Coding indels: PRNP
Introduction
The prion gene has many interesting evolutionary aspects. A few of those -- involving indels with phylogenetic interest - are explored below.
The signal peptide indel establishes Euarchontoglires
The prion gene PRNP exhibits a 6bp indel in its amino-terminal signal peptide that contributed historically to establishing the clade Euarchontoglires. From consideration of outgroups, the indel is a deletion (reducing signal pepide length from 31 to 29) rather than an insertion. It occurs in all species of rodents, rabbits, treeshrews, flying lemurs and primates sequenced to date but not in any other species of mammal.
Remarkably, this indel distribution has held up even as the number of genera sequenced has come to exceed 100. The billions of years of branch length represented by this data suggest that the deletion was a very rare event not subject to independent reoccurence (in effect homoplasy-free). Note it does not occur in a compositionally simple region (strings of leucines are common interiorly). As a typical mammalian gene as of November 08 can only be recovered from about 40 species, meaning similar rare genetic events cannot be as stringently evaluted as in PRNP.
Consequently this data set strongly conflicts with the never-ending computer proposals placing mouse basal relative to dog and human, ie (mouse,(dog,human)), which would require both a global revision of the well-established super-ordinal mammalian tree and in PRNP highly non-parsimonious multiple events both bizarrely located basally at the two unrelated divergence stems (very dense phylogenetic sampling has the effect of squeezing the window on homoplasy).
Signal region indels are not especially rare among orthologs to the 4500-odd human genes with signal peptides of which 595 are experimentally validated, despite steric requirements of the binding pocket of the signal processing complex SRP. In actuality the distribution of signal peptide length is fairly broad. These indels can be rapidly screened in batches of 25 by Blat alignment relative to the 44 available vertebrate genomes.
However few of these indels have any phylogenetic depth. It does not appear that the PRNP indel in euarchontoglires has any significant effect on cell targeting by the signal peptide (or subsequent membrane topology). It is not that indels in signal peptides are so rare but rather narrowly windowed basal events in large clades.
Below is data from 96 species:
MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Homo sapiens MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Pan troglodytes MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Gorilla gorilla MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Pongo pygmaeus MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Nomascus leucogenys MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Hylobates lar MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Symphalangus syndactylus MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Macaca arctoides MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Macaca fascicularis MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Macaca fuscata MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Macaca mulatta MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Macaca nemestrina MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Papio hamadryas MA--NLGCWMLFLFVATWSDLGLCKKRPKPG Callithrix jacchus MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Cebus apella MA--NLGCWMLVVFVATWSDLGLCKKRPKPG Cercopithecus aethiops MA--NLGCWMLVVFVATWSDLGLCKKRPKPG Cercopithecus dianae MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Colobus guereza MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Presbytis francoisi MA--NLGCWMLVLFVATWSDLGLCKKRPKPG Saimiri sciureus MA--KLGYWLLVLFVATWSDVGLCKKRPKPG Tarsius syrichta MA--NLGCWMLVVFVATWSDVGLCKKRPKPG Microcebus murinus MA--RLGCWMLVLFVATWSDIGLCKKRPKPG Otolemur garnettii ME--NLGCWMLILFVATWSDIGLCKKRPKPG Cynocephalus variegatus MA--QLGCWLMVLFVATWSDVGLCKKRPKPG Tupaia belangeri MA--NLGYWLLALFVTMWTDVGLCKKRPKPG Mus musculus MA--NLGYWLLALFVTTCTDVGLCKKRPKPG Rattus norvegicus MA--NLGYWLLALFVTTCTDVGLCKKRPKPG Rattus rattus MA--NAGCWLLVLFVATWSDTGLCKKRPKPG Cavia porcellus MA--NLGYWLLALFVTTWTDVGLCKKRPKPG Apodemus sylvaticus MA--NLGCWLLVLFVATWSDLGLCKKRTKPG Dipodomys ordii MA--NLSYWLLAFFVTTWTDVGLCKKRPKPG Clethrionomys glareolus MA--NLSYWLLALFVATWTDVGLCKKRPKPG Cricetulus griseus MA--NLSYWLLALFVATWTDVGLCKKRPKPG Cricetulus migratorius MA--NLGYWLLALFVTMWTDVGLCKKRPKPG Meriones unguiculatus MA--NLSYWLLALFVAMWTDVGLCKKRPKPG Mesocricetus auratus MA--NLGYWLLALFVATWTDVGLCKKRPKPG Sigmodon fulviventer MA--NLGYWLLALFVATWTDVGLCKKRPKPG Sigmodon hispiedis MV--NPGCWLLVLFVATLSDVGLCKKRPKPG Spermophilus tridecemlineatus MV--NPGYWLLVLFVATLSDVGLCKKRPKPG Sciurus vulgaris MA--HLGYWMLLLFVATWSDVGLCKKRPKPG Oryctolagus cuniculus MA--HLSYWLLVLFVAAWSDVGLCKKRPKPG Ochotona princeps MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Bos taurus MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Bison bison MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Rangifer tarandus MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Alces alces MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Capreolus capreolus MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Kobus megaceros MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Connochaetes taurinus MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Ammotragus lervia MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Hippotragus niger MVKSHMGSWILVLFVVTWSDVGLCKKRPKPG Camelus dromedarius MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Capris hircus MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Cervus elaphus MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Cervus elaphus nelsoni MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Dama dama MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Odocoileus hemionus MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Odocoileus virginianus MVKSHIGSWILVLFVAMWSDVALCKKRPKPG Oryx leucoryx MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Ovibos moschatus MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Ovis aries MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Ovis canadensis MVKSHIGSWILVLFVAMWSDVALCKKRPKPG Tragelaphus strepsiceros MVKSHIGGWILVLFVAAWSDIGLCKKRPKPG Sus scrofa MVKSHMGSWILVLFVVTWSDMGLCKKRPKPG Vicugna vicugna MVKSHVGGWILVLFVATWSDVGLCKKRPKPG Equus caballus MVRSHVGGWILVLFVATWSDVGLCKKRPKPG Diceros bicornis MVKSLVGGWILLLFVATWSDVGLCKKRPKPG Myotis lucifugus MVKNYIGGWILVLFVATWSDVGLCKKRPKPG Pteropus vampyrus MVKSHIANWILVLFVATWSDMGFCKKRPKPG Tursiops truncatus MVKSHIGGWILLLFVATWSDVGLCKKRPKPG Canis lupus familiaris MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Felis catus MVKSHIGSWLLVLFVATWSDIGFCKKRPKPG Mustela putorius MVKSHIGSWLLVLFVATWSDIGFCKKRPKPG Mustela vison MVKSHIGSWILVLFVAMWSDVGLCKKRPKPG Ailuropoda melanoleuca MVKNHVGCWLLVLFVATWSEVGLCKKRPKPG Erinaceus europaeus MVTGHLGCWLLVLFMATWSDVGLCKKRPKPG Sorex araneus MVKSHLGCWIMVLFVATWSEVGLCKKRPKPG Cyclopes didactylus MVRSRVGCWLLLLFVATWSELGLCKKRPKPG Dasypus novemcinctus MVKGTVSCWLLVLVVAACSDMGLCKKRPKPG Echinops telfairi MVKSSLGCWILVLFVATWSDMGLCKKRPKPG Elephas maximus MVKSSLGCWILVLFVATWSDMGLCKKRPKPG Loxodonta africana MVKSSLGCWMLVLFVATWSDVGLCKKRPKPG Procavia capensis MMKSGLGCWILVLFVATWSDVGLCKKRPKPG Orycteropus afer MVKSGLGCWILVLFVATWSDVGVCKKRPKPG Trichechus manatus MAKIQLGYWILALFIVTWSELGLCKKPKTRPG Macropus eugenii MGKIHLGYWFLALFIMTWSDLTLCKKPKPRPG Monodelphis domestica MGKIQLGYWILVLFIVTWSDLGLCKKPKPRPG Trichosurus vulpecular MARLLTTCCLLALLLAACTDVALSKKGKGKPS Gallus gallus MAKLPGTSCLLLLLLLLGADLASCKKGKGKPG Taeniopygia guttata MARLLTTCCLLALLLAACTDVALSKKGKGKPG Meleagris gallopavo MGKHQMTCWLAIFLLLIQANVSLAKK-KPKPS Anolis carolinensis MRRFLVTCWIAVFLILLQTDVSLSKKGKNKPG Gekko gekkko MGRYRLTCWIVVLLVVMWSDVSFSKKGKGKGG Trachemys scripta MGRHLISCWIIVLFVAMWSDVSLAKKGKGKTG Pelodiscus sinensis MPQSLWTCLVLISLICTLTVSSKKSGGGKSKTG Xenopus laevis MLRSLWTSLVLISLVCALTVSSKKSGSGKSKTG Xenopus topicalis
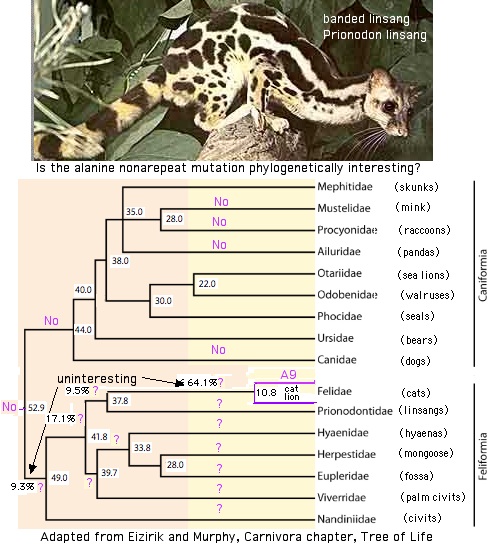
The peculiar prion repeat expansion in Felids
After several false starts involving error-ridden or lab contaminated genBank submissions (eg DQ217930), accurate prion sequences have emerged for 10 species of carnivores. One sees immediately that foxes, dogs and coyotes are united by two short indels upstream of the repeat region that distinguish them from panda, mink, raccoon, lion and cat. The first region is quite homoplasic within laurasiatheres but the second is not: this indel resolve as a deletion in foxes, dogs and coyote.
Of greater interest is the very peculiar nonapeptide expansion in the two felids. This results in an unprecedented alanine insertion in position 3 of repeats 2-5. This cannot have resulted from coincidental separate point mutations but instead must have occured in repeat 2 and then been propagated by replication slippage to the other repeats, obliterating their ancestral octapeptide repeat sequences. This scenario implies felid repeats 3-5 will share synonymous bases of ancestral repeat 2 -- ie the sweep did not propagate from the fifth repeat in the 4-2 direction.
This means repeats 3-5 in felids are not homologous to say repeats 3-5 in human. Only repeats 1 and 2 have common descent. This mode of evolution is reminiscent of gene duplicates in which one copy corrects the other (gene conversion).
This unprecedented insertion of alanine and its propagation may provide a definitive character for all of Felidae if they occured in the stem of this clade. This must be the case according to a recent tree showing lions basal which predicts all Felidae will have alanine nonarepeats.
The issue then concerns the immediate current outgroup to felids, namely lingsangs (Prionodontidae). If the alanine nonarepeat (A9) character occurs there, then hyena, mongoose, suricat, fossa and palrm civit must be considered. Additional species must be sequenced -- and possibly multiple individuals witin each species -- to resolve the timing in these little-studied species.
One uninteresting outcome would be that all feliform species (a well-established wing of carnivores) have this character. Another uninteresting outcome would be occurence restricted to Felidae. However using the calculated tree dates, the probabliy of neither is (52.9-49.0)/(52.9-37.8) = 3.9/15.1 = 25.8% assuming the A9 event is equally likely at any time since divergence from the caniform group up to the divergence of Felidae from Prionodontidae. However stem time within Felidae to lion divergence (the first node known to have A9) should be added. That time has been estimated at 10.8 myr. This improves the odds of a phylogenetically interesting outcome to 92.5%.
It's not clear whether the insertion and propagation resulted within a single event or were temporally separated by millions of years. There may not exist sufficient extant species to resolve a two phase event if such occured. It would be very difficult for the A9 event to completely revert because this would require a deletion of the alanine in repeat 2 followed by its propagation to all the other repeats.
Note lion has 4 repeats whereas cat has 5 (the most abundant ancestral allele). Panda has 6 repeats, again not unusual within mammalian PRNP repeats. Here it cannot be assumed that sequencing one individual animal produces a representative allele for that population; indeed the reference human genome shows 4 repeats even though 98% of the population has 5 repeats.
Neither 4 nor 6 repeat number is associated with the amyloid disease state. A nonapeptide is the norm in marsupials in the downstream repeats and so too is unlikely to predispose to disease.
1 2 3 4 5 6 >PRNP_panLeo KPGGGWNTGG-SRYPGQGSPGGNRYP PQGGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQ GGGTHSQWGK PSKPKTNMKHMAGAAAAGAVVGGLGGYMLG >Felis catus KPGGGWNTGG-SRYPGQGSPGGNRYP PQGGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQ GGGTHGQWGK PSKPKTNMKHMAGAAAAGAVVGGLGGYMLG >cat genom KPGGGWNTGG-SRYPGQGSPGGNRYP PQGGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQ GGGTHGQWGK PSKPKTNMKHMAGAAAAGAVVGGLGGYMLG >Procyon lot KPGGGWNTGG-SRYPGQGNPGGNRYP PQGGGGWGQPH.GGGWGQPH.GGGWGQPH.GGGWGQPHGGGGWGQ GGGSHGQWGK PNKPKTNMKHVAGAAAAGAVVGGLGGYMLG >Mustela KPGGGWNTGG-SRYPGQGSPGGNRYP PQGGGGWGQPH.GGGWGQPH.GGGWGQPH.GGGWGQPHGGGGWGQ GGGSHGQWGK PSKPKTNIKHVAGAASAGAVVGG >Neovison KPGGGWNTGG-SRYPGQGSPGGNRYP PQGGGGWGQPH.GGGWGQPH.GGGWGQPH.GGGWGQPHGGGGWGQ GGGSHGQWGK PSKPKTNMKHVA >Ailuropoda KPGGGWNTGG-SRYPGPGSPGGNRYP PQGGGGWGQPH.GGGWGQPH.GGGWGQPH.GGGWGQPHGGG.WGQPHGGGGWGQGGT.HGQWNK PSKPKTNMKHVAGAAAAGAVVGGLGGYMLG >Vulpes vulp KPGG-WNTGGGSRYPGQGSPGGNRYP PQGGGGWGQPH.GGGWGQPH.GGGWGQPH.GGGWGQPHGGGGWGQ GGGSHG.WGK PNKPKTNMKHVAGAAAAGAVVGGLGGYMLG >Vulpes lag KPGG-WNTGGGSRYPGQGSPGGNRYP PQGGGGWGQPH.GGGWGQPH.GGGWGQPH.GGGWGQPHGGGGWGQ GGGSHG.WGK PNKPKTNMKHVAGAAAAGAVVGGLGGYMLG >Canis fam KPGG-WNTGGGSRYPGQGSPGGNRYP PQGGGGWGQPH.GGGWGQPH.GGGWGQPH.GGGWGQPHGGGGWGQ GGGSHSQWGK >Canis la KPGG-WNTGGGSRYPGQGSPGGNRYP PQGGGGWGQPH.GGGWGQPH.GGGWGQPH.GGGWGQPHGGGGWGQ GGGSHGQWGK PNKPKTNMKHVAGAAAAGAVVGGLGGYMLG
>PRNP_panLeo Panthera leo (lion) EU236260 PMID:18256917 MVKGHIGGWILVLFVATWSDVGLCKKRPKPGGGWNTGGSRYPGQGSPGGNRYPPQGGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQGGGTHSQWGKPSKPKTNMKHMAGAAAAGAVVGGLGGYMLGSAMNRPLIHFGNDYEDRYYRENMYRYPNQVYYRPVDQYSNQNNFVHDCVNITVRQHTVTTTTKGENFTETDMKIMERVVEQMCVTQYQKESEAYYQRGASAILFSPPPVILLLSLLILLIGG >Felis catus (cat) EU588730 MVKGHIGGWILVLFVATWSDVGLCKKRPKPGGGWNTGGSRYPGQGSPGGNRYPPQGGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQGGGTHGQWGKPSKPKTNMKHMAGAAAAGAVVGGLGGYMLGSAMSRPLIHFGNDYEDRYYRENMYRYPNQVYYRPVDQYSNQNNFVHDCVNITVRQHTVTTTTKGENFTETDMKIMERVVEQMCVTQYQKESEAYYQRGASAILFSPPPVILLLSLLILLIGG >Felis catus (cat) genome misassembly but trace ti|662129434 is good MVKGHIGGWILVLFVATWSDVGLCKKRPKPGGGWNTGGSRYPGQGSPGGNRYPPQGGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQPHAGGGWGQGGGTHGQWGKPSKPKTNMKHMAGAAAAGAVVGGLGGYMLG >Vulpes vulpes (fox) EF571898 MVKSHIGGWILLLFVATWSDVGLCKKRPKPGGWNTGGGSRYPGQGSPGGNRYPPQGGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGGWGQGGGSHGWGKPNKPKTNMKHVAGAAAAGAVVGGLGGYMLGSAMSRPLIHFGNDYEDRYYRENMYRYPDQVYYRPVDQYSNQNNFVRDCVNITVKQHTVTTTTKGENFTETDMKIMERVVEQMCVTQYQKESEAYYQRGASAILFSPPPVILLISLLILLIVG >Vulpes lagopus (Arctic fox) EU365392 MVKSHIGGWILLLFVATWSDVGLCKKRPKPGGWNTGGGSRYPGQGSPGGNRYPPQGGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGGWGQGGGSHGWGKPNKPKTNMKHVAGAAAAGAVVGGLGGYMLGSAMSRPLIHFGNDYEDRYYRENMYRYPDQVYYRPVDQYSNQNNFVRDCVNITVKQHTVTTTTKGENFTETDMKIMERVVEQMCVTQYQKESEAYYQRGASAILFSPPPVILLISLLILLIVG >Procyon lotor (raccoon) AY208166 FCKKRPKPGGGWNTGGSRYPGQGNPGGNRYPPQGGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGGWGQGGGSHGQWGKPNKPKTNMKHVAGAAAAGA VVGGLGGYMLGSAMSRPLIHFGNDYEDRYYRENMYRYPNQVYYKPVDQYSNQNNFVHDCVNITVKQHTVTTTTKGENFTETDMKIMERVVEQMCVTQYQRESEAYYQRGASAILFS PPPV >Mustela putorius furo (ferret) GD181110 MVKSHIGSWLLVLFVATWSDIGFCKKRPKPGGGWNTGGSRYPGQGSPGGNRYPPQGGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGGWGQGGGSHGQWGK PSKPKTNIKHVAGAASAGAVVGGCLWF >Neovison vison (ferret) EF508270 MVKSHIGSWLLVLFVATWSDIGFCKKWPKPGGGWNTGGSRYPGQGSPGGNRYPPQGGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGGWGQGGGSHGQWGKPSKPKTNMKHVA >Canis familiaris (dog) genome MVKSHIGGWILLLFVATWSDVGLCKKRPKPGGWNTGGGSRYPGQGSPGGNRYPPQGGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGGWGQGGGSHSQWGK >Canis latrans (coyote) FJ232956 VKSHIGGWILLLFVATWSDVGLCKKRPKPGGWNTGGGSRYPGQGSPGGNRYPPQGGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGGWGQGGGSHGQWGKPNKPKTNMKHVAGAAAAGAVVGGLGGYMLGSAMSRPLIHFGNDYEDRYYRENMYRYPDQVYYRPVDQYSNQNNFVRDCVNITVKQHTVTTTTKGENFTETDMKIMERVVEQMCVTQYQKESEAYYQRGASAI >Ailuropoda melanoleuca (panda) AY327449 MVKSHIGSWILVLFVAMWSDVGLCKKRPKPGGGWNTGGSRYPGPGSPGGNRYPPQGGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGWGQPHGGGGWGQGGTHGQWNK PSKPKTNMKHVAGAAAAGAVVGGLGGYMLGSAMSRPLIHFGSDYEDRY Equus KPGG-WNTGG-SRYPGQGSPGGNRYP Erinaceus KPGG-WNSGG-SRYPGQGSSGSNRY Bos taurus KPGGGWNTGG-SRYPGQGSPGGNRY Lama pacos KPGGGWNTGG-SRYPGQGSPGGK Tursiops KPGGGWNTGG-SRYPGQGSPGGNRYP Myotis KPGGG-NTGG-SRYPGQGSPGGNR Pteropus KPGGGGSSGG-SRYPGQGSPGGNRY
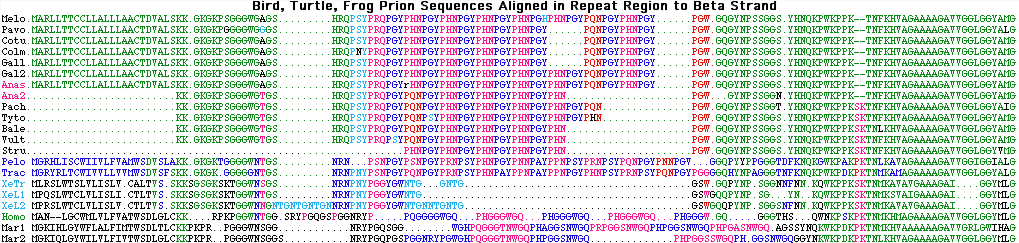
PRNP marsupial and platypus repeat region in transition
The Sarcophilus repeat region is of considerable interest -- the high GC content of this region makes it difficult to sequence and so provides a test of the 454 technology and Newbler assembler. This region consists in placentals a five octapeptide repeat, in marsupials and platypus a five nona- or decapeptide residue repeat that may resolve fine details of the marsupial phylogenetic tree, which in birds, lizards, turtles, frogs and fish is a hexapeptide repeat with trimeric internal substructure. Even though the single exon gene is clearly orthologous in all these species, the repeat regions within it are not directly comparable because they have expanded and contracted through replication slippage, plus experienced the odd repeat length change in marsupials and another in placentals.
The Sarcophilus prion gene has very high coverage that overcomes the occasional problem with frameshifts and allows the gene to be accurately tiled. However familiarity with the gene and reliable fiducial sequences are key to rapid assembly of the full length gene. No sequencing difficulties were observed in the high GC repeat region. The gene is very normal and has no indications whatsoever of abnormal numbers of repeats (4) or prion disease disposition.
Dasypus MVRSRVGCWLLLLFVATWSELGLC KK.RPKPGGGWNTGG SRYPGQ GSPGG NRYP PQGGG WGQ PHGGG WGQ PHGGG WGQ PHGGG WGQ PHGGG WGQ GGAHGQ Trichosurus MGKIQLGYWILVLFIVTWSDLGLC KKPKPRPGGGWNSGGS NRYPGQPGSPGG NRYPGWGH PQGGGTNWGQ PHPGGSNWGQ PHPGGSSWGQ PH GGSNWGQ GG YN Sarcophilus MGKIRLGYWILALFIVTWSDLGLC KKPKPRPGGGWNSGGS NRYPGQPGSAGG NRYPGWGH PQGGGTNWGQ PHPGGSSWGQ PHAGGSNWGQ PH.GGSNWGQ SGSSYNQ Monodelphis MGKIHLGYWFLALFIMTWSDLTLC KKPKPRPGGGWNSGG NRYPGQ SG GWGH PQGGGTNWGQ PHAGGSNWGQ PRPGGSNWGQ PHPGGSNWGQ PHPGGSNWGQ AGSSYNQ Macropus MAKIQLGYWILALFIVTWSELGLC KKPKTRPGGGWNSGGS NRYPGQPGSPGG NRYPGWGH PQGGGTNWGQ PHPGGSSWGQ PHAGGSNWGQ PH.GGSNWGQ GGGSYG Ornithorhynchus ------------------------ -------GGGWNSG NRYPGQPANPG GWGH PQGGGASWGH PQGGGASWGH PQGGGSNWGH PQGGGASWGH PQ GGGYS Dasypus WNKPSKPKTNM KHVAGAAAAGAVVG LGGYLVGSAMSRPLIHFGNDYEDRYYRENMYRYPNQVYYRSVEQYSSEKNFVHD CV MERVVEQMCITQYQ Trichosurus KWKPDKPKTNL KHVAGAAAAGAVVGGLGGYMLGSAMSRPVIHFGNEYEDRYYRENQYRYPNQVMYRPIDQYSSQNNFVHD CVNITVKQHTTTTTTKGENFTETDIKIMERVVEQMCITQYQN Sarcophilus KWKPDKPKTNM KHMAGAAAAGAVLGSLGGYVLGSAMSRPIMHFGNDYEDRYYRENQYRYPNQVMYRPIDQYSSQNNFVHD CVNITVKQHTTTTTTKGENFTETDIKIMERVVEQMCITQYQN Monodelphis KWKPDKPKTNM KHVAGAAAAGAVVGGLGGYMLGSAMSRPIMHFGNDYEDRYYRENQYRYPNQVMYRPIDQYNNQNNFVHD CVNITVKQHTTTTTTKGENFTETDIKIMERVVEQMCITQYQN Macropus KWKPDKPKTNL KHVAGAAAAGAVVGGLGGYMLGSAMSRPVMHFGNEYEDRYYRENQYRYPNQVMYRPIDQYGSQNSFVHD CVNITVKQHTTTTTTKGENFTETDIKIMERVVEQMCITQYQN Ornithorhynchus KYKPDKPKTGM KHVAGAAAAGAVVGGLGGYMIGSAMSRPPMHFGNEFEDRYYRENQNRYPNQVYYRPVDHFCSQDGFVRD CVNITVTQHTVTTT.EGKNLNETDVKIMTRVLEQMC
The signal region of Sarcophilus PRNP is expected to show the same length as the other 3 known marsupial sequences, which is confirmed by the sequence. Placentals exhibit a one residue deletion relative to this ancestral length.
MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Homo sapiens MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Pan troglodytes MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Gorilla gorilla MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Pongo pygmaeus MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Nomascus leucogenys MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Hylobates lar MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Symphalangus syndactylus MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca arctoides MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca fascicularis MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca fuscata MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca mulatta MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Macaca nemestrina MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Papio hamadryas MA--NLGCWMLFLFVATWSDLGLCKK--RPKPG Callithrix jacchus MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Cebus apella MA--NLGCWMLVVFVATWSDLGLCKK--RPKPG Cercopithecus aethiops MA--NLGCWMLVVFVATWSDLGLCKK--RPKPG Cercopithecus dianae MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Colobus guereza MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Presbytis francoisi MA--NLGCWMLVLFVATWSDLGLCKK--RPKPG Saimiri sciureus MA--KLGYWLLVLFVATWSDVGLCKK--RPKPG Tarsius syrichta MA--NLGCWMLVVFVATWSDVGLCKK--RPKPG Microcebus murinus MA--RLGCWMLVLFVATWSDIGLCKK--RPKPG Otolemur garnettii ME--NLGCWMLILFVATWSDIGLCKK--RPKPG Cynocephalus variegatus MA--QLGCWLMVLFVATWSDVGLCKK--RPKPG Tupaia belangeri MA--NLGYWLLALFVTMWTDVGLCKK--RPKPG Mus musculus MA--NLGYWLLALFVTTCTDVGLCKK--RPKPG Rattus norvegicus MA--NAGCWLLVLFVATWSDTGLCKK--RPKPG Cavia porcellus MA--NLGCWLLVLFVATWSDLGLCKK--RTKPG Dipodomys ordii MV--NPGCWLLVLFVATLSDVGLCKK--RPKPG Spermophilus tridecemlineatus MA--HLGYWMLLLFVATWSDVGLCKK--RPKPG Oryctolagus cuniculus MA--HLSYWLLVLFVAAWSDVGLCKK--RPKPG Ochotona princeps MVKSHIGSWILVLFVAMWSDVGLCKK--RPKPG Bos taurus MVKSHIGGWILVLFVAAWSDIGLCKK--RPKPG Sus scrofa MVKSHMGSWILVLFVVTWSDMGLCKK--RPKPG Vicugna vicugna MVKSHVGGWILVLFVATWSDVGLCKK--RPKPG Equus caballus MVRSHVGGWILVLFVATWSDVGLCKK--RPKPG Diceros bicornis MVKSLVGGWILLLFVATWSDVGLCKK--RPKPG Myotis lucifugus MVKNYIGGWILVLFVATWSDVGLCKK--RPKPG Pteropus vampyrus MVKSHIANWILVLFVATWSDMGFCKK--RPKPG Tursiops truncatus MVKSHIGGWILLLFVATWSDVGLCKK--RPKPG Canis lupus familiaris MVKSHIGSWILVLFVAMWSDVGLCKK--RPKPG Felis catus MVKSHIGSWLLVLFVATWSDIGFCKK--RPKPG Mustela putorius MVKSHIGSWLLVLFVATWSDIGFCKK--RPKPG Mustela vison MVKSHIGSWILVLFVAMWSDVGLCKK--RPKPG Ailuropoda melanoleuca MVKNHVGCWLLVLFVATWSEVGLCKK--RPKPG Erinaceus europaeus MVTGHLGCWLLVLFMATWSDVGLCKK--RPKPG Sorex araneus MVKSHLGCWIMVLFVATWSEVGLCKK--RPKPG Cyclopes didactylus MVRSRVGCWLLLLFVATWSELGLCKK--RPKPG Dasypus novemcinctus MVKGTVSCWLLVLVVAACSDMGLCKK--RPKPG Echinops telfairi MVKSSLGCWILVLFVATWSDMGLCKK--RPKPG Loxodonta africana MVKSSLGCWMLVLFVATWSDVGLCKK--RPKPG Procavia capensis MAKIQLGYWILALFIVTWSELGLCKKP-KTRPG Macropus eugenii MGKIHLGYWFLALFIMTWSDLTLCKKP-KPRPG Monodelphis domestica MGKIRLGYWILALFIVTWSDLGLCKKP-KPRPG Sacophilus harrisii MGKIQLGYWILVLFIVTWSDLGLCKKP-KPRPG Trichosurus vulpecular MARLLTTCCLLALLLAACTDVALSKKG-KGKPS Gallus gallus MAKLPGTSCLLLLLLLLGADLASCKKG-KGKPG Taeniopygia guttata MARLLTTCCLLALLLAACTDVALSKKG-KGKPG Meleagris gallopavo MGKHQMTCWLAIFLLLIQANVSLAKK--KPKPS Anolis carolinensis MRRFLVTCWIAVFLILLQTDVSLSKKG-KNKPG Gekko gekkko MGRYRLTCWIVVLLVVMWSDVSFSKKG-KGKGG Trachemys scripta (turtle) MGRHLISCWIIVLFVAMWSDVSLAKKG-KGKTG Pelodiscus sinensis (turtle) MPQSLWTCLVLISLICTLTVSSKKSGGGKSKTG Xenopus laevis MLRSLWTSLVLISLVCALTVSSKKSGSGKSKTG Xenopus topicalis
>PRNP_sacHar Sarcophilus harrisii (tasmanian_devil) single exon gene YVLG like Dasypus MGKIRLGYWILALFIVTWSDLGLCKKPKPRPGGGWNSGGSNRYPGQPGSAGGNRYPGWGHPQGGGTNWGQPHPGGSSWGQPHAGGSNWGQPHGGSNWGQ SGSSYNQKWKPDKPKTNMKHMAGAAAAGAVLGGVGGYVLGSAMSRPIMHFGNDYEDRYYRENQYRYPNQVMYRPIDQYSSQNNFVHDCVNITVKQHTTTTTT KGENFTETDIKIMERVVEQMCITQYQNEYRAAQYSYNMAFFSAPPVTLLLLGFLIFLIVS* >PRNP_mdo Monodelphis domestica opossum, from frameshifted genomic MGKIHLGYWFLALFIMTWSDLTLCKKPKPRPGGGWNSGGNRYPGQSGGWGHPQGGGTNWGQPHAGGSNWGQPRPGGSNWGQPHPGGSNWGQPHPGGSNWG QAGSSYNQKWKPDKPKTNMKHVAGAAAAGAVVGGLGGYMLGSAMSRPIMHFGNDYEDRYYRENQYRYPNQVMYRPIDQYNNQNNFVHDCVNITVKQHTTT TTTKGENFTETDIKIMERVVEQMCITQYQNEYRSAYSVAFFSAPPVTLLLLSFLIFLIVS* >PRNP_tvu Trichosurus vulpecular brushtail opossum MGKIQLGYWILVLFIVTWSDLGLCKKPKPRPGGGWNSGGSNRYPGQPGSPGGNRYPGWGHPQGGGTNWGQPHPGGSNWGQPHPGGSSWGQPHGGSNWGQGGY NKWKPDKPKTNLKHVAGAAAAGAVVGGLGGYMLGSAMSRPVIHFGNEYEDRYYRENQYRYPNQVMYRPIDQYSSQNNFVHDCVNITVKQHTTTTTTKGENFTETDIKIMERVVEQM CITQYQAEYEAAAQRAYNMAFFSAPPVTLLFLSFLIFLIVS* >PRNP_meu Macropus eugenii (tammar wallaby) MAKIQLGYWILALFIVTWSELGLCKKPKTRPGGGWNSGGSNRYPGQPGSPGGNRYPGWGHPQGGGTNWGQPHPGGSSWGQPHAGGSNWGQPHGGSNWGQ GGGSYGKWKPDKPKTNLKHVAGAAAAGAVVGGLGGYMLGSAMSRPVMHFGNEYEDRYYRENQYRYPNQVMYRPIDQYGSQNSFVHDCVNITVKQHTTTTTT KGENFTETDIKIMERVVEQMCITQYQNEYQAAQRYYNMAFFSAPPVTLLLLSFLIFLIVS* >PRNP_oan Ornithorhynchus anatinus platypus fragment PHWGKSPVHHWIIDICVVHLERRCRGHLHPNPCPGGRCVQQQPNRYPGQPATPGGWGHPQGGGASWGHPQGGGSNWGHPQGGGASWGHPQGGGYSKYKPDKPKTG MKHVAGAAAAGAVVGGLGGYMIGSAMSRPPMHFGNEFEDRYYRENQNRYSNQVYYRPVDQYGSQDGFVRDCVNITVTQHTVTTTEGKNLNETDVKIMTRVLEQMCVNLY