Opsins underground
Introduction
The genome article for the naked mole-rat (Heterocephalus glaber) appeared in Nature in October, 2011. That paper makes some remarkable observations about aging in this species, the longest-lived of all rodents/ The mole-rat is thus an appropriate model species for genomic adaptations to aging processes.
Because the mole-rat has a very unusual life history, in effect spending its entire life underground darkness, the question arises as to what has happened to opsins and other components of the visual repertoire. Such questions have been studied before in cave fish and crickets, with the outcome supporting "use it or lose it" -- genes are quickly lost to mutation as selective pressure for their maintainence ceases.
Thus like most placental mammals, mole-rat has the genetic potential for two-color cone vision plus dim-light rhodopsin, in addition to other opsins whose anatomical sites of expression (in the absence of mole-rat data) are likely the same as in mouse. The functions of these other opsins -- with the exception of rhabdomeric melanopsin -- are poorly understood but must be important because of their long history of conservation. The types and distribution of cone cells in the mole-rat retina has not yet been determined.
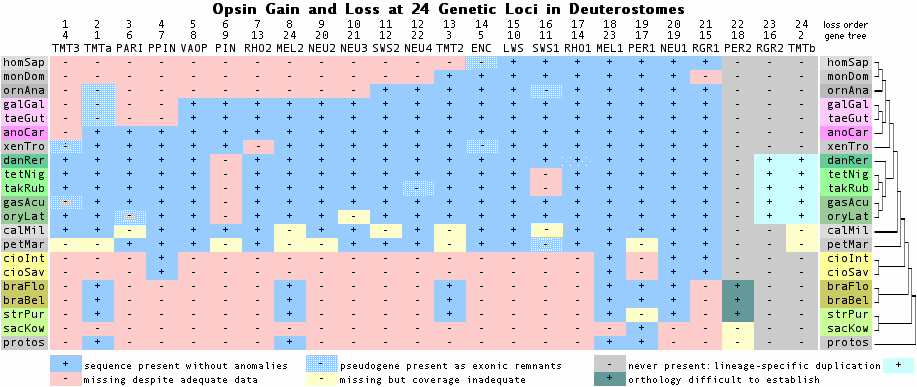
The genome of last common ancestor of living amniotes contained a remarkable 21 opsins (defined as full length paralogs to rhodopsin with covalently bound retinoid at lysine-296). However by the time of earliest mammals, 9 of these opsins had already been lost; an additional two were lost in placental mammals. In terms of rod and cone opsins, mammals originally had 4-color vision along with colored oil droplet wavelength filters like lizards, turtles and birds still do.
Degeneration of the mammalian retina was the subject of a 1942 book by Gordon Walls. Although opsin were not sequenced until 50 years later, he anticipated their loss from detailed anatomical considerations. Walls proposed that mammals experienced a multi-million year bottleneck of deep forest nocturnality (rather than cave, marine or subterranean lifestyle) during their evolution and attributed opsin loss to loss of darwinian selection. Cone opsin loss has been previously studied in marine mammals (dolphin SWS1 opsin pseudogenized) and fruit bats (again SWS1 loss).
The figure above -- derived from exhaustive opsin gene family annotation in all available genome projects -- shows that the immediate rodent ancestor of naked mole-rat (NMR) had eight phylogenetically expected opsins, as mouse does today. The Nature article addresses the opsin portfolio of mole-rat but constrained by article size limits and the many other genes deserving commentary.
According to the article,
"We further examined the molecular basis for poor visual function and small eyes in the NMR (naked mole-rat). Of the four vertebrate opsin genes (RHO, OPN1LW, OPN1MW and OPN1SW), two (OPN1LW and OPN1MW) were missing; this distinguishes the NMR from other rodents with dichromatic color vision, such as mice, rats and guinea pigs. However, the NMR has intact RHO (rhodopsin) and OPN4 (melanopsin), supporting the presence of rod-dominated retina and the capacity to distinguish light/dark cues."
"Of about 200 genes associated with visual perception (GO:0007601) in humans and mice, almost 10% were inactivated or missing. These mammalian genes participate in crystallin formation, phototransduction in the retina, retinal development, dark adaptation, night blindness and colour vision.... Inactivation of CRYBA4, a microphthalmia-related gene, may be associated with the small-sized eyes, whereas inactivation of CRYBA4 and CRYBB3 and a NMR-specific mutation in CRYGS may be associated with abnormal eye morphology. Thus, while some genes responsible for vision are preserved, poor visual function may be explained by deterioration of genes coding for various critical components of the visual system."
Gene Protein Description Inactivation event Time of gene loss OPN1MW MWS mid wavelength complete loss detected with synteny within NMR stem lineage OPN1LW LWS long wavelength complete loss detected with synteny within NMR stem lineage OPN1SW SWS1 short wavelength none retained to present RHO RHO1 dim rhodopsin none retained to present OPN3 ENC encephalopsin none retained to present OPN4 MEL1 melanopsin none retained to present RRH PER peropsin none retained to present OPN5 NEUR1 neuropsin none retained to present RGR RGR rgropsin none retained to present
The first two lines of the article summary table are reproduced above. The other lines are derived here using Blat of mouse opsin repertoire queries. Note the first line OPN1MW is an outright error as this gene is well-known to be a tandem duplication restricted to higher primates with no applicability to other mammals. These duplicates never existed in any ancestor of mole-rat and so was not lost in mole-rat. The genome assembly has good quality in this region; no tandem duplication of opsins specific to the mole-rat lineage occurs.
The second line is also in error. The terminal exon of mole-rat OPN1LW (LWS) is readily recovered by Blat of mouse query against mole-rat assembly. This gene fragment occurs in mole-rat in the expected syntenic location IRAK1 MECP2 OPN1LW TEX28 with conserved strand orientations. Because of assembly gaps, this exon could be either part of an intact gene or be all that is left after a large deletion, as discussed in depth below.
The article does not discuss OPN1SW, the short wavelength opsin lost in marine mammal and bat. This cone opsin is however present in mole-rat and little diverged from available rodent orthologs. This suggests functionality, apart from a six residue deletion in exon 4 associated with a frameshift in a run of eight thymidines TTTTTTTT that should be nine. This may represent inability of short read technology to precede past a compositionally simple region and allow read tiling rather than an actual deletion. No assembly gap is shown in the relevant region (JH170314:3551709-3551938).
The other six opsins expected in mole-rat are readily located and appear intact apart from a small number of missing exons. As in all genome assemblies, that of mole-rat has incomplete coverage. Here eight genes were expected and eight can be detected. However only 11 of the 50 expected exons are represented (78% exonic coverage). However if a deletion has taken out 5 exons of OPN1LW (LWS) then coverage is better -- 6 out of 50 missing of 88%.
This article will use standard protein acronyms when assigned gene names have little mneumonic value, do not follow HGNC rules for paralog nomenclature, and do not exist at all for ancestral opsins still present in other amniotes. The table above provides nomenclatural correspondences.
Does mole-rat have OPN1LW (LWS) or just a residual exon?
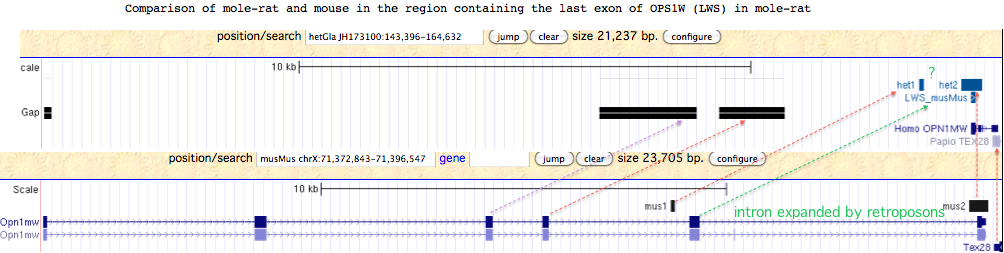
Since the only detectable OPN1LW exon of mole-rat lies in a region with several large assembly gaps corresponding to the missing exon placement in mouse genome, mole-rat may have a complete functional copy of OPN1LW. Alternatively, the observed genomic compression of this region relative to mouse suggests a large deletion occurred in mole-rat.
Although exons 3 and 4 of mole-rat OPN1LW may lie in assembly gaps, it is not clear why the penultimate exon 5 cannot be recovered by blastx from the region indicated by the green arrow in the figure below. Rapid lineage-specific gain and loss of retroposons can cause intronic and inter-genic regions to diverge very rapidly. Here, the sole correspondences of RepeatMasked sequence between mole-rat and mouse are indicted by the two small regions denoted het1/2 and mus1/2.
Conservation of the amino acid sequence of the sole exon present is high; only very recent pseudogenization is compatible with 6 seemingly mild substitutions and no internal stop codons or frameshifts:
OPN1LW LWS_hetGla FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA* last mole-rat exon aligned to mouse
F.NCIL.LFGKKV.DS.ELSS.SKTE.SSVSSVSPA* 6 differance out of 36 residues
OPN1LW LWS_musMus FRNCILHLFGKKVDDSSELSSTSKTEVSSVSSVSPA*
Sole exon of mole-rat OPN1LW (LWS) Difference alignment relative to other mammals Explanation of genus species codes LWS_hetGla FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA LWS_hetGla FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA Heterocephalus glaber (naked_molerat) LWS_musMus FRNCILHLFGKKVDDSSELSSTSKTEVSSVSSVSPA LWS_musMus .R....H......D..S....T....V......... Mus musculus (mouse) LWS_ratNor FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA LWS_ratNor .R...........D..S....T....V......... Rattus norvegicus (rat) LWS_nanEhr FQNCILQLFGKKVDDSSELSSTSKTEASSVSSVSPA LWS_nanEhr .Q...........D..S....T.............. Nannospalax ehrenbergi (mole_rat) LWS_speTri FRNCILQLFGKKVDDTSELSSASRTEASSVSSVSPA LWS_speTri .R...........D.TS......R............ Spermophilus tridecemlineatus (squirrel) LWS_cavPor FRNCILQLFGKKVEDSSELSSTSRTEASSVSSVSPA LWS_cavPor .R..............S....T.R............ Cavia porcellus (guinea_pig) LWS_oryCun FRNCILQLFGKKVEDSSELSSASRTEASSVSSVSPA LWS_oryCun .R..............S......R............ Oryctolagus cuniculus (rabbit) LWS_homSap FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_homSap .R...........D.GS.........V......... Homo sapiens (human) LWS_panTro FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_panTro .R...........D.GS.........V......... Pan troglodytes (chimp) LWS_gorGor FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_gorGor .R...........D.GS.........V......... Gorilla gorilla (gorilla) LWS_ponPyg FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_ponPyg .R...........D.GS.........V......... Pongo pygmaeus (orang_sumatran) LWS_nomLeu FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_nomLeu .R...........D.GS.........V......... Nomascus leucogenys (gibbon) LWS_macMul FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_macMul .R...........D.GS.........V......... Macaca mulatta (rhesus) LWS_macFas FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_macFas .R...........D.GS.........V......... Macaca fascicularis (macaque) LWS_papHam FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_papHam .R...........D.GS.........V......... Papio hamadras (baboon) LWS_calJac FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_calJac .R...........D.GS.........V......... Callithrix jacchus (marmoset) LWS_cebCap FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_cebCap .R...........D.GS.........V......... Cebus capucinus (monkey) LWS_ateJef FRNCILQLFGKKVDDGSELSSASKTEVSSVSSVSPA LWS_ateJef .R...........D.GS.........V......... Ateles geoffroyi (monkey) LWS_otoGar FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA LWS_otoGar .R...........D..S....T....V......... Otolemur garnettii (bushbaby) LWS_otoCra FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA LWS_otoCra .R...........D..S....T....V......... Otolemur crassicaudatus (bushbaby)full LWS_micMur FRNCILQLFGKKVDDGSELSSTSKTEVSSVSSVSPA LWS_micMur .R...........D.GS....T....V......... Microcebus murinus (mouse_lemur) LWS_tupBel FRNCILQLFGKKVDDSSELSSASRTEASSVSSVSPA LWS_tupBel .R...........D..S......R............ Tupaia belangeri (tree_shrew)full LWS_canFam FRNCILQLFGKKVDDSSELSSASRTEASSVSSVSPA LWS_canFam .R...........D..S......R............ Canis familiaris (dog) LWS_phoVit FRNCILQLFGKKVDDSSELSSASKTEASSVSSVSPA LWS_phoVit .R...........D..S................... Phoca vitulina (seal) LWS_felCat FRNCIMQLFGKKVDDGSELSSASRTEASSVSSVSPA LWS_felCat .R...M.......D.GS......R............ Felis catus (cat) LWS_bosTau FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_bosTau .R...........D..S....V.............. Bos taurus (cow) LWS_turTru FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_turTru .R...........D..S....V.............. Tursiops truncatus (dolphin) LWS_delDel FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_delDel .R...........D..S....V.............. Delphinus delphis (dolphin) LWS_gloMel FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVTPA LWS_gloMel .R...........D..S....V...........T.. Globicephala melas (pilot_whale) LWS_susScr FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_susScr .R...........D..S....V.............. Sus scrofa (pig) LWS_equCab FRNCILQLFGKKVDDSSELSSVSKTEASSVSSVSPA LWS_equCab .R...........D..S....V.............. Equus caballus (horse) LWS_myoLuc FRNCILQLFGKRVDDSSELSSTSRTEASSVSSVSPA LWS_myoLuc .R.........R.D..S....T.R............ Myotis lucifugus (brown LWS_pteVam FRNCILQLFGKKVDDSSELSSSSKTEASSVSSVSPA LWS_pteVam .R...........D..S....S.............. Pteropus vampyrus (flying_fox) LWS_loxAfr FRNCILQLFGKKVDDSSELSSASKTEASSVSSVSPA LWS_loxAfr .R...........D..S................... Loxodonta africana (elephant) LWS_triMan FRNCILQLFGKKVDDGSELSSASKTEASSVSSVSPA LWS_triMan .R...........D.GS................... Trichechus manatus (manatee) LWS_echTel FRNCILQLFGKKVDDSSELSSTSRTEASSVSSVSPA LWS_echTel .R...........D..S....T.R............ Echinops telfairi (tenrec) LWS_proCap FRNCILQLFGKKVDDSSELSSTSKMEASSASSVSPA LWS_proCap .R...........D..S....T..M....A...... Procavia capensis (hyrax) LWS_monDom FRTCILQLFGKKVDDGSEVSSTSKTEGSSVSSVAPA LWS_monDom .RT..........D.GS.V..T....G......A.. Monodelphis domesticus (opossum) LWS_didAur FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA LWS_didAur .RT..........D.GS.V..T....V......A.. Didelphis aurita (big-eared LWS_tarRos FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_tarRos .RT..........D.GS.V..T.R..V......A.. Tarsipes rostratus (honey LWS_macEug FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_macEug .RT..........D.GS.V..T.R..V......A.. Macropus eugenii (wallaby) LWS_setBra FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVPA- LWS_setBra .RT..........D.GS.V..T.R..V......PA- Setonix brachyurus (quokka) LWS_cerCon FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_cerCon .RT..........D.GS.V..T.R..V......A.. Cercartetus concinnus (pygmy LWS_smiCra FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_smiCra .RT..........D.GS.V..T.R..V......A.. Sminthopsis crassicaudata (dunnart) LWS_myrFas FRTCILQLFGKKVDDGSEVSSTSRTEVSSVSSVAPA LWS_myrFas .RT..........D.GS.V..T.R..V......A.. Myrmecobius fasciatus (numbat) LWS_isoObe FRTCILQLFGKKVDDGSEVSGTSRTEVSSVSSVAPA LWS_isoObe .RT..........D.GS.V.GT.R..V......A.. Isoodon obesulus (bandicoot) LWS_ornAna FRNCIMQLFGKKVDDGSELSSTSRTEVSSVSSVSPA LWS_ornAna .R...M.......D.GS....T.R..V......... Ornithorhynchus anatinus (platypus) LWS_tacAcu FRTCILQLFGKKVDDGSEVSSTSKTEVSSVSSVAPA LWS_tacAcu .RT..........D.GS.V..T....V......A.. Tachyglossus aculeatus (echidna)
Rodent opsin reference sequences
The comparative genomics of opsins have been studied to exhaustion: no variation in exon number or intron location or phasing occurs in any terrestrial vertebrate.)
The sequences below have one exon per row. The numbers 0 1 2 indicate the observed codon overhang (reading frame). These are deeply conserved within eumetazoans; mole-rat presents no new issues. Processed pseudogenes are absent, consistent with no gene expression of opsins in terminal germline cell lineages.
>ENC_hetGla Heterocephalus glaber (naked molerat) OPN3 88% identical musMus, missing first exon 0 1 2 GIVSITTLTVLAYERYIRVVHARVINFSWAWKAITYIWLYSLAWAGAPLLGWNRYILDVHGLSCTVDWKSKDANDSSFVLFLFLGCLVVPMGVIAHCYGHILYSIRM 0 0 FSMLQLRCVEDLQTIQVMKILRYEKKVAKMCFLMVFIFLVCWMPYIVICFLVVNGYGHRVTPTVSVVSYLFAKSSTVYNPVIYTIMIRK 0 0 FRRSLLQLLCFRLLRCQQPAKDLPAVESEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDRTSGSKVDTIQVRPL* 0 >LWS_hetGla Heterocephalus glaber (naked molerat) OPN1LW syn(-IRAK1 -MECP2 -TEX28) 88% identical musMus, missing exons 1-5 because of assembly gaps 0 1 2 1 2 2 1 2 0 0 0 FPNCILQLFGKKVEDSLELSSASKTEASSVSSVSPA* 0 >SWS1_hetGla Heterocephalus glaber (naked molerat) OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC) 89% identical musMus, frameshifting deletion 6aa exon 4 0 MSGEEDFYLFKNTSSVGPWDGSQYHIAPIWAFHLQAAFMGLVFFVGTPLNAIVLVATLQYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFFFGRHVCALEAFLGSVA 1 2 GLVTGWSLAFLAFERYLVICKPFGNFRFSSKHALIVVLATWVIGIGVSIPPFFGWSR 2 1 YIPEGLQCSCGPDWYTVGTKYRSEYYSWFLFIFCFIVPLSLICFSYSQLLRTLRA 0 0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAALAMYIVNNRNHGLDLRLFTIPAFFSPKSCVYNPIIYCFMNKQ 0 0 FRACIMELVCRKSMADESDMSSSQKTEVSAVSSSKVGPN* 0 >RHO1_hetGla Heterocephalus glaber (naked molerat) RHO syn(-MBD4 +IFT122 +H1FOO -PLXND1) MNGTEGPNFYVPFSNVTGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIVIFFCYGQLVFTVKE AAAQQQESATTQKAEKEVTRMVIIMVIFFLICWLPYASVAFYIFTHQGSNFGPIFMTLPAFFAKSSSIYNPVIYIMLNKQ FRNCMLTTLCCGKNPLGDDDASATASKTETSQVAPA >MEL1_hetGla Heterocephalus glaber (naked molerat) OPN4 86% identical musMus, missing exons 1,8,9 0 0 0 AAGTQAAVRVPFSTADVPAHVHYTLGAVILLVGLTGMLGNLTVIYTFFR 2 1 RGLRTPANMFVINLAVSDFLMSFTQAPVFFTSSLYKRWLFGEA 2 2 GCEFYAFCGALFGITSMITLTAITLDRYLVITRPLATIGLASKRQAALVLLGIWLYALAWSLPPFFGW 1 2 sAYVPEGLLTSCSWDYMTFTPSVRSYTMLLFCFVFFLPLLIIIYCYVFIFRAIRETGR 2 1 AFESCGESPSRRRQWQRLRSEWKMAKIALLVILLFVLSWAPYSTVALVAFAG 2 1 HAHILTPSMSSVPAVIAKASAIHNPIIYAIAHPKYR 2 1 MAIVQHLPCLGLLLGVSGQRSLPSLSYRSTHRSRLSSQASDLSWISGRRRQESLGSESEV 0 0 0 0 * 0 >PER_hetGla Heterocephalus glaber (naked molerat) RRH syn(-CFI +NOLA1 +EGF -ELOVL6) 79% identical musMus, missing exons 1,7 0 1 2 GLISILSNIIVLGIFVKYKELRTPTNAIIMNLALTDIGVSSIGYPMSAASYLHGSWKFGYPGCQ 0 0 VYAGLNIFFGMASIGLLTVIAVDRYLTICRPDI 1 2 GRRLTSTSYVSMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDt 2 1 SFVSYTMTVIAVNFTGPLAVMFYCYFHVTWSIKHHATGNCPTFPNRDWSDQVDVTK 0 0 MSVVMILMFLVAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 * 0 >NEUR1_hetGla Heterocephalus glaber (naked molerat) OPN5 94% identical musMus 0 MALNHTAPPQDERLPHYLQDEDPFVSKLSWEADLVAGFYLTII 1 2 GILSTCGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1 2 VVGKPFTIISCFRHRWVFGWIGCRWYGWAGFFFGCGSLTTMTVVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAAIWAYVSFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASARGQVFILNILFFCLLLPMATIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQNGGLKATKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEV* 0 >RGR_hetGla Heterocephalus glaber (naked molerat) RGR syn(+PCDH21 -LRIT1 -GRID1 -WAPAL) 74% identical musMus, missing exons 1 0 1 2 ALCGLSLNGLTIVSFCKIPELWTPRHLLVLSLALADSGLSLNTLIAAVSSLLR 2 1 RWPYGSGGCQTHGFQGFVTALASISGSAAIAWGHHQHYCT 1 2 GSQLAWSTAIRLVLFVWLSSAFWAALPLLGWGHYDYEPLGTCCTLDYSKRDR 2 1 NSTSFLFTMGFFSFLIPLFITFTSYQLMEQKLARSSRLQ 0 0 VNTSLPARTLMLCWGPYVFLHLYAVIADASSLSPKLQM 0 0 VPALIAKTVPTINAINYALGSKMGSRRPWQCLSLQRSKRD* 0 >ENC_musMus Mus musculus (mouse) OPN3 encephalopsin panopsin 2aa del 0 MYSGNRSGDQGYWEDGAGAEGAAPAGTRSPAPLFSPTAYERLALLLGCLALLGVGGNLLVLLLYSKFPRLRTPTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1 2 GFVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWRSKDANDSSFVLFLFLGCLVVPVGIIAHCYGHILYSVRM 0 0 LRCVEDLQTIQVIKMLRYEKKVAKMCFLMAFVFLTCWMPYIVTRFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMNRK 0 0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMHIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVEDSDRSSASKVDVIQVRPL* 0 >LWS_nanEhr Nannospalax ehrenbergi (mole_rat) OPN1LW syn(-IRAK1 -MECP2 -TEX28) 0 MAQQWAPQRLAGGQTQDSYEDSTQASLFTYTNSNSTR 1 2 GPFEGPNYHIAPRWVYHLTTTWMILVVVASIFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYTVSLC 1 2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLATMGITFAWVWAAVWTAPPVFGWSR 2 1 YWPYGLKTSCGPDVFSGTSYPGVQSYMVVLMITCCIIPLSIILLCYLQVWLAIRT 2 0 VAKQQKESESTQKAEKEVTHMVVVMVFAYCLCWGPYTFFVCFATAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0 0 FQNCILQLFGKKVDDSSELSSTSKTEASSVSSVSPA* 0 >SWS1_musMus Mus musculus (mouse) OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC) 0 MSGEDDFYLFQNISSVGPWDGPQYHLAPVWAFRLQAAFMGFVFFVGTPLNAIVLVATLHYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFLFGRHVCALEAFLGSVA 1 2 GLVTGWSLAFLAFERYVVICKPFGSIRFNSKHALMVVLATWIIGIGVSIPPFFGWSR 2 1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIIPLSLICFSYSQLLRTLRA 0 0 VAAQQQESATTQKAEREVSHMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0 0 FRACILEMVCRKPMADESDVSGSQKTEVSTVSSSKVGPH* 0 >RHO1_musMus Mus musculus (mouse) RHO syn(-MBD4 +IFT122 +H1FOO -PLXND1) 0 MNGTEGPNFYVPFSNVTGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1 2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR 2 1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIVIFFCYGQLVFTVKE 0 0 AAAQQQESATTQKAEKEVTRMVIIMVIFFLICWLPYASVAFYIFTHQGSNFGPIFMTLPAFFAKSSSIYNPVIYIMLNKQ 0 0 FRNCMLTTLCCGKNPLGDDDASATASKTETSQVAPA* 0 >MEL1_nanEhr Nannospalax ehrenbergi (molerat) OPN4 0 MNSPSGPRVPPGLAQKPSFMVTPVLPNQWISFQKNVSVGIQLPPASAT 0 0 ATGAQAASWVPFPTVDVPVHAHYTLGTVILLVGLTGMLGNLIVIYTFCR 2 1 SRGLRTRANMFTVNLAVSDFLMSFTQAPVFFASSLYKRWLFGEA 2 2 GCEFYAFCGAVSGITSMTTLTAIALDRYLVITRPLATIGVASKRRTALVLLGVWLYALAWSLPPFFGW 1 2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFLPLLIIIFCYIFIFKAIRETGR 2 1 ACEGCGESPQRRRQWQRLQNEWKMAKVALLVIFLFVLSWAPYSTVALVAFAG 2 1 YSHILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2 1 LAISQHLPCLGVLIGVSSQRSHPSLSYRSTHRSTLSSQASDLSWISGRKRQESLGSESEV 0 0 GWTDTEVTAAWGVAQEASGWSPYRHSLEDGEVKASPSPQGQEAKTSR 0 0 TKGQLPSLNLRM* 0 >PER_musMus Mus musculus (mouse) RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6) 0 MLSEASDNSSGSRSEGSVFSRTEHSVIAAYLIVA 1 2 GITSILSNVVVLGIFIKYKELRTPTNAVIINLAFTDIGVSSIGYPMSAASDLHGSWKFGHAGCQ 0 0 IYAGLNIFFGMVSIGLLTVVAMDRYLTISCPDV 1 2 GRRMTTNTYLSMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRNNDT 2 1 SFVSYTMMVIVVNFIVPLTVMFYCYYHVSRSLRLYAASDCTAHLHRDWADQADVTK 0 0 MSVIMILMFLLAWSPYSIVCLWACFGNPKKIPPSMAIIAPLFAKSSTFYNPCIYVAAHKK 2 1 FRKAMLAMFKCQPHLAVPEPSTLPMDMPQSSLAPVRI* 0 >NEUR1_musMus Mus musculus (mouse) OPN5 neuropsin 0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1 2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1 2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1 2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0 0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQAGGLRGTKKKSLEDFR 2 1 LHTVTTVRKSSAVLEIHQEV* 0 >RGR_musMus Mus musculus (mouse) RGR syn(+PCDH21 -LRIT1 -GRID1 -WAPAL) 0 MAATRALPAGLGELEVLAVGTVLLME 1 2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2 1 RWPHGSEGCQVHGFQGFATALASICGSAAVAWGRYHHYCT 1 2 GRQLAWDTAIPLVLFVWMSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2 1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKFSRSGHLP 0 0 VNTTLPGRMLLLGWGPYALLYLYAAIADVSFISPKLQM 0 0 VPALIAKTMPTINAINYALHREMVCRGTWQCLSPQKSKKDRTQ* 0
Glires reference sequence sets
>ENC_musMus Mus musculus (mouse) OPN3 encephalopsin panopsin 2aa del
0 MYSGNRSGDQGYWEDGAGAEGAAPAGTRSPAPLFSPTAYERLALLLGCLALLGVGGNLLVLLLYSKFPRLRTPTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1
2 GFVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWRSKDANDSSFVLFLFLGCLVVPVGIIAHCYGHILYSVRM 0
0 LRCVEDLQTIQVIKMLRYEKKVAKMCFLMAFVFLTCWMPYIVTRFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMNRK 0
0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMHIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVEDSDRSSASKVDVIQVRPL* 0
>ENC_ratNor Rattus norvegicus (rat) XP_573517 2aa del
0 MYSGNRSGGQGYWEDGAGAEGAAPAGTRSPAPLFSPTAYERLALLLGCLALLGVGGNLLVLLLYSKFPRLRTPTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1
2 GFVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPMGIIAHCYGHILYSVRM 0
0 LRCVEDLQTIQVIKMLRYEKKVAKMCFLMAFVFLTCWMPYVVTRFLVVNGYGHLVTPTVSIVSYLFAKSSTVYNPVIYIFMIRK 0
0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVEDSDRSSASKVDVIQVRPL* 0
>ENC_speTri Spermophilus tridecemlineatus (squirrel)
0 MYSGNRSGSQGSWEGDGSAGAEGSAPEGTLSPTPLFSPGTNERLALLFRSVGLLGAGSNLLVLVLYYKFQGSAHPLTFFLVNISLGDLLMSLFGVTFTFVSCLRNRWVWDTVACVWDGFSSSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDVHGLGCTVEWKSKDANDSSFVLFLFLGCLVVPVGVIAHCYGHILYSIRM 0
0 LRCVEDLQIFQVIKILRYEKKLAKMCFVMVFTFLICWMPYIVVCFLVANGYGQRVTPTVSIVSNLFAKSSTVYNPVIYIFMIRK 0
0 FRRSLLQLLCSRLLRCQQPAKDLPAVGNEMQIRPIVISQKDGERPKKKVTFNSSSIVFIITSDESLSVDDSNRTSGSKADVIQVRPL* 0
>ENC_criGri Cricetulus griseus (hamster) XP_003499904=pathetic
0 PTHLFLVNLSLGDLLVSLFGVTFTFASCLRNGWVWDAVGCAWDGFSGSLF 1
2 GFVSITTLTVLAYERYIRVVHSRVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWKSKDANDSSFVLFLFLGCLVVPLGIITHCYGHILYSVRM 0
0 LRCVEDLQTIQVIKILRCEKKVAKMSFAMVFVFLTCWMPYIVTRFLVVNGYGHLVTPTVSIVSSLFAKSSTVYNPIIYIFMIRK 0
0 FRRSLLQLLCFRLLRCQRPAKNLPAAESEMQIRPIVMSQKDRDRPKKKVTFNSSSIIFIITSDESLSVDDSDRTNASKANVIQVRPL
>ENC_dipOrd Dipodomys ordii (kangaroo_rat)
0 MYSGNRSGGQEYWEDGGAAGSEGPAPAGTLSPAPLFSAGAYERLALLLGSAGLLGVGNNLLVLVLYYKFQRLRTPTHLLLVNISLSDLLVSLFGVTFTFVSCLRNGWVWDTVGCVWDGFSRSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFTWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWKAKDANDSSFVLFLFIGCLVVPVGIIAHCYGHILYSIRM 0
0 LRCVEDLQTIQIIKILQYEKKLAKMCFLMALTFLMCWMPYIVTCFLVVNSHGHLVTPTISIVSHLLAKSSTIYNPVIYIFMIRK 0
0 FRRSLLQLLCFRLLRCQRPAKDLPAAGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSVRSSGSKADVIQVRPL* 0
>ENC_cavPor Cavia porcellus (guinea pig) 3 aa del
0 MYSGNRSSGQGYWEGGGPEDPAPAGTLSPAPLFSPGAYERLALLLGSLGLLGVGNNLLVLVLYYKFQRLRSPTHLFLANISLSDLLGSLFGVTFTFVSCLKNGWVWDAVGCVWDGFSRSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLSCTVDWKSKDANDSSFVLFLFLGCLVVPVGVIVHCYGHILYSIRM 0
0 LRGVEDLQTIQVMKILRSENKVAIMCFLMVFIFLVCWMPYIVICFLLVNGYRHRVTPTVSIVSYLFTKSSTVYNPVIYVLMIRK 0
0 FRRSLLQLHCLRLLRCQQPAKDLPAVEREMHIRPIVMSQKDGDRPKKKVTFNSSSIIFIITSDESLSVDDSDRTSGSKVDTIQVRPL* 0
>ENC_oryCun Oryctolagus cuniculus (rabbit)
0 MYSGNRSGEQGYWEGGGAAGAEGPGPAGTLSPAPLFSPSTYERLALLLGSIGLLGVGSNLLVLVLYYKFQRLRTPTLLFLVNISLSDLLVSVFGVTFTFVSCLRNGWVWDTVGCVWDGFSSSLF 1
2 GIVSITTLTVLAYERYIRVVHARVINFSWAWRAITYIWLYSLAWAGAPLLGWNRYILDIHGLGCTVDWKSKNANDSSFVLFLFLGCLVVPVGVIAHCYGHILYSVRM 0
0 LRCVEDLQTIQVIKILRYEKKVAKMCFFMVFTFLICWMPYVVICFLVVNGYGHLVTPTLSIVSYLFCKSSTAYNPIIYIFMIRK 0
0 FRRSLLQLLCFQPLRCQQPPKDLPTVGSEMQIRPIVMSQKDGDRPKKKVTFNSSSIIFIIASDESLAVDDNEKASGPKVDVIQVRPL* 0+
>LWS_musMus Mus musculus (mouse) Opn1mw = OPN1LW syn(-IRAK1 -MECP2 -TEX28)
0 MAQRLTGEQTLDHYEDSTHASIFTYTNSNSTK 1
2 GPFEGPNYHIAPRWVYHLTSTWMILVVVASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYIVSLC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLATVGIVFSWVWAAIWTAPPIFGWSR 0
0 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIFPLSIIVLCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATAHPGYAFHPLVASLPSYFAKSATIYNPIIYVFMNRQ 0
0 FRNCILHLFGKKVDDSSELSSTSKTEVSSVSSVSPA* 0
>LWS_ratNor Rattus norvegicus (rat)
0 MAQQLTGEQTLDHYEDSTQASIFTYTNSNSTR 1
2 GPFEGPNYHIAPRWVYHLTSTWMILVVIASVFTNGLVLAATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYIVSLC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLATVGIVFSWVWAAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIFPLSIIVLCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATAHPGYAFHPLVASLPSYFAKSATIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVDDSSELSSTSKTEVSSVSSVSPA* 0
>LWS_nanEhr Nannospalax ehrenbergi (mole_rat)
0 MAQQWAPQRLAGGQTQDSYEDSTQASLFTYTNSNSTR 1
2 GPFEGPNYHIAPRWVYHLTTTWMILVVVASIFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETIIASTISVVNQIYGYFVLGHPLCVIEGYTVSLC 1
2 GITGLWSLAIISWERWMVVCKPFGNVRFDAKLATMGITFAWVWAAVWTAPPVFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGVQSYMVVLMITCCIIPLSIILLCYLQVWLAIRT 2
0 VAKQQKESESTQKAEKEVTHMVVVMVFAYCLCWGPYTFFVCFATAHPGYAFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0
0 FQNCILQLFGKKVDDSSELSSTSKTEASSVSSVSPA* 0
>LWS_speTri Spermophilus tridecemlineatus (squirrel) Ictidomys
0 MAQRWDPQRLAGGQPQDSHEDSTQSSIFTYTNSNATR 1
2 GPFEGPNYHIAPRWVYHITSIWMIIVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLAETVIASTISVVNQFFGYFVLGHPLCVVEGYTVSVC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIAFAWTWSAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGNSYPGVQSYMMVLMVTCCIIPLSIIILCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYASFACFATANPGYAFHPLVAAVPAYFAKSATIYNPVIYVFMNRQ 0
0 FRNCILQLFGKKVDDTSELSSASRTEASSVSSVSPA* 0
>LWS_sciCar Sciurus carolinensis (squirrel) frag
0 mAQRWDPQRLAGGQPQDSHEDSTQSSIFTYTNSNATR 1
2 GPFEGPNYHIAPRWVYHITSTWMIIVVIASVFTNGLVLVATMKFKKLRHPLNWILVNLAIADLAETVIASTISVVNQLYGYFVLGHPLCVVEGYTVSVC 1
2 GITGLWSLAIISWERWLVVCKPFGNMRFDAKLAIVGIAFSWIWSAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGMQSYMMVLMVTCCIIPLSIIILCYLQVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATANPGYAFHPLVAALPAYFAKSATIYNPIyvfmnrq 0
0 * 0
>LWS_cavPor Cavia porcellus (guinea_pig)
0 MAQRWGPHALSGVQAQDAYEDSTQASLFTYTNSNNTR 1
2 GPFEGPNYHIAPRWVYHLTSAWMTIVVIASIFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETVIASTISVVNQVYGYFVLGHPLCVVEGYTVSLC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIVGIVFSWVWSAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCITPLSIIVLCYLHVWLAIRA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVLAYCLCWGPYAFFACFATANPGYSFHPLVAALPAYFAKSATIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVEDSSELSSTSRTEASSVSSVSPA* 0
>LWS_oryCun Oryctolagus cuniculus (rabbit)
0 MTQPWGPQMLAGGQPPESHEDSTQASIFTYTNSNSTR 1
2 GPFEGPNFHIAPRWVYHLTSAWMILVVIASVFTNGLVLVATMRFKKLRHPLNWILVNLAVADLAETVIASTISVVNQFYGYFVLGHPLCVVEGYTVSLC 1
2 GITGLWSLAIISWERWLVVCKPFGNVRFDAKLAIAGIAFSWIWAAVWTAPPIFGWSR 2
1 YWPYGLKTSCGPDVFSGTSYPGVQSYMMVLMVTCCIIPLSVIVLCYLQVWMAIPA 0
0 VAKQQKESESTQKAEKEVTRMVVVMVFAYCLCWGPYTFFACFATAHPGYSFHPLVAAIPSYFAKSATIYNPIIYVFMNRQ 0
0 FRNCILQLFGKKVEDSSELSSASRTEASSVSSVSPA* 0
>SWS1_musMus Mus musculus (mouse) OPN1SW syn(-FAM137A +SWS1 -CALU -NAG6 -FLNC)
0 MSGEDDFYLFQNISSVGPWDGPQYHLAPVWAFRLQAAFMGFVFFVGTPLNAIVLVATLHYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFLFGRHVCALEAFLGSVA 1
2 GLVTGWSLAFLAFERYVVICKPFGSIRFNSKHALMVVLATWIIGIGVSIPPFFGWSR 2
1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFIFCFIIPLSLICFSYSQLLRTLRA 0
0 VAAQQQESATTQKAEREVSHMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0
0 FRACILEMVCRKPMADESDVSGSQKTEVSTVSSSKVGPH* 0
>SWS1_ratNor Rattus norvegicus (rat)
0 MSGEDEFYLFQNISSVGPWDGPQYHIAPVWAFHLQAAFMGFVFFAGTPLNATVLVATLHYKKLRQPLNYILVNVSLGGFLFCIFSVFTVFIASCHGYFLFGRHVCALEAFLGSVA 1
2 GLVTGWSLAFLAFERYLVICKPFGNIRFNSKHALTVVLITWTIGIGVSIPPFFGWSR 2
1 FIPEGLQCSCGPDWYTVGTKYRSEHYTWFLFIFCFIIPLSLICFSYFQLLRTLRA 0
0 VAAQQQESATTQKAEREVSHMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0
0 FRACILEMVCRKPMTDESDMSGSQKTEVSTVSSSKVGPH
>SWS1_criGri Cricetulus griseus (hamster) XP_003497566
0 MSGEDEFYLFKNISSVGPWDGPQYHIAPVWAFHLQAAFMGFVFLAGTPLNATVLVVTLRYKKLRQPLNYILVNVSLGGFLFCSFSVFTVFIASCHGYFLFGRHVCALEAFLGSVA 1
2 GLVTGWSLAFLAFERYIVICKPFGNIRFSSKHALMVVLVTWTIGIGVSIPPFFGWSR 2
1 FIPEGLQCSCGPDWYTVGTKYRSEYYTWFLFLFCFIVPLSLICFSYSQLLRTLRA 0
0 VAAQQQESATTQKAEREVSHMVVVMVGSFCLCYVPYAALAMYMVNNRNHGLDLRLVTIPAFFSKSSCVYNPIIYCFMNKQ 0
0 FRACIMEMVCRKPVTDESDMSGSQKTEASTVSSSKVGPH* 0
>SWS1_cavPor Cavia porcellus (guinea_pig)
0 MSEEEDFYLFKNASSVGPWDGPQYHVAPVWAFRLQAAFMGIVFCIGTPLNGIVLVATLLYKKLRQPLNYILVNVSLGGFLVCIFS
MVTGWSLAFLAFERYLVICKPFGNFRFSSKHALIVVLATWVIGIGVSIPPFFGWSR 2
1 YMPEGLQCSCGPDWYTMGTKYRSEYFAWFLFIFCFIVPLSLICFSYCQLLRTLRT 0
0 VAAQQQESATTQKAEREVSRMVVVMVGSFCVCYVPYAALAMYIVNNRNHGLDLRLVTIPAFFSKSSCIYNPIIYCFMNKQ 0
0 FRACIMELVCRKPMADESDMSTSQKTEVSAVSSSKVGPN*
>RHO1_musMus Mus musculus (mouse) RHO syn(-MBD4 +IFT122 +H1FOO -PLXND1)
0 MNGTEGPNFYVPFSNVTGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIVIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIFFLICWLPYASVAFYIFTHQGSNFGPIFMTLPAFFAKSSSIYNPVIYIMLNKQ 0
0 FRNCMLTTLCCGKNPLGDDDASATASKTETSQVAPA* 0
>RHO1_ratNor Rattus norvegicus (rat)
0 MNGTEGPNFYVPFSNITGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIGLWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIVIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIFFLICWLPYASVAMYIFTHQGSNFGPIFMTLPAFFAKTASIYNPIIYIMMNKQ 0
0 FRNCMLTTLCCGKNPLGDDEASATASKTETSQVAPA* 0
>RHO1_cavPor Cavia porcellus (guinea_pig)
0 MNGTEGENFYIPFSNATGVVRSPFEYPQYYLAEPWQFSILAAYMFMLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVANLFMVLGGFTTTLYTSMNGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR 2
1 YIPEGMQCSCGVDYYTLKSEVNHESFVIYMFVVYFTIPMIIIFFCYEQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAAYIFTHQGSNFGPIFMTVPAFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEASTTVSKTETSQVAPA* 0
>RHO1_nanEhr Nannospalax ehrenbergi (mole-rat) AAG25707
MNGTEGPNFYVPFSNGTGVVRSPFEQPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLALADLFMVIGGFTTTFYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAITGVAFTWVMALACAVPPLVGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAMYIFTHQGSNFGPIFMTLPAFFAKSASIYNPVIYIMMNKQ 0
0 FRNCMLTTLCCGKNLLGDEEASPTASKTETSQVAPA* 0
>RHO1_criGri Cricetulus griseus (hamster) EGW12630
MNGTEGPNFYVPFSNATGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVVFTWIMALACAAPPLVGWSR 2
1 YIPEGMQCSCGVDYYTLKPEVNNESFVIYMFVVHFTIPLIVIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVILMVVFFLICWVPYAGVAFYIFTHQGSNFGPIFMTLPAFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTLCCGKNILGDDEASATASKTETSQVAPA* 0
>RHO1_oryCun Oryctolagus cuniculus (rabbit)
MNGTEGPDFYIPMSNQTGVVRSPFEYPQYYLAEPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTTTLYTSLHGYFVFGPTGCNVEGFFATLG 1
2 GEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWIMALACAAPPLVGWSR 2
1 YIPEGMQCSCGIDYYTLKPEVNNESFVIYMFVVHFTIPLIIIFFCYGQLVFTVKE 0
0 AAAQQQESATTQKAEKEVTRMVIIMVIAFLICWVPYASVAFYIFTHQGSNFGPIFMTIPAFFAKSSSIYNPVIYIMMNKQ 0
0 FRNCMLTTICCGKNPLGDDEASATASKTETSQVAPA* 0
>MEL1_musMus Mus musculus (mouse) OPN4 melanopsin
0 MDSPSGPRVLSSLTQDPSFTTSPALQGIWNGTQNVSVRAQLLSVSPT 0
0 TSAHQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 NRGLRTPANMFIINLAVSDFLMSVTQAPVFFASSLYKKWLFGET 1
2 GCEFYAFCGAVFGITSMITLTAIAMDRYLVITRPLATIGRGSKRRTALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPQVRAYTMLLFCFVFFLPLLIIIFCYIFIFRAIRETGR 2
1 ACEGCGESPLRQRRQWQRLQSEWKMAKVALIVILLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 VAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGRKRQESLGSESEV 0
0 GWTDTETTAAWGAAQQASGQSFCSQNLEDGELKASSSPQVQRSKTPK 0
0 VPGPSTCRPMKGQGARPSSLRGDQKGRLAVCTGLSECPHPHTSQFPLAFLEDDVTLRHL* 0
>MEL1_ratNor Rattus norvegicus (rat) AY072689
0 MNSPSESRVPSSLTQDPSFTASPALLQGIWNSTQNISVRVQLLSVSPT 0
0 TPGLQAAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 NRGLRTPANMLIINLAVSDFLMSFTQAPVFFASSLYKKWLFGET 1
2 GCKFYAFCGAVFGIVSMITLTAIAMDRYLVITRPLATIGMRSKRRTALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYVTFTPLVRAYTMLLFCFVFFLPLLIIIFCYIFIFRAIRETGR 2
1 ACEGCGESPLRRRQWQRLQSEWKMAKVALIVILLFVLSWAPYSTVALVGFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIIYAITHPKYR 2
1 AAIAQHLPCLGVLLGVSGQRSHPSLSYRSTHRSTLSSQSSDLSWISGQKRQESLGSESEV 0
0 GWTDTETTAAWGAAQQASGQSFCSHDLEDGEVKAPSSPQEQKSKTPK 0
0 TKRHLPSLDRRM* 0
>MEL1_nanEhr Nannospalax ehrenbergi (molerat) OPN4
0 MNSPSGPRVPPGLAQKPSFMVTPVLPNQWISFQKNVSVGIQLPPASAT 0
0 ATGAQAASWVPFPTVDVPVHAHYTLGTVILLVGLTGMLGNLIVIYTFCR 2
1 SRGLRTRANMFTVNLAVSDFLMSFTQAPVFFASSLYKRWLFGEA 2
2 GCEFYAFCGAVSGITSMTTLTAIALDRYLVITRPLATIGVASKRRTALVLLGVWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYMTFTPSVRAYTMLLFCFVFFLPLLIIIFCYIFIFKAIRETGR 2
1 ACEGCGESPQRRRQWQRLQNEWKMAKVALLVIFLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMNSVPAVIAKASAIHNPIIYAITHPKYR 2
1 LAISQHLPCLGVLIGVSSQRSHPSLSYRSTHRSTLSSQASDLSWISGRKRQESLGSESEV 0
0 GWTDTEVTAAWGVAQEASGWSPYRHSLEDGEVKASPSPQGQEAKTSR 0
0 TKGQLPSLNLRM* 0
>MEL1_phoSun Phodopus sungorus (hamster) AY726733 ends in error: AKGQLPSLDLGMQDAP
0 MDSPPGPTAPPGLTQGPSFMASTTLHSHWNSTQKVSTRAQLLAVSPT 0
0 ASGPEAAAWVPFPTVDVPDHAHYILGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRSLRTPANMLIINLAVSDFLMSFTQAPVFFASSLYKKWLFGET 2
1 GCEFYAFCGAVLGITSMITLTAIALDRYLVITRPLATIGMGSKRRTALVLLGIWLYALAWSLPPFFGW 1
2 SAYVPEGLLTSCSWDYVTFTPQVRAYTMLLFCFVFFLPLLVIIFCYISIFRAIRETGR 2
1 ACEGWSESPQRRRQWHRLQSEWKMAKVALIVILLFVLSWAPYSTVALVAFAG 2
1 YSHILTPYMSSVPAVIAKASAIHNPIVYAITHPKYR 2
1 AAIAQHLPCLGVLLGVSSQRNRPSLSYRSTHRSTLSSQSSDLSWISAPKRQESLGSESEV 0
0 GWTDTEATAVWGAAQPASGQSSCGQNLEDGMVKAPSSPQ
0 * 0
>MEL1_oryCun Oryctolagus cuniculus (rabbit)
0 MNSPWGSRVPPGPAQEPRSTAPPSRWDGSESISSPGQLTPGSPT 0
0 APGAQEAAWVPFPTVDVPDHAHYTLGTVILLVGLTGMLGNLTVIYTFCR 2
1 SRSLRTPANMFIINLAVSDFLMSFTQAPVFFASSLYKRWLFGET 2
1 GCEFYAFCGALFGISSMITLTAIALDRYLVITRPLAAVGMVSKKRAGLVLLGVWLYALAWSLPPLFGW 1
2 SAYVPEGLLTSCSWDYVTFTPSVRSYTMLLFCFVFFLPLLVIVYCYIFIFRAIRETGR 2
1 ACQGSHESRQWRRLQSEWKMAKVALLVILLFLLSWAPYSTVALVAFAG 2
1 YAHSLSPYMNSVPAVIAKASAIHNPIIYAITHPKY 2
1 0
0 GWTDTEAAAAWGAALQLSGRYLCGQGLEDGEIKATPRRQ 0
0 GPEAETPKVILSTCQSVKGVGAEASYQDSGQKRGFVLGAGFWESPHPPPCLFFAHGVSVSPSHSE
>RGR_musMus Mus musculus (mouse) RGR syn(+PCDH21 -LRIT1 -GRID1 -WAPAL)
0 MAATRALPAGLGELEVLAVGTVLLME 1
2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2
1 RWPHGSEGCQVHGFQGFATALASICGSAAVAWGRYHHYCT 1
2 GRQLAWDTAIPLVLFVWMSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2
1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKFSRSGHLP 0
0 VNTTLPGRMLLLGWGPYALLYLYAAIADVSFISPKLQM 0
0 VPALIAKTMPTINAINYALHREMVCRGTWQCLSPQKSKKDRTQ* 0
>RGR_ratNor Rattus norvegicus (rat)
0 MTATRALPAGFGELEVLAIGIVLLME 1
2 ALSGISLNGLTIFSFCKTPDLRTPSNLLVLSLALADTGISLNALVAAVSSLLR 2
1 RWPHGSEGCRVHGFQGFATALASICGSAAIAWGRYHHYCT 1
2 GRQLAWDTAIPLVLFVWLSSAFWASLPLMGWGHYDYEPVGTCCTLDYSRGDR 2
1 NFISFLFTMAFFNFLVPLFITHTSYRFMEQKLSRSGHLQ 0
0 VNTTLPGRMLLLGWGPYALLYLYAAVADVSFISPKLQM 0
0 VPALIAKTMPTINAINYALRSEMVCRGTWQCRSAQKSKQDRTQ* 0
>RGR_speTri Spermophilus tridecemlineatus (ground_squirrel)
0 MAETAALPAGFGELEVLAVGTVLLVE 1
2 ALSGLSLNGLTIFSFCKTPELRTPNHLLLLSLAVADSGISLNALIAAISSLLR 2
1 RWPYGSDGCQAHGFQGFVTALSSICGSAAIAWGRYHHYCT 1
2 GSQLAWNTAIPLVLFVWLSSTFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2
1 NFISFLFTMAFFNFFVPLFITLTSYRLMEQKLARSGHLQ 0
0 0
0 * 0
>RGR_dipOrd Dipodomys ordii (kangaroo_rat)
0 MATSGDLPTGFGELEVLTVGTVLLVE 1
2 ALSGLSLNTLTIFSFCKTPELRTPIHLLDLSLAVADSGISLNALIAAISSLEW 2
1 HWPYGLEGCQAHGFQGFVTALASISGSAAIAWGRCHHHCT 1
2 GSLLGWDTAVSLVIFVWLSSAFWAALPLLGWGHYNYEPLGTCCTLDYSRGDR 2
1 NFTSFLFTMAFFNFLVPLFITLTSYQLMKQKFARSGRLQ 0
0 VNTTLPTRTLLLGWGPYALLYFYAAIMDVNSISPKLQM 0
0 VPALIAKMVPTVNAINYALCNELLCGGFSLGLLPQKGKQDRTQ* 0
>RGR_cavPor Cavia porcellus (guinea_pig)
0 MATSEALPAGFGELEVLAVGTVLLLE 1
2 GLCGLSLNGLTVVSFWKSPALRTPNHLLVLSLALADSGLSLNALVAAGSSLLR 2
1 HWPGSGHCQALGFQGFTTALASISGTAALSWGRHQQCCT 1
2 RGRLTWSTAVPLVLFVWLSSAFWAALPLLGWGRYDYEPLGTCCTLDYSTGDR 2
1 NFTSFLFTMAFFNFLVPLFITVTSCQLMERHLARSSRLQ 0
0 VSVRQPARTLLLCWSPYALLYLYAVLADAHTLSPRLQM 0
0 VPALIAKTVPTIYSLGRGPWQSLEMQRSKQD* 0
>RGR_oryCun Oryctolagus cuniculus (rabbit)
0 MAEPGTLPPGFEELEVLAVGTVLLVE 1
2 ALSGLSLNGLTIFSFCKTPELWTPSHLLVLSLAVADSGISLNALIAAVSSLLR 2
1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1
2 GSQLAWNTAVLLVLFVWLSSVFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2
1 NFISFLITMAFFNFLMPLFITLTSYSLMEQKLSKSGRLQ 0
0 VNTTLPGRTLLFCWGPYAVLYLCAAVADMSSITLKLQM 0
0 VPALIAKTVPTVNAVNYALGSEVIRRGIWQCLLPQRSVRGRAQ* 0
>RGR_ochPri Ochotona princeps (pika)
0 MAEPGTLPPGFEELEVLAVGTVLLVE 1
2 ALTGLSLNSLTIFSFCTSPELRTPSHLLVLSLALADSGVSLNALAAATASLLR 2
1 RWPYGSDGCQAHGFQGFATALASICGSAAIAWGRYHHYCT 1
2 GSQLAWNTAVLLVLFVWLSSVFWAALPLLGWGHYDYEPLGTCCTLDYSRGDR 2
1 NFISFLVTMAFFNFLMPLFIMLTSYSLMEQKLAKSGRLQ 0
0 VNTTLPARTLLFCWGPYAILCLCATVMDMSTVSPKLLM 0
0 VPALIAKAVPTVNAINYALGSEVIRRGIWQCLLPQRSVRDRAQ* 0
>NEUR1_musMus Mus musculus (mouse) OPN5 neuropsin
0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQAGGLRGTKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHQEV* 0
>NEUR1_ratNor Rattus norvegicus (rat)
0 MALNHTALPQDERLPHYLRDEDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWFGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 gVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASGGGQVFILSILFFCLLLPTAVIVFSYAKIIAKVKSSSKEVAHFDSRIHSSHVLEVKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPNSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYRFACCQTGGLRATKKKSLEDFR 2
1 LHTVTAVRKSSAVLEIHPEv* 0
>NEUR1_speTri Spermophilus tridecemlineatus (squirrel)
0 MALNHTALPQDEHLPHYLRDEDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAFICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFINILFFCLLLPTAVIEFSYVKIIAKVKSSSEEVAHFDSRIHSSHV 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPSLLAKSAAMYNPIIYQVIDYKFACCQTGGLKATKKKSLEDFR 2
1 LHTVTAVRKSSAVVEIHQEv* 0
>NEUR1_dipOrd Dipodomys ordii (kangaroo_rat)
0 MAFNHTAGTQGQGLPHYLPEEDPFTSKLSWEADIVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAVIWAYASFWTTMPLVGLGDYVPEPFGTSCTLDWWLAQASLAGQVFILNILFFCLLLPTSVIVFSYVKIIAKVKSSSKEVAHFDSRIPSSHVLEMKLTK 0
0 2
1 * 0
>NEUR1_cavPor Cavia porcellus (guinea_pig)
0 MALNHTAPPQNEHLPRYLQDEDPFVSKLSWEADLVAGFYLTII 1
2 GILSTVGNGYVLYMSSRRKKKLRPAEIMTINLAICDLGIS 1
2 vVGKPFTIISCFRHRWVFGWIGCRWYGWAGFFFGCGSLITMTVVSLDRYLKICYLSY 1
2 GVWLKRKHAYICLAAIWAYVSFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASAGGQIFILHILFFCLLLPTAMIVFSYVKIIAKVKSSSKEIAHFDSRIHSSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDSRFACCQNAGLKATKKKSLEDFR 2
1 LHTVTTDRKSAVLEIHQEV* 0
>NEUR1_oryCun Oryctolagus cuniculus (rabbit)
0 MALNHTALPQDEHLPHYLREGDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRWYGWAGFFFGCGSLITMTAVSLDRYLKICYLSY 1
2 GVWLKRRHAYICLALIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHSSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFSCCRTSGLKATKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHQEv* 0
>NEUR1_ochPri Ochotona princeps (pika)
0 MALNDTALPQDEHLPHYFRDGDPFASKLSWEADLVAGFYLTII 1
2 GILSTFGNGYVLYMSSRRKKKLRPAEIMTINLAVCDLGIS 1
2 VVGKPFTIISCFCHRWVFGWIGCRLYGWADFFFGCGSLITMTAVSLDRYLK 1
2 GVWLKRRHAYICLAVIWAYASFWTTMPLVGLGDYAPEPFGTSCTLDWWLAQASVGGQVFILNILFFCLLLPTAVIVFSYVKIIAKVKSSSKEVAHFDSRIHGSHVLEMKLTK 0
0 VAMLICAGFLIAWIPYAVVSVWSAFGRPDSIPIQLSVVPTLLAKSAAMYNPIIYQVIDYKFSCCRTGGLKQTKKKSLEDFR 2
1 LHTVTTVRKSSAVLEIHQEv* 0
>PER_musMus Mus musculus (mouse) RRH peropsin syn(-CFI +NOLA1 +EGF -ELOVL6)
0 MLSEASDNSSGSRSEGSVFSRTEHSVIAAYLIVA 1
2 GITSILSNVVVLGIFIKYKELRTPTNAVIINLAFTDIGVSSIGYPMSAASDLHGSWKFGHAGCQ 0
0 IYAGLNIFFGMVSIGLLTVVAMDRYLTISCPDV 1
2 GRRMTTNTYLSMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRNNDT 2
1 SFVSYTMMVIVVNFIVPLTVMFYCYYHVSRSLRLYAASDCTAHLHRDWADQADVTK 0
0 MSVIMILMFLLAWSPYSIVCLWACFGNPKKIPPSMAIIAPLFAKSSTFYNPCIYVAAHKK 2
1 FRKAMLAMFKCQPHLAVPEPSTLPMDMPQSSLAPVRI* 0
>PER_ratNor Rattus norvegicus (rat)
0 MLRDALDNSSGSGSE-GSVFTKSEHSIIAAYLIVA 1
2 GIISILSNIIVLGIFIKYKELRTPTNAVIINLAFTDIGVSSIGYPMSAASDLHGSWKFGHAGCQ 0
0 VYAGLNIFFGMVSIGLLTVVALDRYLTISCPDV 1
2 GRRMTGNTYLSMVLGAWINGLFWALMPIVGWASYAPDPTGATCTINWRKNDT 2
1 SFVSYTMMVIVVNFIVPLTVMFYCYYHVSQSMRLSAASNCTTHLNRDWAHQADVTK 0
0 MSVMMILMFLLAWSPYSVVCLWACFGNPKKIPPSLAIIAPLFAKSSTFYNPCIYVAANKK 2
1 FRKAMFAMLKCQPHQAMPEPSTLAMGVPHSPLAPARI* 0
>PER_dipOrd Dipodomys ordii (kangaroo_rat)
0 MLRNNVGNSSGSRNEDGSVFSQTEHNIVATYLITA 1
2 GVISILSNLIVLGIFIKYKELRTPTNAIIINLALTDIGVSSIGYPMSAASDLYGRWKFGYAGCQ 0
0 IYAGLNIFFGMASIGLLTVVAIDRYLTICHPDI 1
2 GRGMTTRTYVTMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDT 2
1 0
0 MSVVMILMFLVAWSPYSIVCLWASFGDPKEIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2
1 FRRAMLAMLKCQTHQAMPVTSILPMDVSQNPLASGRI* 0
>PER_cavPor Cavia porcellus (guinea_pig)
0 MLRHSLGNSSDSKNEDGSVFSQTEHNIVAAYLILA 1
2 GLISILSNIIVLGIFIKYKELRTPTNAIIMNLALTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0
0 VYAGLNIFFGMASIGLLTVVAVDRYLTICRPDI 1
2 GRRMTSHSYVGMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDV 2
1 AFVSYTMTVIAINFIVPLAVMFYCYLHITRAIRRHVAGDRPPNLSGDWSDQVDVTK 0
0 MSVVMILMFLVAWSPYSIVCLWASFGDPRRISPSMAIIAPLFAKSSTFYNPCIYVIANKK 2
1 FRRAMFAMFQCQTHQAVPVASILPMDASQSPLASGRI* 0
>PER_oryCun Oryctolagus cuniculus (rabbit)
0 MLRNNLSNSSDFKHEDGSVFSQTEHNIVATYLILA 1
2 GMISILSNLIVLGIFIKYKELRTPTNAIIINLAFTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0
0 IYAGLNIFFGMASIGLLTVVAMDRYLTICHPDV 1
2 GRRMTTRTYLGLILGAWVNGLFWALMPIAGWASYAPDPTGATCTINWRKNDT 2
1 SFVSFTMAVIAINFVVPLTVMFYCYYHVTQSIKQHRASDCTEYLNRDWSDQVDVTK 0
0 MSVIMIFMFLVAWSPYSIVCLWASFGDPKKIPPAMAIIAPLFAKSSTFYNPCIYVAANKR 2
1 FRRAMFAMFKCQTHQAMPVTSVLPMDVSQNPLPSGII* 0
>PER_ochPri Ochotona princeps (pika)
0 MLRHNLGNSSEAKVEAGSVFSQTEHNIVAAYLILA 1
2 GMISILSNLIVLGIFIKYKELRTPTNAIIINLAFTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0
0 IYAGLNIFFGMASIGLLTVVAMDRYLTICQPDI 1
2 GRRMTTHTYFGMILGAWINGLFWALMPIVGWASYAPDPTGATCTINWRKNDK 2
1 SFVSFTMAVIMVNFVVPLTVMFYCYYYVTQSIKHHTASDCTKSLNRDWSDQVDVTK 0
0 MSVIMILMFLVAWSPYSIVCLWASFGDPQKIPPSMAIIAPLFAKSSTFYNPCIYVAANKR 2
1 SRRAMFAMFKCQIPQAKPVTSLSPRDVSQSPLSSGRT* 0