTCAAATGTTGCTTCTTTTCTCAAACGTCTGGAGGTCAGTGGTCGACTGAAAAAGAAGC CCCTGTTGAAATGTGAATCTGTTATAAGATTTTAAAGTTAGAGGAGGCAGAGAGGAAAGAACCAGGTCAAAGCCCCAAGTATGGGAAAATACTAGTGCAT CTTTGGAGTTTTGCTCTTCTAGGGTCACTATAGATCTCTACACTCAGTGTAATTAGGGCACCCCCCAGAGTTGTGCAGTGCACACCTGCACAACTGTATG TGGCAGTACTAGAAAGTAGTGTTTAAATCTTCTTTGGAGGAAAAAGTTGGCATTAATCCATTGTTGTCTCCTTTCCCTTTCCTGATCTACAGTACACTTT TCAGCCCACCCCTGAGGACGTGTCCATATCCTACCTCCCCTTGGCTCATATGTTTGATAGGATCGTACAG >INT391_ACSL5_Peg_vicVic -MER91 284-337 24% No L1MA9 GDPKGAMITHENVVSNVAAFLKFME YSFEPTPEDVAISYLPLAHMFERVVQ ti|1970855441 GGTGACCCCAAAGGAGCCATGATAACCCATGAAAATGTTGTTTCAAATGTTGCTGCTTTTCTCAAATTTATGGAGGTCAGTGATCAACTGAAAAAGACAC CCTCGTTAAAATGTGAATCTGTCATAAGACTTTAATCTTCAGGTTAGAGGAGGCAGAGAGGGAAAATGACAGGTTTAAAGCCTGAGGGTTGACAAAGACT GGTGCATCTTTGTCTGGAGGACTGTGCGTTTCCAAGTTTTACTCTTAAGAATCACTGCCGGTCTCTCCACCCAGTGCAGTTAGGGCATCTCTTAGATTTG CGCAGTGCACACTTGTGCAACTGTATGTGGCGGTCCTAGAAAGAAGTAGTGCTTAAATCTTCTTTGGAAGAGAAAGTTGGCATTAATCGAATGTTGTCTT CCTCCCATTCCCTGATCTCTAGTATTCTTTCGAGCCCACCCCTGAGGATGTGGCCATATCCTACCTCCCCTTGGCTCATATGTTTGAGAGGGTTGTACAG >INT391_ACSL5_Peg_sorAra -SOR1SINE No L1MA9 WGPKGAKITHEILSSKAZAFLNSVE YAFEPTPEDVSISYLPLAHMFERVVQ AALT01576933 GGGCCTAAGTGGTGCTGAGGATGGAACCCAGGCCTTCTGCAGCTCCAACCCCCTGGGCCAGCTCTCCAGCTCTAAAGTGCCCCTAATGTAAGGGGAT GCAGGAAATATGGCAGAGCTGAAGTCATGAACCCAGAAACAACAGGAGGAGGTGATGGGCTTTTCTTTGTAACTGCATCTGTGATTGTGGTCTTGTGGAA TGTCGCTGCACATTGCAAAGCCAAAGACGGGCTGTGTGCTTTATAAAGGGTCTTTCTCTCCACCTCTTGTCTCCTCCAGGTGACCCCAAAGGAGCCATGA TCACGCATGAAAATATTGTTTCAAACGCCTCTGCTTTCCTCAAGTGTGTGGAGGTCAGTGGATGTGGGAAAAGAGGTCCTAGCAAAAGGGTGGATGCCAC AAAGTTCAGAAGTGGAAGTTAGAGCAGCAGCAGGGCTGGAGGGTGGCGTTCAAAGGGCTGTGTGTGTGCAGATGCCCCGACAGCTTGGGACATCAGTGTT ATCATTATCATTATTATTATTACCATTTTGGTTTTTGGGGTACACTTGGGAATGGACAGGGGGCACTTCTGGCTTATGCACTCAGGAATTACTCCTGGTG GTGCTCAGGGAACCATGTGGGATGCTGGGAATCAAGCCACATGCAAGGCAAATGCCCTACCCACTGTGCTATTGCTCCAGTCTCATCAGTGTTTTAGGAA GCTGTGTATGTTGCTGCCTTGATATCCAGCACCTCTCTGCTCTCGGCGTGTAACAGCGCCCCTCAGAGCTCCACGGGGGGTCTAGCCTGCACACCCAGGT GTGGCCCTGCTGGAAATGCCTGGTCTTTAGGTCTTCTTTGTCTGGGGAAATTTGGCATTGATCGATGGTCTCTTTCCTCTGTGCCCTGATCTGTAGTATG CGTTCGAGCCCACGCCTGAGGATGTGAGCATCTCCTACCTCCCCTTGGCACACATGTTTGAGAGGGTCGTGCAG
Phylogenetically informative coding insertions and deletions
Mammalian genomes contain about 20,000 genes comprised of some 190,000 exons. Insertions and deletions (indels) accrue slowly in these exons over time as species diverge from one another. These supplement information that can be derived from amino acid substitution analysis. In some instances, the initial indel occurs and is fixed across a stem population (a stem is an interval of evolutionary time not giving rise to lineages surviving to the present day).
Such rare genomic events can be phylogenetically informative. However they are potentially subject to re-occurence and reversion in some descendent clades which could confuse the issue. For this reason, it is imperative to screen out indels in regions of dna prone to this type of event by virtue of their repetitive nature (conducive to repeated replication slippage) or compositional simplicity.
Lineage-sorting is another issue but provided the stem persisted long enough for informative indels to arise in adequate numbers, those of the true topology should significantly outnumber anomalies arising from the two alleles exisitng at the time of speciation sorting out differently in descendent clades. However in the situation when several mammalian orders arise over a very short period of time (in effect a polytomy) or when the speciation process itself is blurred by millions of years of hybridization or genetic mixing at population boundaries, the whole concept of discrete topological tree branching may be inapplicable.
Analysis of a potentially informative indel in CHML
A loss of one amino acid can be observed within an exon of the gene CHML in carnivores, bats, and horse but not in other Laurasiatheres (or other placental mammals. This gene is similar enough to a second gene CHM that it too needed full-on phylogenetic annotation -- it has no indel in any species so must not be cross-annotated for CHML in species without assemblies. The key to that is including upstream amino acids where sufficient divergence between the two genes resides.
The background story here is a common one in gene dosage compensation. In the amniote ancestor and birds and lizards today there is but one multi-exonic gene CHM. In mammals, as the sex chromosomes underwent upheaval, the ortholog ended up on chrX which has implications for reduced levels of expression compared to an autosomal gene. That may have favored persistence of a retroprocessed (intronless) copy on chr1. In marsupials, the two copies remained quite similar. In placentals, the second gene CHML diverged extensively from the first while remaining well conserved within its orthology class.
The one-residue indel appears cleanly restricted to Pegasoferae. There are 5 Laurasiatheres with it and 5 without it, which needs further buttressing by PCR (adding more basal species has the effect of localizing the event farther back on the stem). The parental gene did not develop the indel in any bioinformatically accessible species.
Interestingly, the retrogene CHML landed within intron 1 of encephalopsin OPN3 with the same direction of transcription. This raises some questions about how CHML is translated (as it would apparently be spliced out). Encephalopsin has an interesting history in mammals involving multiple independent losses.
The region annotated below corresponds to a known domain called GDI (for GDP dissociation inhibitor: pfam00996). The 3D structure of this region is available in the parent gene in rat, PDB 1LTX so the structural location of the indel could be determined. Very likely it lies within a loop and has minimal functional impact.
CHML chr1:239,864,567-239,864,746 gene_genSpp AFRQCSFSEYLKTKKLTPNLQHFVLHSIAMTSESSCTTIDGLNATKNFLQCLGRFGNTPF CHML_homSap Homo sapiens (human) AFRQCSFSEYLKTKKLTPNLQHFVLHSIAMTSESSCTTIDGLNATKNFLQCLGRFGNTPF CHML_panTro Pan troglodytes (chimp) AFRQCSFSEYLKTKKLTPNLQHFVLHSIAMTSESSCTTIDGLNATKNFLQCLGRFGNTPF CHML_ponPyg Pongo pygmaeus (orang_sumatran) AFRQCSFSEYLKTKKLTPNLQHFVLHSIAMTSESSCTTIDGLNAIKNFLQCLGRFGNTPF CHML_macMul Macaca mulatta (rhesus) AFRQCSFSEYLKTKKLTPNLQHFVLHSIAMTSESSCTTIDGLNATKNFLQCLGRFGNTPF CHML_calJac Callithrix jacchus (marmoset) AFEQCLFSEYLKTKKLTPNLQHFILHSIAMTSESSCTTIDGLKATKNFLQCLGRFGNTPF CHML_otoGar Otolemur garnettii (bushbaby) AFKQCSFSEYLKTKKLTPNLQHFVLHSIAMTSESSCTTVDGLKATKNFLQCLGRFGDTPF CHML_micMur Microcebus murinus (mouse_lemur) AFEQCLFSEYLKTKKLTPNLRHFILHSIAMTSESSCSTLDGLKATKTFLQCLGRFGNTPF CHML_tupBel Tupaia belangeri (tree_shrew) DFKQCSFSDYLKTKKLTPNLQHFILHSIAMTSESSCTTLDGLQATKTFLQCLGRFGNTPF CHML_musMus Mus musculus (mouse) DFKQCSFSDYLKTKKLTPNLQHFILHSIAMSSDSSCTTLDGLQATKNFLRCLGRFGNTPF CHML_ratNor Rattus norvegicus (rat) DFQQCLFSEYLKTKRLTPNLQHFILHSIAMTSESSCTTLDGLKATKNFLQCLGRFGNTPF CHML_cavPor Cavia porcellus (guinea_pig) DFKQCSFSEYLKAKKLTPNLQHFVLHSIAMTSETSCTTLDGLKATKIFLQCLGRFGNTPF CHML_oryCun Oryctolagus cuniculus (rabbit) DFKQCSFSEYLKTKKLTPNLQHFILHSIAMTSESSCTTLDGLRATKNFLQCLGRFGNTPF CHML_ochPri Ochotona princeps (pika) AFVHCSFSDYLKTKKLTPNLQHFVLHSIAMT-ESSCTTIDGLKATKNFLRCLGRFGNTPF CHML_canFam Canis familiaris (dog) AFMQCSFSEYLKTKKLTPNLQHFVLHSIAMT-ESSCTTIDGLKATKNFLQCLGRFGNTPF CHML_felCat Felis catus (cat) AFMQCSFSEYLKTKKLTPNLQHFVLHSIAMT-ESSCTTIDGLKATKNFLQCLGRFGNTPF CHML_equCab Equus caballus (horse) DFTQRPFSEYLKTQKLTPNLQHFILHSIAMT-EPSCLTVDGLKATKHFLQCLGRYGNTPF CHML_myoLuc Myotis lucifugus (microbat) DFTQCSFSEYLKTKKLTPNLQHFVLYSIAMT-ESSCTTVDGLKAAKNFLRCLGRFGNTPF CHML_pteVam Pteropus vampyrus (macrobat) AFTQCSFSEYLKTKNLTPSLQHFILHSIAMMSESSCTTVDGLKATKTFLQCLGRFGNTPF CHML_bosTau Bos taurus (cow) AFTQCSFSEYLKTKKLTPSLQHFVLHSIAMMSESSCTTIEGLKATKNFLQCLGKFGNTPF CHML_turTru Tursiops truncatus (dolphin) DFMQCSFSEYLKAKKLTPSLQHFVLHSIAMTSESSCTTIDGLKATKNFLQCLGRFGNTPF CHML_susScr Sus scrofa (pig) AFVQSSFSEYLKTKKLTPNLQHFVLHSIAMMSESPCTTIDGLKATKNFLQCLGRFGNTPF CHML_vicVic Vicugna vicugna (vicugna) AFVQSSFSEYLKTKKLTPNLQHYILHSISMTSESSCTTLDGLKATKKFLQCLGRFGNTPF CHML_eriEur Erinaceus europaeus (hedgehog) AFIQCSFSDYLKTKKLTPNLQHFILHSIAMTPEASCSTVDGLKATKIFLQCLGRFGNTPF CHML_sorAra Sorex araneus (shrew) TFKQCSFSEYLKTKRLTPNIHHFVLHSIAITSQSSCTIIDGLKATKTFLWCLGWFSKNPF CHML_dasNov Dasypus novemcinctus (armadillo) AFEQCSFSEYLKTKKLTPNLQHFILHSIAMTSQSSCTTLDGLKATKNFLQCLGRFGNTPF CHML_choHof Choloepus hoffmanni (sloth) AFKQCSFSEYLKTKKLTPNLQHFVLHSIAMTSESSCTTIDGLKATKNFLQCLGRFGNTPF CHML_loxAfr Loxodonta africana (elephant) AFKHCSFSEYLKTKKLTPNLQHFVLHSIAMTSESSCTTIDGLKATKTFLQCLGRFGNTPF CHML_proCap Procavia capensis (hyrax) ... CHML_echTel Echinops telfairi (tenrec) AYEESTFSEYLKTQKLTPILRHFVLHSIAMASETSTSTLDGLRGTKNFLQCLGRYGNTPF CHML_monDom Monodelphis domestica (opossum) ... CHML_ornAna Ornithorhynchus anatinus (platypus) AQKECTFSDYLKTQKLTPNLQHFILHSIAMVSEVNCCTIDGLKATQRFLQCLGRYGNTPF CHML_anoCar Anolis carolinensis (lizard) NYKNSTFAQFLKTRKLTPSLQHFILHSIAMVSEKDCNTLEGLQATRKFLQCLGRYGNTPF CHML_galGal Gallus gallus (chicken) CHM chrX:85,097,861-85,098,040 genSpp GYEEITFYEYLKTQKLTPNLQYIVMHSIAMTSETASSTIDGLKATKNFLHCLGRYGNTPF CHM_homSap Homo sapiens (human) GYEEITFYEYLKTQKLTPNLQYIVMHSIAMTSETASSTIDGLKATKNFLHCLGRYGNTPF CHM_panTro Pan troglodytes (chimp) GYEEITFYEYLKTQKLTPNLQYIVLHSIAMTSETTSSTMDGLKATKNFLHCLGRYGNTPF CHM_ponPyg Pongo pygmaeus (orang_sumatran) GYEDITFYEYLKTQKLTPNLQYIVLHSIAMTSETASSTIDGLKATRNFLHCLGRYGNTPF CHM_macMul Macaca mulatta (rhesus) GYEEITFCEYLKTQKLTPNLQYIVLHSIAMTSQTASSTIDGLKATKNFLHCLGRYGNTPF CHM_calJac Callithrix jacchus (marmoset) ... CHM_otoGar Otolemur garnettii (bushbaby) ... CHM_micMur Microcebus murinus (mouse_lemur) AYEEITFSEYLKTQKLTPNLQYFVLHSIAMTSELASSTLDGLKATKNFLRCLGRYGNTPF CHM_tupBel Tupaia belangeri (tree_shrew) AYEETTFSEYLKTQKLTPNLQYFVLHSIAMTSETTSSTVDGLKATKKFLQCLGRYGNTPF CHM_musMus Mus musculus (mouse) AYEGTTFSEYLKTQKLTPNLQYFVLHSIAMTSETTSCTVDGLKATKKFLQCLGRYGNTPF CHM_ratNor Rattus norvegicus (rat) AYETITFSEFLKTQKLTPNLQYFVLHSIAMTSETTSSTIDGLKATKNFLHCLGRYGNTPF CHM_cavPor Cavia porcellus (guinea_pig) GYEEIAFSEYLKTQKLTPNLQYFVLHSIAMTSETTTTTLDGLKATKNFLHCLGRYGNTPF CHM_oryCun Oryctolagus cuniculus (rabbit) ... CHM_ochPri Ochotona princeps (pika) AYEEITFSEYLKTQKLTPNLQYFVLHSIAMTSETASNTIDGLKATKNFLHCLGRYGNTPF CHM_canFam Canis familiaris (dog) ... CHM_felCat Felis catus (cat) AYEEITFSEYLKTQKLTPNLQYFVLHSIAMTSETASSTIDGLKATKNFLHCLGRYGNTPF CHM_equCab Equus caballus (horse) AYEDITFSEYLKTQKLTPNLQHFVLHSIAMTSKTTSSTIDGLKATRKFLHCLGRYGNTPF CHM_myoLuc Myotis lucifugus (microbat) AYEEITFSEYLKTQKLTPNLQYFVLHSIAMISERASSTIDGLKATKNFLCCLGRYGNTPF CHM_pteVam Pteropus vampyrus (macrobat) AYEEITFSEYLKTQKLTPNLQYFVLHSMAMTSETGSSTIDGLKATKNFLHSLGRYGNTPF CHM_bosTau Bos taurus (cow) AYEEITFSEYLKTQKLTPNLQYFVLHSIAMTSETASSTIDGLRATKNFLHCLGRYGNTPF CHM_turTru Tursiops truncatus (dolphin) ... CHM_susScr Sus scrofa (pig) AYEEISFSEYLKTQKLTPNLQYFVLHSIAMTSETASSTIDGLKATRNFLHCLGRYGNTPF CHM_vicVic Vicugna vicugna (vicugna) ... CHM_eriEur Erinaceus europaeus (hedgehog) AYEKMTFSEYLKTQNLTPNLQYFVLHSIAMASETGSSTIDGLKATKNFLQCLGRYGNTPF CHM_sorAra Sorex araneus (shrew) ... CHM_dasNov Dasypus novemcinctus (armadillo) ... CHM_choHof Choloepus hoffmanni (sloth) ... CHM_loxAfr Loxodonta africana (elephant) ... CHM_proCap Procavia capensis (hyrax) AYEEITFSEYLKMRKLTPNLQYFVLHSIAMTSETTSTTIDGLKATRNFLHCLGRYGNSPF CHM_echTel Echinops telfairi (tenrec) AYEDCTFSEYLKTQRLTPNLQHFVLHSIAMVSETSTSTLDGLRETRNFLQCLGRYGNTPF CHM_monDom Monodelphis domestica (opossum) ... CHM_ornAna Ornithorhynchus anatinus (platypus) AQKECTFSDYLKTQKLTPNLQHFILHSIAMVSEVNCCTIDGLKATQRFLQCLGRYGNTPF CHM_anoCar Anolis carolinensis (lizard) NYKNSTFAQFLKTRKLTPSLQHFILHSIAMVSEKDCNTLEGLQATRKFLQCLGRYGNTPF CHM_galGal Gallus gallus (chicken)
Analysis of a five residue deletion in DIO1
The five amino acid deletion here occurs in first exon of iodothyronine deiodinase, a much-studied gene. A consistent picture is seen in dog, bears, cat, horse, macrobat, and microbat with artiodactyls (cow, dolphin, pig, vicuna) and eulipotyphlya (hedgehog and shrew) lacking the deletion, which is evidently the ancestral condition judging by marsupials, more basal Atlantogenata, and Euarchontoglires.
This event supports -- but does not by itself establish -- the topology ((((dog horse) bat) cow) eulipotyphla) at the expensive of ((((dog horse) cow) bat) eulipotyphla). It does not speak to various possible internal resolutions of ((dog horse) bat). Longer indels are uncommon however so it is difficult to avoid the conclusion that artiodactyls are an outgroup (without invoking lineage-sorting).
Mammals have two other iodothyronine deiodinase paralogs but these are sufficiently diverged at this exon to prevent any annotational confusion. They too are full-length in this region, suggesting that length is a very ancient character indeed. No 3D structure is available for any member of the family but they can be supposed to have the basic ferredoxin fold (like many other selenoproteins).
Note two species within Afrothera have an 8 residue deletion in a similar region of the protein, suggesting it is a loop region inessential to protein function. No data is available for tenrec or other species in this clade, so it cannot be more precisely dated without PCR of additional species. However armadillo and sloth lack the deletion, bounding its timeframe somewhat.
The alignment below shows the deletion in 'difference' mode relative to human. Here dots mean the residue matches human. This mode of display results in less visual clutter while emphasizing degree of conservation, which is an important quality parameter for phylogeneticall informative indels.
DIO1_homSa MGLPQPGLWLKRLWVLLEVAVHVVVGKVLLILFPDRVKRNILAMGEKTGMTRNPHFSHDNWIPTFFSTQYFWFVLKVRWQRLEDTTELGGLAPNCPVVRLSGQRCNIWEFMQ DIO1_panTr ..................................................................................................H............. DIO1_macMu .....S...V.K................................................................................................D... DIO1_calJa ....G..................A......T.......K......D........N...........................................H.........D... DIO1_otoGa ....R..........F.......A...M..........SQ.....QQ.V.AK.............LQHPV.LVCPEGPL.....M..R.........SAS...K....D... DIO1_musMu .....LW......VIF.Q..LE.A.....MT...G...QS.....Q....A...R.AP...V.....I................RA.F.......T..C....K....D.I. DIOI_ratNo ...S.LW......VIF.Q..LE.AT....MT...E...Q......Q........R.AP...V.....I................RA.Y.......T.......K..V.D.I. DIO1_oryCu ....R...........VQ...E.A.....MT...E...Q......Q...IAQ..N.AQ.S........................A..P.......S.......Q.S..D..R DIOI_cavPo ...TW...........VQ...E.AM....MT...E.I.KS..............Q..................I.........E.A.......D.S..C...EKRT..D..H DIO1_canFa ....R.V...R......Q...Q.A....F.K...A...QH.V..NGN-----K....Y...A..LY.M.........Q......R..P....................D... DIO1_ursAr ....R.V...R......Q..M..A..........E...QQV...NK.-----.....Y...L...Y.M................R..P....................D... DIO1_felCa ...S.L....R.....FQ..LQ.A....F.....S...QH.V..NR.-----.....Y...A..LY.V................R..P.................S..D..K DIO1_equCa ....RA...........Q..LQ.A......T.......QH.V..NQ.-----.....Y...V..LY...........H........KR......S.............D... DIO1_myoLu ..............I..Q..L..TL...Q.K...R...QH....NR.-----.....Y...A..L...P....I..........K..E.S...............H..D... DIO1_pteVa .E..W..R.........Q..L..A....Q.T...R...Q..V..NR.-----.....F...L..L...............Q.....KE..........C.........D... DIO1_bosTa ....S...........FQ..L..AI.....T...R...Q...................E..............I..........M..Q..............E..S..D... DIO1_turTr ....L...........FQ.GL..AM.....T...R...Q.....S.....AK.....YE........A.....I..........M..Q..R.................D... DIO1_susSc .E..L...........FQ..L..AM....MT...G...QD....SQ....AK......E........A................K..E..........S......H..D... DIO1_vicVi ...SL...........FQ.VL..AL.....T...G...QD....SQR...AQ.....YE..............I.............Q.....D....C..-D.VH..D... DIO1_eriEu ....S...........FQ..L..AI.....T...R...Q...................E..............I..........M..Q..............E..S..D... DIO1_sunMu ....GL..L...FG..VR..LK.A......T.W.SAIRPHL...S.....AK..R.TYED.A...............N..Q...R.KQ.DI..DS...H.....ARL.D... DIO1_dasNo ...S..........I.FQ..L..A...T..T...G...Q....KSQ.SHKAE....PY...G....N......L..IG......K..Q..........H.........D... DIO1_choHo ...SW............Q..L..AM..I..T...G...Q.....SRRANN.KD.Q.PY...G....N.................K..Q..........H..R......D... DIO1_loxAf ..............IF.K..L..AM.........G...K....Q--------....AY.M.GS.L..IP....I...Y......K..E..P..D....C........SD... DIO1_proCa ......V..........R..L..AM.....A...G...K....Q--------....AY.M.CS.L..VP........Y......K..E..........H.....R...D... DIO1_monDo .LRLWLW.........Q.VG..LM..LMKM.S...M.QH..G..Q.SSIFQ..N.KYE..G....TLP..L...R........QALQ..P..D....S.R..PRRL.D..HA DIO1_triVu . AG.L..VR.F.A..Q..F......L.KT...NMM.KH..SL.QRSSISQ.TQ.AYE..G.....I...F............QALQ......P...T.K.ESRH..D..H DIO1_anoCa . FKA.RLVLKT.L..Q.CLSTA...LFM....ATA..Y..KQS.RSS.G...N.VYE..G.....F..LL.....K.K....KALQ.CP...T...DFD.KIHH.LD...
Possibly informative whole gene duplication in bat opsin
An apparental functional duplication of a visual opsin gene has been reported previously in the megabat Haplonycteris fischeri that is missing in Pteropus and Myotis. This is another form of rare genomic event, a large insertion indel since most mammals have but two imaging opsins.
This gene duplication might simply be restricted to this one genus (or even this one species). Alternatively it might have greater phylogenetic depth and be somewhat informative in resolving intra-bat taxonomy. This would require PCR of many additional bat species since no further genomic work is anticipated any time soon. It is not relevent to Pegasoferae per se since bats are clearly monophyletic overall, even as the macro/micro distinction remains equivocal as a taxonomic character.
The key paragraph in the article:
"When we sequenced the region from exon 4 to exon 5 of the M/L opsin gene in an individual from H. fischeri, an additional copy was found. While exons 4 and 5 of the two copies were identical at nonsynonymous sites, the two copies of intron 4 differed by 20 nucleotides and 11 indels, including one large indel of ~750 bp and involving a total of 794 sites. These differences appeared to be too large to represent two alleles of the same locus, indicating the possibility of two duplicate genes. Indeed, the sequence data from intron 5 revealed three different sequences, denoted as 5–1, 5–2, and 5–3 (fig. 4). Whereas introns 5–1 and 5–2 may represent two polymorphic alleles from the same locus, intron 5–3 differs from intron 5–1 at 10 sites and should represent another locus. (Because a locus can have at most only two different sequences, the presence of three sequences should indicate a gene duplication.)"
Possibly informative indel in OPN4 bat melanopsin
Another potentially informative indel for bats -- a one residue deletion -- separates Myotis from everything else. Its phylogenetic depth can only be determined by sequencing additional species of bats. It might contribute to establishing microbates are polyphyletic (or it could simply be an idiosyncracy of the genus Myotis). The indel occurs in a well-understood region of melanopsin, the only rhabdomeric opsin that has persisted into mammals. This gene is not directly involved in imaging vision but the structural/functional implications of the indel still might be determinable.
Melanopsin OPN4 chr10:88,408,220-88,408,423 indel in cytoplasmic loop between transmembrane segments 3 and 4 .....FYAFCGALFGISSMITLTAIA.......................FVLLGVWLYALAWSLPPFFGW 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 MEL1_homSap Homo sapiens (human) melanopsin OPN4 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATFGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 MEL1_panTro Pan troglodytes (chimp) 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATIGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 MEL1_ponpyg Pongo pygmaeus (orang_sumatran) 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLATIGVASKRRAAFVLLGVWLYALAWSLPPFFGW 1 MEL1_macMul Macaca mulatta (rhesus) 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLTTVGVASKRRAALVLLGVWLYSLAWSLPPFFGW 1 MEL1_otoGar Otolemur garnettii (bushbaby) 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLASVGTASKRRAGLVLLGVWLYALAWSLPPFFGW 1 MEL1_micMar Microcebus murinus (mouse_lemur) 2 GCEFYAFCGAVFGITSMITLTAIAMDRYLVITRPLATIGRGSKRRTALVLLGVWLYALAWSLPPFFGW 1 MEL1_musMus Mus musculus (mouse) 2 GCKFYAFCGAVFGIVSMITLTAIAMDRYLVITRPLATIGMRSKRRTALVLLGVWLYALAWSLPPFFGW 1 MEL1_ratNor Rattus norvegicus (rat) 2 GCEFYAFCGAVLGITSMITLTAIALDRYLVITRPLATIGMGSKRRTALVLLGIWLYALAWSLPPFFGW 1 MEL1_phoSun Phodopus sungorus (hamster) 2 GCEFYAFCGAVFGISSMITLTAIALDRYLVITRPLATIGMASKKRAAFFLLGVWFYALAWSLPPFFGW 1 MEL1_speTri Spermophilus tridecemlineatus (squirrel) 2 GCEFYAFCGAVSGITSMTTLTAIALDRYLVITRPLATIGVASKRRTALVLLGVWLYALAWSLPPFFGW 1 MEL1_nanSpa Nannospalax ehrenbergi (mole-rat) 2 GCEFYAFCGALFGITSMITLTAITLDRYLVITRPLATIGVASKRQAALVLLGVWLYALAWSLPPFFGW 1 MEL1_cavPor Cavia porcellus (guinea_pig) 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLAAVGMVSKKRAGLVLLGVWLYALAWSLPPLFGW 1 MEL1_oryCun Oryctolagus cuniculus (rabbit) 2 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLAAVGMVSKRRTGLVLLGVWLYSLACSLPPLFGw 1 MEL1_ochPri Ochotona princeps (pika) 2 GCEFYAFCGALFGITSMITLTAIALDRYLVITHPLAAVGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1 MEL1_canfam Canis familiaris (dog) 2 GCEFYAFCGALFGITSMITLMAIALDRYLVITHPLATIGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1 MEL1_felCat Felis catus (cat) 2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATVGVVSKRWAALVLLGIWLYALAWSLPPFFGW 1 MEL1_equCab Equus caballus (horse) 2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLA-IGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1 MEL1_myoLuc Myotis lucifugus (microbat) 7/7 traces 2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLAAIGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1 MEL1_pteVam Pteropus vampyrus (macrobat) 3/3 traces 2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATVGMVSKRRAALVLLGVWLYALAWSLPPFFGW 1 MEL1_bosTau Bos taurus (cow) 2 GCEFYAFCGAVFGITSMITLTAIALDRYLVITRPLATVGMVSKRRAALVLLGVWLYALAWSLPPFFGW 1 MEL1_turTru Tursiops truncatus (dolphin) 2 GCEFYAFCGAVFGITSMITLTAIALDRYLVITYPLATVGMVSKRRAALVLLGVWLYALAWSLPPFFGW 1 MEL1_susScr Sus scrofa (pig) VPT 2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRVALVLLGVWLYSLAWSLPPFFGW 1 MEL1_eriEur Erinaceus europaeus (hedgehog) 2 GCEFYAFCGALFGITSMMTLTAIALDRYLVITRPLASIGVVSKRRAALVLLGVWLYALAWSLPPFFGW 1 MEL1_sorAra Sorex araneus (shrew) 2 GCKFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRAALVLLGIWLYALAWSLPPFFGW 1 MEL1_loxAfr Loxodonta africana (elephant) 2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRAALVLLVIWLYALAWSLPPFFGW 1 MEL1_echTel Echinops telfairi (tenrec) 2 GCEFYAFCGALFGITSMITLTAIALDRYLVITRPLATIGVVSKRRTALVLLGTWLYALAWSLPPFFGW 1 MEL1_proCap Procavia_capensis (hyrax) 2 GCEIYAFCGALFGIASMMTLLAISLDRYLVITRPLAATGVVSRRRALLALPGIWLYALAwSLPPFFGW 1 MEL1_dasNov Dasypus novemcinctus (armadillo) 2 ACEFYAFCGALFGITSMITLMAIALDRYFVITRPLASIGVISKKKTGFILLGVWLYSLAWSLPPFFGW 1 MEL1_monDom Monodelphis domestica (opossum) 2 GCEFYAFCGALFGITSMITLMVIALDRYFVITRPLASIGMISKKKTGLILLGVWLYSLAWSLPPFFGW 1 MEL1_smiCra Sminthopsis crassicaudata (fat-tailed dunnart) 2 GCEFYAFCGALFGITSMITLMVIALDRYFVITRPLASIGVVSKKKTGLILLGVWLYSLAWSLPPFFGW 1 MEL1_macEug Macropus eugenii (wallaby) 2 GCEFYAFCGALFGITSMITLMVIALDRYFVITRPLASIGMISKKKTGLILLGVWLYSLAWSLPPFFGW 1 MEL1_smiCra Sminthopsis crassicaudata (dunnart) 2 GCQLYAFCGALFGITSMITLTVIALDRYFVITRPLASIGVISKKRALLILTGVWFYSLAWSLPPFFGW 1 MEL1_ornAna Ornithorhynchus anatinus (platypus) 2 GCELYAFCGALFGITSMITLMVIALDRYFVITKPLASVRVMSKKKALIILVGVWLYSLAWSLPPFFGW 1 MEL1_galGal Gallus gallus (chicken) 2 GCELYAFCGALFGITSMITLMVIALDRYFVITKPLASVGVTSKKKALIILVGVWLYSLAWSLPPFFGW 1 MEL1_taeGut Taeniopygia guttata (finch) 2 GCELYAFCGALFGIASMITLTVIALDRYFVITRPLASIGAMSTKKALLILSGVWLYSLAWSLPPFFGW 1 MEL1_anoCar Anolis carolinensis (lizard) 2 GCELYAFCGALFGITSMITLMVIAVDRYFVITRPLTSIGVMSKKRAVLILSGVWLYSLAWSLPPFFGW 1 MEL1_xenTro Xenopus tropicalis (frog)
These sequences may be relevent to primer design:
>MEL1_myoLuc Myotis lucifugus (microbat)
aggctgtgagttctatgccttctgtggggctctctttggcatcacctccatgatcaccctgacggccattgccctggaccgctacctggtgatcacgcgccctctggc catcggg
gtggtgtccaaaaggcgggcggccctcgtcctgctgggcgtctggctctacgccctggcctggagt
>MEL1_pteVam Pteropus vampyrus (macrobat)
aggctgcgagttctatgccttctgtggtgctctctttggcatcacctccatgattaccctgacggctatcgccctggaccgctacctggtgatcacacgcccactggctgccatcggg
gtggtgtccaagaggcgggcagcgcttgttctgctgggtgtctggctctacgccctggcctggagtctgccacccttctttggctggag
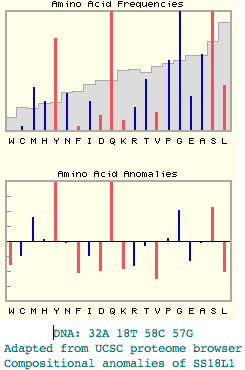
Possibly informative indel in SS18L1 supporting chiroptera + artiodactyl
This autosomal indel, at face value, groups artiodactyls and bats to the exclusion of dog, horse, shrew, and hedgehog. Note the ambient gene SS18L1 has rather anomalous composition in its distal half and the exon containing the indel falls under standard masking filters such as seg. It is anomalous in the mammalian proteomewide context in glutamine and tyrosine, no doubt because the dna composition of the exon exhibits very high GC, 32A 18T 58C 57G. The exon has some self-similarity upon blast to a distal region of the gene:
SNPPSQQGSSQQYLGQEEYYGEQYSHSQGAAEPMGQQ S+ S QG SQ Y GQ SQG++ MGQ+ SHYSSAQGGSQHYQGQSSI-AMMGQGSQGSSM-MGQR
The alignment shows the indel phylo-consistently in 3/3 available artiodactyls and 2/2 available bats but not in 4/4 other Laurasiatheres or other vertebrates. The percent identity of human to artiodactyls fallsoff fairly dramatically relative to percent identity to horse and dog (or even marsupial and platypus). It's most unusual for a human/cow ortholog to be more diverged than human/lizard. Some Atlantogenata also appear to be diverging rapidly but without the indel. This cannot be attributed to misalignment or paralog/pseudogene confusion.
In summary, this indel demonstrates some of the subtle issues in using rare genomic events in phylogeny. The indel supports cow + bat topology but not so strongly given the compositional propensity to homoplasy in the context of rapid sequence divergence in some clades.
SMMQQQAATSHYSSAQGGSQHYQGQSSIAMMGQGSQGSSMMGQRPMAPYRPSQQ Homo sapiens SMMQQQAATSHYSSAQGGSQHYQGQSSIAMMGQGSQGSSMMGQRPMAPYRPSQQ Pan troglodytes 100% SMMQQQAATSHYSSAQGGSQHYQGQSSIAMMGQGSQGSSMMGQRPMAPYRPSQQ Macaca mulatta 100% SMMHQQAATSHYNSAQGGSQHYQGQAPIAMMGQGGQGGSMMGQRPMAPYRPSQQ Mus musculus 88% SMMHQQAATSHYNSAQGGSQHYQGQAPIAMMGQGGQGGSMMGQRPMAPYRPSQQ Rattus rattus 88% SMMHQQAASSHYNSAQGGSQHYQGQSSIAMMGQSGQGSSMMGQRPMAPYRPSQQ Canis familiaris 90% SMMHQQAASSHYNSAQGGSQHYQGQSSIAMMGQSGQGSSMMGQRPMAPYRPSQQ Equus caballus 90% SMMHQQAASSHYNSAQGGSQHYQGQP-IAMMGQSGQGSSMMGQRPMAPYRPSQQ Myotis lucifugus 87% SMMHQQAASSHYASAQGGSQHYQGQP-IAMMGQSGQGSSMMGPRPLAPYRPSQQ Pteropus vampyrus 83% SMMHQQAASSHYSAAQGGSQHYQGQS-MAMMGQSGQGGGVMGQRPMAPYRPSQQ Bos taurus 81% SMMHQQAASSHYSAAQGGAQHYQGQS-MAMMGQSGQGSGMIGQRPMAPYRPSQQ Sus scrofa 81% SMMHQQAASSHYSAAQGGSQHYQGQS-MAMMGQSGQAGSMMGQRPMAPYRPSQQ Tursiops truncatus 83% SMMHQQAASSHYNSAQGGSQHYQGQSSIAMMGPSGQGNSMMGQRPLAPYRPSQQ Erinaceus europaeus 85% SMMHQQAASSHYSSAQGGG.HYQNQLALAMMGSGGQGSSLMSQRLLALYWPSQQ Sorex araneus 70% poor quality traces SMMHQPSATPHYSSAPGGGPHYQGQASLAAMGQGTQGSSMMGQRPMAPYRSSQQ Dasypus novemcinctus 77% SMMHQQTATSHYSSTQSGSQHYQGQSSIAMMGQSGQGSSLMGQR Loxodonta africana SIMHPQEAQTHFSSVQGGSQHYQGPSPVAMMGQGGQGGGLMGQRPMAPYRASQQ Procavia capensis SMMHQQAATSHYSSAQGGSQHYQGQSSIAMMGQGGQGSGLMGQRPMAPYRASQQ Echinops telfairi 90% SMMHQQAATSHYNSAQGGSQHYQGQSSIAMMSQSNQGSSMMGQRPMGPYRPSQQ Monodelphis domestica 88% SMMHQQAATSHYNSAQGGTQHYQGQSSIAMMSQSNQGNSMMGQRPMGPYRPSQQ Ornithorhynchus anatinus 85% SMMHQQAATSHYNSAQAGTQHYQGQSSIAMMSQSNQGNSMMGQRPMGPYRPSQQ Anolis carolinensis 83% SMMHQQAATSHYNSAQGGSQHYQGQSSIAMMSQSNQGNSMMGQRPMGPYRASQQ Gallus gallus 85% SMMHQQATGSHYTSAQAGSQHYQGQPSISMMNQSSQGSGMIGQRPLGPYRPSQQ Xenopus tropicalis 75% not available Felis catus not available Vicugna vicugna not available Choloepus hoffmanni
Possibly informative indel in ZNF622 supporting chiroptera + artiodactyl
This 3 residue deletion in exon 1 of ZNF622 seems informative but has some troubling side issues. First, two species seem to have a second somewhat diverged copy, rat and mouse lemur. With rat, it is on another chromosome but mouse has no counterpart. If this gene is prone to duplication, it becomes difficult in species without global assemblies (ie bats) to establish orthology.
Second, only dog is available for carnivores and its sequence is disturbingly diverged in percent identity -- 72% in a gene where cow is 82% with the 3 residue indel and 88% if the indel is filled by consensus sequence. The dog gene is in syntenic position with human. Meanwhile earlier diverged and usually fast evolving species like armadillo and rabbit are over 98% identical to human. Horse at 94% is about what is expected. Third, horse dna exhibits slight tandem repeat at both dna and protein level at the site of the indel that might be conducive to repeated replication slippage events.
horse 8 bp tandem repeat in indel region
aaggccgtgcaggccgtgagc
A V Q A V S
This indel would benefit from PCR of additional carnivores, pangolin, perissodactyl, and bat species to expand representation of those divisions. It is necessary to develop an understanding on when and why the carnivore gene began to diverge anomalously. It is unusual to see 3 aa deletion in a gene that is fairly well conserved to depth but it may be no coincidence that laurasiatheres other than horse have both rapid divergence and the indel in some species.
SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGVDSV ZNF622_homSap Homo sapiens (human) SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGVDSV ZNF622_panTro Pan troglodytes (chimp) 100% SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGVDSV ZNF622_ponPyg Pongo pygmaeus (orang_sumatran) 100% SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGLDSV ZNF622_macMul Macaca mulatta (rhesus) 100% SKKFASFNAYENHLKSRRHAELEKKAVQAVNRKVEMMNEKNLEKGLGVDGV ZNF622_calJac Callithrix jacchus (marmoset) SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGGDSL ZNF622_otoGar Otolemur garnettii (bushbaby) 96% SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGVDSV ZNF622_micMur Microcebus murinus (mouse_lemur) 100% ABDC01026332 NKKFASFNAYENHLKSWRHIDLEKKVVQAVNRKVEMMNEKNLEKILGMDSV ZNF622_micMur Microcebus murinus (mouse_lemur) 86% ABDC01455616 SKKFASFNAYENHLKSRRHVKLEKKAVQAVNRKVEMMNEKNLQKGLGVESV ZNF622_tupBel Tupaia belangeri (tree_shrew) weak trace GKKFATFNAYENHLGSRRHAELERKAVRAASRRVELLNAKNLEKGLGADGV ZNF622_musMus Mus musculus (mouse) 74% SKKFATFNAYENHLKSRRHVELEKKAVQAVSRQVEMMNEKNLEKGLGVDSV ZNF622_ratNor Rattus norvegicus (rat) 94% AAHX01014435 chr2 ++ 77572184 77572336 SKKFATFNAYENYLKSRLHVELEKKTVQAVSRQVEMMNEKNLEKGLGVDSV ZNF622_ratNor Rattus norvegicus (rat) 88% AAHX01059254 chr9 +- 31986229 31986381 153 SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLDVDSV ZNF622_speTri Spermophilus tridecemlineatus (squirrel) SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGVDSV ZNF622_cavPor Cavia porcellus (guinea_pig) SKKFASFNAYENHLKSRRHVELEKRAVQAVNRKVELMNEKNLEKGLGVDSV ZNF622_oryCun Oryctolagus cuniculus (rabbit) 100% .KKFASLNAYENHLRSRRHLELEKKAVQAVNRQVELMNEKNLEKGLGADGV ZNF622_ochPri Ochotona princeps (pika) 86% GKRFASFNAYENHLQSRRHAELERAAVRAVSRQVQLRNAKNLEKGLGADGV ZNF622_canFam Canis familiaris (dog) 72% SKKFASFNAYENHLKSRRHVELEKKAVQAVSRKVEVMNEKNLEKGLSVDSV ZNF622_equCab Equus caballus (horse) 94% SKKFACANAYENHLRSRRHVELERK---AVSRRVEMMNEKNLEKGLGVDRV ZNF622_myoLuc Myotis lucifugus (microbat) 80%/86% SKKFACFNAYENHLKSRRHMELEKK---AVNRKVEMMNEKNLEKGLGVDSL ZNF622_pteVam Pteropus vampyrus (macrobat) 88%/94% SKKFASFKAYENHLRSRRHVELEKQ---AVSRKVALMNEKNLEKGLGVDSV ZNF622_bosTau Bos taurus (cow) 82%/88% SKKFASFKAYENHLRSRRHVELEKQ---AVSRKVALMNEKNLEKGLGVDSV ZNF622_oviAri Ovis aries (sheep) 84% SKKFASFKAYENHLKSRRHVELEKR---AVSRKVAILNEKNLEKGLGVDSV ZNF622_susScr Sus scrofa (pig) 82% SKKFASFKAYENHLKSRRHVELEKK---AVSRKVAIMNEKNLEKGLGVDSV ZNF622_vicVic Vicugna vicugna (vicugna) 86%/92% GKRFASLNAFENHLRSRRHLELEKKAVQAASRRVQMLNAKNLEKGLAADGL ZNF622_eriEur Erinaceus europaeus (hedgehog) ...FASFNAYDNHLRSRRHVELEARAVQAVSRRVQRLNEKNLEKGL..... ZNF622_sorAra Sorex araneus (shrew) SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGADSV ZNF622_dasNov Dasypus novemcinctus (armadillo) 98% SKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGADSV ZNF622_choHof Choloepus hoffmanni (sloth) ............HLKSRRHVELEKKAVQAVSRRVEMMNEKNLEKGLGADGV ZNF622_loxAfr Loxodonta africana (elephant) SKKFASFNAYENHLKSRRHVELEKKAVQAVSRRVEMMNEKNLEKGLDAGGV ZNF622_proCap Procavia capensis (hyrax) SKKFASFNAYENHLQSRRHVELEKKAVQAVNRRVERMNEKNLEKGLDADNV ZNF622_echTel Echinops telfairi (tenrec) 88% SKKFATFNAYENHLKSRRHLELEKKAVQAVSRKVEMLNEKNLEKGLAPDGL ZNF622_monDom Monodelphis domestica (opossum) 84% SKRFSTFNAYENHLKSKKHLELEKKAVQAVSKKVKILNEKNLEKGLAVESV ZNF622_galGal Gallus gallus (chicken) SKRFSNFNAYENHLKSKKHLELEKKAVQAVSKKVELMNEKNLEKGLAQESV ZNF622_anoCar Anolis carolinensis (lizard) not available ZNF622_felCat Felis catus (cat) not available ZNF622_turTru Tursiops truncatus (dolphin) not available ZNF622_ornAna Ornithorhynchus anatinus (platypus)
>homSap cagtaagaagtttgcctctttcaacgcctacgagaaccacctcaagtcccggcgtcacgttgagctggagaagaaggccgtgcaggcagtgaatcggaaagtggagatgatgaatgaaaagaacttggagaaaggactgggcgtggacagtgtgga >uc003jfq.1_hg18_1_6 208 0 1 chr5:16518150-16518774 - ZNF622 MATYTCITCRVAFRDADMQRAHYKTDWHRYNLRRKVASMAPVTAEGFQERVRAQRAVAEEESKGSATYCTVCSKKFASFNAYENHLKSRRHVELEKKAVQAVNRKVEMMNEKNLEKGLGVDSVDKDAMNAAIQQAIKAQPSMSPKKAPPAPAKEARNV VAVGTGGRGTHDRDPSEKPPRLQWFEQQAKKLAKQQEEDSEEEEEDLDGD
Two residue insert in cow and bat CD5 illustrates homoplasy
Exon 5 of gene CD5 exhibts a two residue insertion in cow, sheep, and bat relative to other placental mammals. However, the insertion is not found in other artiodactyls (dolphin, pig, and vicugna) and so is homoplasic. Grouping bovids with microbats to the exclusion of other even-hoofed animals is a ridiculous concept.
Macrobat has no available data but this locus is unsuitable as a phylogenetic marker within bats. It is not so clear from dna alignement that the insertion occurs in quite the same spot in cows and microbat. This cell surface receptor CD5 is evolving too fast for all but the narrowest phylogenetic purposes -- the percent identity within Laurasiatheres is 70% relative to dog, far too low for an informative rare genomic event.
human chr11 60645699 60645759 6 human AGCATCTGTGAAGGCACCGTGGAGGTGCGC------CAGGGGGCTCAGTGGGCAGCCCTGTGTGAC ...cow GACGTGTGCGAAGGCTCCGTGGAAGTGCGCAGTGGGAAGGGCCAGAAGTGGGACACGCTATGTGAC
human AGCATCTGTGAAGGCACCGTGGAGGTGCGCCAGGG----GGCT--CAGTGGGCAGCCCTGTGTGACA mcbat AGCGTGTGCGCGGGCTCCGTGGAGGTGCGCCAGGGCCAGGGCCGGCAGTGGGAAGCCCTGTGCCACA
FQPKVQSRLVGGSSICEGTVEVR--QGAQWAALCDSSSARSSLRWEEVCREQQCGSVNSYRV CD5_homSap Homo sapiens (human) FQPKVQSRLVGGSSICEGTVEVR--QGAQWAALCDSSSARSSLRWEEVCREQQCGSVNSYRV CD5_panTro Pan troglodytes (chimp) FQPKVQSRLVGGSSICEGTVEVR--QGAQWAALCDSSSFRSSLRWEEVCREQQCGGVNSYRV CD5_ponPyg Pongo pygmaeus (orang_sumatran) FQPKVQSRLVGGSSICEGTVEVR--QGAQWAALCDSSSAKSSLRWEEVCREQQCGSFNSYQA CD5_macMul Macaca mulatta (rhesus) FQPKVQSRLVGGSSMCEGTVEVR--QGAQWAALCDSSSARSPLRWEEVCQEQRCGGVNSYRV CD5_calJac Callithrix jacchus (marmoset) FQPKVQSRLVGGSGMCEGTVEVR--QSTQWAPLCDSSPLKGTARWEEVCQEQQCGSVNSYHM CD5_otoGar Otolemur garnettii (bushbaby) FQPKVQSRLVGGSSVCEGSVEVR--QGTQWAALCDSSPAKGTARWEEVCQEQQCGSVNSYHV CD5_micMur Microcebus murinus (mouse_lemur) FQPKVQSRLVRGSGVCEGAVEVR--QGLQWAALCDSSGARRMERWNEVCQEQQCGNASSYRL CD5_tupBel Tupaia belangeri (tree_shrew) FQPKVQSRLVGGSSVCEGIAEVR--QRSQWEALCDSSAARGRGRWEELCREQQCGDLISFHT CD5_musMus Mus musculus (mouse) FQPKVQSRLVGGSSVCEGIAEVR--QRSQWAALCDSSAARGPGRWEELCQEQQCGNLISFHV CD5_ratNor Rattus norvegicus (rat) FQPKVQSRLVGGGSTCQGTVEVR--QGGPEAQWVALCHSTRSSARWEALCQEQQCGRFLTYH. CD5_cavPor Cavia porcellus (guinea_pig) FQPKVQSRLVGGGSVCKGTVEVR--QATQWAALCDSHVSRGPARWAELCQEQKCNGLVSYH. CD5_speTri Spermophilus tridecemlineatus (squirrel) FQPKVQSRLVGGSSMCEGTAEVR--QGPRWAALCHNSSAKGTARWEELCQEQQCGIVNSYYV CD5_oryCun Oryctolagus cuniculus (rabbit) FQPKVQSRLVGGSGTCEGTVEVR--QRTQWAALCHSAPAKGMVRWEELCQEQQCGRVSSYHL CD5_ochPri Ochotona princeps (pika) FQPKVQSRLVGGSGLCEGSVEVR--QGRQWEVLCDVPRAKGTARWEEVCQEQRCGKLNSFRV CD5_canFam Canis familiaris (dog) FQPKVQSRLVGRRSLCEGSVEVR--QGKQWEVLCDSPRPKGTARWEEVCQELQCGSVASYRV CD5_felCat Felis catus (cat) FQPKVQSRLVGGSSLCEGSVEVR--QGKQWKTLCDIPSPKGAVRWEEVCQEQQCGKVNSYVH CD5_equCab Equus caballus (horse) 79% to dog FQPQVQSRLVGGSSVCAGSVEVRQGQGRQWEALCHSPWAKSMARWAEVCREQQCGPANSYQV CD5_myoLuc Myotis lucifugus (microbat) 69% to dog FQPKVQSRLVGGSDVCEGSVEVRSGKGQKWDTLCDDSWAKGTARWEEVCREQQCGNVSSYRG CD5_bosTau Bos taurus (cow) FQPKVQSRLVGGSDMCEGSVEVRSGKGQQWDTLCDSSWAKGTARWEEVCREQQCGNVSFYQ. CD5_oviAri Ovis aries (sheep) 68% to dog FQPKVQSRLVGGSGTCEGSVEVR--QGKQWDALCDSSSAKGMARWEDVCREQQCGNVSSYQL CD5_turTru Tursiops truncatus (dolphin) FQPKVQSRLVGHSGTCEGSVEVR--QGKQWDTLCDSSSAKSMARWEEVCREQQCGNVSSYQI CD5_susScr Sus scrofa (pig) AK239283 transcript 70% to dog FQPKVQSRLVGGSSSCEGAVEVR--HGKQWDPLCDSSSAKGAAARWEEVCQEQQCGNVSSYQ CD5_vicVic Vicugna vicugna (vicugna) SQPKVQSRLVGSSDPCAGTVEVR--QDGQWATLCSSNSAKSRALWEEVCHEQQCGRLSSYRE CD5_sorAra Sorex araneus (shrew) FQPKVRSRLAGGSSMCEGTVEVR--QGKQWGALCSSSLAKGMAPWEEVCQEQQCGHVRSYHM CD5_dasNov Dasypus novemcinctus (armadillo) FQPKVQSRLVGGRSMCEGTVEVR--QGGQWAALCDTSPAGTTARWEEVCQEQQCGGVSAYRV CD5_proCap Procavia capensis (hyrax) FQPKVQSRLVEGHSLCEGVVEVR--QGGRWAALCHNSMAT..ARWNEVCQEQQCGSVVTYGK CD5_echTel Echinops telfairi (tenrec) FQPKVRSRLKGGSHACAGTVEVN--FEGQWRALCS...QKN...WEEVCHEQHCGRLLT... CD5_monDom Monodelphis domestica (opossum) FQPEVKSRLTGGTSHCSGVVEVF--HKSNWKTLG....LNGASLWADVCQRQECGALISQKT CD5_ornAna Ornithorhynchus anatinus (platypus) not available CD5_pteVam Pteropus vampyrus (macrobat)
Deletion in SLC44A1 in bat and cow again homoplasic
This loss of one amino acid in the conserved transport gene SLC44A1 is limited to cow, sheep, and microbat. It is absent in dog, cat, horse, pig, vicugna, and macrobat. A moderately close paralog (at least in this exon) SLC44A3 is not an actual source of confusion; it has no indel in any species. This rare genomic event is not suited for within-bat or within-artiodactyl tree refinement because of this homoplasy, as well as an unrelated 3 aa loss in guinea pig.
SALCSYNLKPSEYTTSPKSSVLCPKLPVPA SLC44A1_homSap Homo sapiens (human) SFLCVYSLNSFNYTHSPKADSLCPRLPVP SLC44A3 SALCSYNLKPSEYTTSPKSSVLCPKLPVPA SLC44A1_panTro Pan troglodytes (chimp) SALCSYNLKPSEYTTSPKSSVLCPKLPVPA SLC44A1_ponPyg Pongo pygmaeus (orang_sumatran) SALCSYNLKPSEYTTSPKSSVLCPKLPVPA SLC44A1_macMul Macaca mulatta (rhesus) SALCSYNLKPSEYTTSSKSSVLCPKLPVPA SLC44A1_otoGar Otolemur garnettii (bushbaby) SALCSYNLKPSEYTTSSKSSVLCPKLPVPA SLC44A1_micMur Microcebus murinus (mouse_lemur) SALCSYNLKPSEYTTSSKSSVLCPKLPVPA SLC44A1_tupBel Tupaia belangeri (tree_shrew) SALCSYNIKPSEYTLTSKSSGFCPKLPVPA SLC44A1_musMus Mus musculus (mouse) SALCSYNIKPSEYTLTAKSSAFCPKLPVPA SLC44A1_ratNor Rattus norvegicus (rat) SALCSYHIKPSEYT---KSSDFCPKLPVPA SLC44A1_cavPor Cavia porcellus (guinea_pig) SALCSYDLKPSEYTTSPKSSVLCPKLPVPA SLC44A1_oryCun Oryctolagus cuniculus (rabbit) SALCSYNLKPSEYTTSSKASVLCPKLPVPA SLC44A1_canFam Canis familiaris (dog) SALCTYDLKPSEYTTSSKASALCPKLPVPA SLC44A1_equCab Equus caballus (horse) SALCSYNLKHSEYTTSSKASVLCPKLPVPA SLC44A1_felCat Felis catus (cat) SALCSYTLKPSEYTIPSKASGVCPKLPVPA SLC44A1_pteVam Pteropus vampyrus (macrobat) SALCHYKLKPSEYTSS-KASSLCPKLPVPA SLC44A1_myoLuc Myotis lucifugus (microbat) SALCSYDLKPSEYASS-KAKGLCPKLPVPA SLC44A1_bosTau Bos taurus (cow) SFLCVYNLNSFNYTQIPNADLLCPRLPVP SLC44A3 SALCSYDLKPSEYASS-KAKGLCPKLPVPA SLC44A1_oviAri Ovis aries (sheep) SALCSYSLKPSEYTTSSKAPVLCPKLPVPA SLC44A1_susScr Sus scrofa (pig) SALCSYDLKPSEYTTSSKASVLCPKLPVPA SLC44A1_vicVic Vicugna vicugna (vicugna) SLLCSYNLKPSEYTTSSKASVLCPKLPVPA SLC44A1_eriEur Erinaceus europaeus (hedgehog) SALCSYNLKPSEYTTSSKASVLCPKLPVPA SLC44A1_sorAra Sorex araneus (shrew) SALCNYNLKPSEYTTSSKASALCPKLPVPA SLC44A1_dasNov Dasypus novemcinctus (armadillo) SALCSYNLKPSEYTTSSKASVLCPKLPVPA SLC44A1_loxAfr Loxodonta africana (elephant) SALCTYNLKPSEYVTHSKASVFCPKLPVP. SLC44A1_echTel Echinops telfairi (tenrec) SALCTYNLKPSEYVTHSKASVFCPKLPVP. SLC44A1_monDom Monodelphis domestica (opossum) SALCVYDLKPSEYSTHPKASVRCPKLPVP. SLC44A1_triVul Trichosurus vulpecula (possum) SALCSYHISPSAYTSDPSASKLCPKLPVP. SLC44A1_ornAna Ornithorhynchus anatinus (platypus) not available SLC44A1_turTru Tursiops truncatus (dolphin) chr9 107150470 107150533 -3 human TGTAGCTACAACCTAAAGCCTTCTGAATACACTACATCTCCAAAATCTTCTGTTCTCTGCCCC ..cow TGCAGCTATGACCTAAAGCCTTCTGAATAT---GCATCATCAAAAGCAAAAGGTCTTTGCCCC human TGTAGCTACAACCTAAAGCCTTCTGAATACACTACATCTCCAAAATCTTCTGTTCTCTGCCCCA mcbat TGTCACTACAAGCTAAAGCCTTCTGAATACAC---ATCTTCAAAAGCGTCTTCTCTTTGCCCCA
Deletion in MLL3 in bat and cow not strongly informative
The very large gene MLL3 (4911 aa) is not strongly informative because the lack of conservation around the site of the one residue deletion seen in cow, dolphin, and pig makes comparison to bats too difficult. Both micro and macrobats have a similarly sited deletion but one that nucleotide-based alignment places somewhat offset. The site is also quasi-homoplasous in mouse lemur and nearby deletions are seen in pig and marsupial. The protein is too diverged in eulipotyphyla to allow meaningful comparison. Here again Laurasiatheres are anomalously non-conserved within amniotes suggesting a lack of selective pressure which is conducive to rapid change including in length and so homoplasy.
NSRPPSPMDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSVSRPLQMNETTANRPS MLL3_homSap Homo sapiens (human) NSRPPSPMDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSVSRPLQMNETTANRPS MLL3_panTro Pan troglodytes (chimp) NSRPPSPVDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSVSRPLQMNETTANRPS MLL3_ponPyg Pongo pygmaeus (orang_sumatran) NSRPPSPMDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSVSRPLQMNETTANRPS MLL3_macMul Macaca mulatta (rhesus) NSRPPSPMDPYAKMVGTPRPPPGGHSFPRRNS-APMENCTPLSSVTRPIQMNETTANRPS MLL3_micMur Microcebus murinus (mouse_lemur) NSRPPSPADPYAKMVGTPRPPPGGHSFSRRNSVAPMENCAPLSSVSRPVQMSETAAGRPS MLL3_tupBel Tupaia belangeri (tree_shrew) HSRPPSPVDPYAKMVGTPRPPPGGHSFPRRNSVTPVENCVPLSSVPRPIHMNETSATRPS MLL3_musMus Mus musculus (mouse) HSRPPSPVDPYAKMVGTPRPPPVGHSFPRRNSVTPVENCVPLSSVPRPIHMNETSATRPS MLL3_ratNor Rattus norvegicus (rat) NSRPPSPIDPYAKMVGTPRPPPGGHSFCRRNSVAPVENCVPLSSVPRPIQMSETPANRPS MLL3_cavPor Cavia porcellus (guinea_pig) .SRPPSPMDPYAKMVGTPRPPPGGHSFPRRNSVAQVENCTPLSSVSRPIQMNETTTNRPS MLL3_oryCun Oryctolagus cuniculus (rabbit) NSRPPSPVDPYAKMVGTPRPPPVGHSFPRRNSVPPVENCAPLSSVSRPMQVNETTTNRPS MLL3_ochPri Ochotona princeps (pika) HSRPPSPMDPYAKMVGTPRPAPGGHSFSRRNSAAPADSCTPLPSVSRPAQMTDTTANRPS MLL3_canFam Canis familiaris (dog) HSRPPSPVDPYAKMVGTPRPPPGGHGFSRRNSTALVENCAPLPPVPRPAPVSEATANRPS MLL3_felCat Felis catus (cat) NSRPPSPVDPYAKMVGTPRPPPGAHCFSRRNSAALVENCAPLSSVSRPAQMSETTANRPS MLL3_equCab Equus caballus (horse) NSRPPSPVDPYAKMVGTPRPPPGGHSFSRRN-SALVESCVSLSSAPRPPQMGEGTANRPS MLL3_myoLuc Myotis lucifugus (microbat) .SRPPSPMDPYAKMVGTPRPPPGGHSFPRRT-SALAETRVPLPPAPRPTQTSEATANRPS MLL3_pteVam Pteropus vampyrus (macrobat) NSRPPSPMDPYAKMVGTPRPPPGGHSFPRRN-AALAESCGPLPSGSRPPQMGEAAANRPS MLL3_bosTau Bos taurus (cow) NSRPPSPADPYAKMVGTPRPPAGGHGFSRRN-SALVENCAPLPSVSRPTQMSETTASRPS MLL3_turTru Tursiops truncatus (dolphin) NPRPPSPVDPYAKMVGTPRPPPGGHGFSRRN-SALVETSAPLSSVSRP--MSETPASRPS MLL3_susScr Sus scrofa (pig) NSRPPSPMDPYAKMVGTPRPPPEVHSFSRRNSVASVENCAPLSSVSRSIQISETAANRPS MLL3_dasNov Dasypus novemcinctus (armadillo) NSRPPSPVDPYAKMVGTPRPSAAGHSFPRRNSVTPVENCAPLSSVSRPIQISETTANRPS MLL3_loxAfr Loxodonta africana (elephant) NSRSPSPMDPYAKMVGTPRPPAGGHSFPRRNSVAPGENCTSLSSASRPIQISETATNRPS MLL3_echTel Echinops telfairi (tenrec) SSRPPSPVDPYAKMVGTPRPLPAGPNFPRRNSVGPVETCT---TPSRSIQVTETTASRLS MLL3_monDom Monodelphis domestica (opossum) .SRPPSPLDPYAKMVGTPRPPPVGANFARRNSIAPLESCGPPPSAARPVPSPETTGRRPS MLL3_ornAna Ornithorhynchus anatinus (platypus) NTRPPSPMDPYAKMVGTPRPAPTNQNFVRRGSIAPLDTCAPQSSISRPVQVTEPSGSRPS MLL3_galGal Gallus gallus (chicken) SARPPSPIDPYAKMVGTPRPLPVNQNYIRRNSVPSVDFCPPPSPISRPAQAPEIAGKRPS MLL3_anoCar Anolis carolinensis (lizard) not available MLL3_vicVic Vicugna vicugna (vicugna) not available MLL3_eriEur Erinaceus europaeus (hedgehog) not available MLL3_sorAra Sorex araneus (shrew)
chr7 151510091 151510154 -3 human AAAGGTGTACAGTTTTCCACTGGTGCAGCAGAATTTCTTCTGGAAAAACTATGGCCCACAGGA cow AGTGGCCCACAGCTCTCCGCCAGTGCAGCG---TTTCTTCTGGGAAAGCTGTGGCCCCCGGGA human AAAGGTGTACAGTTTTCCACTGGTGCAGCAGAATTTCTTCTGGAAAAACTATGGCCCACAGGAG mibat AAAGATACACAGCTTTCCACCAGT---GCAGAATTCCTTCTGGAAAAGCTGTGGCCCCCAGGAG
Deletion in KIAA0240 in bat and cow is terminal-leaf homoplasic
The deletion here is found solely in cow and microbat. It is lacking in dolphin, pig, and vicugna as well as macrobat. The gene KIAA0240 is otherwise well-suited for phylogeny. This is an example of where the screen worked but a genuine case of terminal leaf homoplasy occured.
LSPGQSSVSQGRPGFATMPSVTSMSGPSRFPAVSSASTAHPSLGSAVQSGSSGSNFTGDQ KIAA0240_homSap Homo sapiens (human) LSPGQSSVSQGRPGFATMPSVTSMSGPSRFPAVSSASTAHPSLGSAVQSGSSGSNFTGDQ KIAA0240_panTro Pan troglodytes (chimp) LSPGQSSVSQGRPGFTTMPSVTSMSGPSRFPAVSSASTAHPSLGSAVQSGSSGSNFTGDQ KIAA0240_ponPyg Pongo pygmaeus (orang_sumatran) LSPGQSSVSQGRPGFTTMPSVTSMSGPSRFPAVSSASTAHPSLGSAVQSGSSGSNFTGDQ KIAA0240_macMul Macaca mulatta (rhesus) LSPGQSSVSQGRPGFTTMPSVTSMSGPSRFPAVSSASTAHPSLGSAVQSGSSGSNFTGDQ KIAA0240_calJac Callithrix jacchus (marmoset) LSPGQSSVSQGRPGFTAMPSVTSMSGPSRFPAVSSTSTAHPSLGSVVQSGASGSNFTGDQ KIAA0240_otoGar Otolemur garnettii (bushbaby) LSPGQSSVSQGRPGFTAMPSVTSMSGPSRFPAVSSASTAHPSLGSVVQSGASGSNFTGDQ KIAA0240_micMur Microcebus murinus (mouse_lemur) LSPGQSSVSQGRPVFTPMSSVTSMSGPSRFPAVSSASTAHPSLGSAVQSGASGSNFTGEQ KIAA0240_tupBel Tupaia belangeri (tree_shrew) LSPGQSSVSQGRPGFATMPAVSGMAGPARFPAVSSASTAHPTLGPTVQSGAPGSNFTGDQ KIAA0240_musMus Mus musculus (mouse) LSPGQSSVSQGRPGFATMASVSSMSGPARFPAVSSASTAHPTLGPAVQSGASGSNFTGDQ KIAA0240_ratNor Rattus norvegicus (rat) LSPGQSSVSQGRPGFTTMPSVTSMAGPSRFPAASSASTAHPSLGPAAQSGPSGANFTGDQ KIAA0240_cavPor Cavia porcellus (guinea_pig) LSPGQSSVSQGRPGFTTMPSATSMSGPSRFPAVSSGNTAHPSLGSVVQSGASGSNFTGD. KIAA0240_speTri Spermophilus tridecemlineatus (squirrel) LSPGQSSVSQGRPAFTTMPSATSLSGPSRFPAVSSASTAHPSLGSAVQSAASGSNFAGDQ KIAA0240_oryCun Oryctolagus cuniculus (rabbit) LSPGQSNVSQGRPGFTTMASATSMSGPSRFPAVSSASTAHPSVGSAVQSAASGSNFTGDQ KIAA0240_ochPri Ochotona princeps (pika) LSPGQSNVSQGRPGFTTMPSVTSMSGPSRFPSVSSSSTAHPSLGSVVQSGASGSNFTGDQ KIAA0240_canFam Canis familiaris (dog) LSPGQSSVSQGRPGFTTMPSVTNMSGPSRFPVVSSSSTAHPSLGSAVQSGASGSNFTGDQ KIAA0240_felCat Felis catus (cat) LSPGQSSVSQGRPGFTTMPSVTSMAGPSRFPVVSSTSTAHPSLGSAVQSGASGSNFTGDQ KIAA0240_equCab Equus caballus (horse) LSPGQSNVSQGRPGFTTVPSVTSMSGPSRFPVVSSSSTAHPSLGSAVQSGASGSNFAGD. KIAA0240_pteVam Pteropus vampyrus (macrobat) LSPGQSSVSQGRPGFTPMPSVTSMSGPSRFP-VSSSSTVHPSLGSAVQSGASGSNFTGDQ KIAA0240_myoLuc Myotis lucifugus (microbat) LSPGQSSVSQGRPGFTTMPSVTSMAGPSRFP-VSSSSTAHPSLGSAVQSGASGSNFTGDQ KIAA0240_bosTau Bos taurus (cow) LSPGQSSVSQGRPGFTTMPSVTSMSGPSRFPAVSSSSTAHPSLGSAVQSGASGSNFTGDQ KIAA0240_susScr Sus scrofa (pig) LSPGQGSVSQGRPGFAAMPSVTSMSGPSRFPVVSSSSTAHPSLGAAVQSGASGSNFTGDQ KIAA0240_turTru Tursiops truncatus (dolphin) LSPGQSSVSQGRPGFTTMPSVTSMSGPSRFPVVSSSSTAHPSLGSAVQSGATGSNFTGDQ KIAA0240_vicVic Vicugna vicugna (vicugna) LSPGQSSVSQGRPGFTTMPSVTSMSGSNRFPAVSSSSTAHPSLGSSVQSGASGSNFMGDQ KIAA0240_eriEur Erinaceus europaeus (hedgehog) LSPGQGSVSQGRPGFAAMPSVPSMSGPSRFPAVSSSGTAHQNLGSATQAGAPGSSFTPDQ KIAA0240_sorAra Sorex araneus (shrew) LSPGQSSVSQGRPGFTTMPSVTSMSGPSRFPNVSSSSTAHPSLGSAVQSGASGSNFTGDQ KIAA0240_dasNov Dasypus novemcinctus (armadillo) LSPGQSSVSQGRPGFTTMPSVTNMSGPSRFPNVSSSSTAHPSLGSAVQSGMSGSNFTGDQ KIAA0240_choHof Choloepus hoffmanni (sloth) LLPGQSGISQGRPGVTTMPSVIPMSEPSQFLAVNSPKTAHPRLGSAPQSGALGSKFPGD. KIAA0240_loxAfr Loxodonta africana (elephant) LSPGZNVLTRGKPGFTTMPSVTSMAGPSRFPVVSSSSTAHPSLGSTVQAGVSGSNFPGDQ KIAA0240_echTel Echinops telfairi (tenrec) LSPGQSSVSQGRPGFPGQSAVTSVSAPGRFTVVSSSSTALPNLGPSVQSATSATNFNGDQ KIAA0240_monDom Monodelphis domestica (opossum) LSPGQTGVSQGRPGFTSMPAVANVSVPNRFTVVSSSATVHPSPGSAVQSVASGSNFTGDQ KIAA0240_ornAna Ornithorhynchus anatinus (platypus) LSPSQANASQGRSSFTTMSTVSNMSASNRFAVVSSSGSVHPSLGPSVQSVASGGNFTGDQ KIAA0240_galGal Gallus gallus (chicken) LSPSQGNISQSRTSFSTMSSSSNMSSNSRFTAVSSSAVVHPSIGPSAQSVASGGSFAGDQ KIAA0240_anoCar Anolis carolinensis (lizard)
not available KIAA0240_oviAri Ovis aries (sheep) not sought KIAA0240_tarSyr Tarsius syrichta (tarsier) not sought KIAA0240_proCap Procavia capensis (hyrax) not sought KIAA0240_macEug Macropus eugenii (wallaby) not sought KIAA0240_taeGut Taeniopygia guttata (finch) chr6 42905641 42905704 -3 human GACAAGCATGTCAGGACCTAGTCGGTTCCCTGCTGTCAGCTCAGCCAGCACTGCCCATCCTAG cow GACAAGCATGGCAGGACCTAGTCGGTTCCC---TGTTAGTTCATCCAGCACTGCCCATCCTAG human GACAAGCATGTCAGGACCTAGTCGGTTCCCTGCTGTCAGCTCAGCCAGCACTGCCCATCCTAGT mibat GACAAGCATGTCAGGACCTAGTCGGTTCC---CTGTTAGCTCGTCCAGCACTGTCCATCCTAGT
EIPEDPLVAEEYYADAFDSYCEESDEEEEE-IALERPEKEIRNEGSQPAYRTNQQ NEK11_homSap Homo sapiens (human) EIPEDPLVAEEYYADAFDSYCEESDEEEEE-IALERPEKEIRNEGSQPAYRTNQQ NEK11_panTro Pan troglodytes (chimp) EIPEDPLVAEEYYADAFDSYCEESDEEEEE-IVLAGPEKEIKNEGSQPTYRTNQQ NEK11_ponPyg Pongo pygmaeus (orang_sumatran) EIPEDPLVAEEYYADAFDSYCEESDEEEEE-IVLAGPEKEIKNEGSQPTYRTNQQ NEK11_macMul Macaca mulatta (rhesus) EIPEDPLVAEEYYADTFDSYCEESDEEEEG-TVLSGPEKEIKKES-QPAYRTNQQ NEK11_calJac Callithrix jacchus (marmoset) EIPEDPLVAEEYYADVFDS-CSE-EEEEEE-IVFSASEGEVKDEGPQPPYRTHQQ NEK11_otoGar Otolemur garnettii (bushbaby) EIPEDPLVAEQYYSDVFDSCSEDSEEQEEE-MIFSEAGGDTKEEESPSVYRTNQQ NEK11_musMus Mus musculus (mouse) EIPEDPLVAEQYYADVFDSCSEDSGEQEEE-MAFSEAGGDMREEGSPPTYRTNQQ NEK11_ratNor Rattus norvegicus (rat) EIPEDPLVAEVYYADAFDSCSEDSEEQEGE-TGSLGPEVEAQDEGSQPTSRTNHQ NEK11_cavPor Cavia porcellus (guinea_pig) .IPEDPFVAEEYYADVFDSCSEESEEQEEE-ILFSGPEREVKD------YRTNQQ NEK11_oryCun Oryctolagus cuniculus (rabbit) EIPEDPLVAEEYYTDVFDSCSEESEEQEED-TLFSGPDRETKD-----GNRINQQ NEK11_ochPri Ochotona princeps (pika) EIPEDPLVAEEYYADAFDSCSEESEEEEEE-IMFSAPEEEVKDEGPRPAHRIFQQ NEK11_canFam Canis familiaris (dog) DIPEDPIVAEEYYADAFDSCSEESEEEEEE-IVFSGPEEEVKDEGPQPAYRTIQQ NEK11_felCat Felis catus (cat) EIPEDPLVAEEYYADAFDSCSEESEDEEEE-IVFSGPEEEVKDEVQQPAYRTNQQ NEK11_equCab Equus caballus (horse) .IPEDPRVAEEYYADTFDSCSEESEEEEEEEMVLAGLEAEVRDKGPQPDCRANQQ NEK11_myoLuc Myotis lucifugus (microbat) EIPEDPLVAEEYYADVFDSCSEESEESEEGKTVFSVKDEEVEDEGPQPVYRTNQQ NEK11_bosTau Bos taurus (cow) EIPADPLVAEEYYADVFDSCSEESEESEEEKTVFSVKDEEVEDEGPQPVYRTNQQ NEK11_oviAri Ovis aries (sheep) EIPEDPLVAEEYYTDAFDSCSEESEEEEEN-MVFSAGEEEVQDEGSQPAYRTNQQ NEK11_susScr Sus scrofa (pig) EIPEDPLVAEEYYADAFDSCSEESEEEEEN-TVFSAGEEEVKDEEPQPAYRTNQQ NEK11_vicVic Vicugna vicugna (vicugna) EIPEDPLKAEEYYADVFDSCSEESEEEGEE-TVFSGSE-EVSDDGLQPAHRTNQQ NEK11_eriEur Erinaceus europaeus (hedgehog) EIPEDPLVAEEYYADAFDSCSEESEDRGEE-ALFSGSGEDCEEEEPQLSYRTNQQ NEK11_sorAra Sorex araneus (shrew) EIPEDPLVAEEYYADVFDSCSEESEEEEEE-TVFSGPEEKAKDEGPQPVYRTNQQ NEK11_choHof Choloepus hoffmanni (sloth) EIPEDPLVAEEYYADTFDSCSEESEEEEEE-IVFSGPEEEVKDKEPQPAYRTNQ. NEK11_loxAfr Loxodonta africana (elephant) EIPEDPLVAEEYYADTFDSCSEESEEEEDE-TVFSGPGEEVKDEESQPASRTDQQ NEK11_echTel Echinops telfairi (tenrec) EIPEDPLVAEEYYDDVFDSCSEESEEEAEE-----KSEEMVEYETSQTVVKTNQQ NEK11_ornAna Ornithorhynchus anatinus (platypus) .IPEDPLIAEEYYNDVFDSCSETSEDQEEE-VVEADEVKEDEEEDTLFTYTTNQQ NEK11_anoCar Anolis carolinensis (lizard) not available NEK11_micMur Microcebus murinus (mouse_lemur) not available NEK11_tupBel Tupaia belangeri (tree_shrew) not available NEK11_speTri Spermophilus tridecemlineatus (squirrel) not available NEK11_pteVam Pteropus vampyrus (macrobat) not available NEK11_turTru Tursiops truncatus (dolphin) not available NEK11_dasNov Dasypus novemcinctus (armadillo) not available NEK11_monDom Monodelphis domestica (opossum) not sought NEK11_tarSyr Tarsius syrichta (tarsier) not sought NEK11_proCap Procavia capensis (hyrax) not sought NEK11_macEug Macropus eugenii (wallaby) not sought NEK11_galGal Gallus gallus (chicken) not sought NEK11_taeGut Taeniopygia guttata (finch)
Query: 13 EIPEDPLVAEEYYADAFDSYCEESDEEEEEIALERPEKEIRNEGSQPAYRTNQQ genome
EIPEDPLVAEEYYADAFDSYC ESDEEEEEIALERPEKEIRNEGSQPAYRTNQQ
Sbjct: 467 EIPEDPLVAEEYYADAFDSYCVESDEEEEEIALERPEKEIRNEGSQPAYRTNQQ webb
chr3 132430108 132430168 3
human TTTGATTCCTATTGTGAAGAGAGTGATGAG---GAGGAAGAAGAAATAGCGTTAGAAAGACCA
cow TTTGATTCCTGTTCTGAAGAAAGTGAGGAGAGTGAGGAAGGAAAAACAGTATTCTCAGTCAAG
human TTTGATTCCTATTGTGAAGAGAGT---GATGAGGAGGAAGAAGAAATAGCGTTAGAAAGACCAG
mibat TTTGACTCCTGTTCTGAAGAGAGTGAGGAGGAGGAGGAGGAAGAAATGGTGTTGGCAGGACTGG