Vertebrate Gene Origins
The Origin of Vertebrate Genes
Introduction to gene families
Contemporary vertebrates genomes contain about 20,000 coding genes, not many more than pre-Bilatera and far fewer than was believed prior to the genomic era. These genes fall into perhaps 3000 gene families each with its own history of expansion and contraction, so sizes today vary from singletons to over a thousand members We wish to track each of these gene histories back to an ancestral genome in which a parental gene was present without additional homologs.
There can be no single sweeping explanation of gene family histories because several gene duplication mechanisms have been continuously at work over an extended time frame. However histories can overlap to a degree when unrelated gene family members are duplicated or deleted in a contiguous block. Shared history can also be less than the full length gene in the case of ubiquitous domains, internal tandem expansions and multi-domain chimeric proteins, complexities not considered further here.
Because convergent homology of sequences and folds is immensely implausible, gene families with multiple members clearly experienced a history of net gene duplication (gains outpacing losses). But singleton genes too may have a significant duplication history when gene loss of their paralogs is considered. For completeness, we should also consider ghosted proteins present in humans today only as pseudogene debris (or gone entirely and only inferred in an ancester from continuing presence in other extant mammals, amniotes, or vertebrates).
Gene loss processes are quite significant, with human a net loser in gene count over the last 310 myr, contrary to cultural narratives taking 'progress' towards 'complexity' as a given. It's well-known that human sensory genes (olfaction and vision) have been decimated but the list hardly stops there.
Definition of gene family
The specification of gene family is quite subtle because any practical operational definition is limited by sensitivity and the arbitrariness of phylogenetic depth for coalescence cut-off. This is illustrated by a simple gene family, the sulfatases. The 17 paralogs in human, surely the final count because of completeness of that genome and definitive probes, cluster quite distinctly within all-vs-all blastp searches and are readily organized by sequence and exon pattern conservation into a conventional gene tree whose duplications can be timed relative to various ancestral divergences of the metazoan lineage relative to human.
Yet when 3D structures at PDB are consulted, it becomes undeniable that sulfatases and alkaline phosphatases are structurally homologous over their entire length despite a complete lack of convincing primary sequence alignability even by profile. Fold convergence can and does occur (eg TIM beta barrels) but here the fold is so complex, unconventional and isolated in fold space that coincidental arrival at this common structure can be dismissed out of hand. That conclusion is independently validated by colocalization of active sites and similarity of catalytic mechanism (hydrolysis of ester) despite lack of post-translational modification to formylglycine in any phosphatase.
Consequently, the 4 phosphatase paralogs (comprised of a tandem triple on chr2 and an isolated gene on chr1) bring the human gene family size to 21 if we admit gene convergence back to common ancester of human with bacteria. The cut off at Ur-metazoan would keep these gene families separate (and even slightly partition the phosphatase group). Future methods might require coalescence with ever more subtly related gene families. Once a genome is sequenced, blastp becomes available across it; however tertiary structure determination of entire proteomes is not yet in sight.
The small gene family consisting of PRNP and PRND is also instructive. Although in parallel tandem position and clearly homologous despite deletions of the 40 residue repeat in the latter, the relatively short length of 255aa, compositional simplicity in some regions, and percent blastp identity in the 20's invariably causes pipeline clustering procedures to miss the family association.
This gene family also illustrates the total folly of attempting to date duplications -- it's not particularly ancient despite the great divergence. Compare this to a histone that differs at one residue between human and garden pea. PRND can only be traced to amniote by all-out hand curation; it is cleanly deleted in a gapless chicken contig but fortunately a second node representative (lizard) retained it.
PRNP tracks back to teleost fish but again only using tools such as syntenic flanking genes and secondary structure can establish this because fish experienced an astonishing internal expansion amino terminally. The gene didn't necessarily originate in teleosts but to date it has proven impossible to find it chondricthyhes or earlier diverging species because conserved residues have been swamped out.
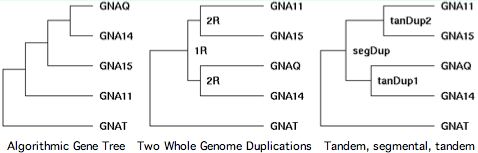
Chromosomal adjacency and broader synteny are thus be important in determining the gene tree. Sequence algorithms alone can get it wrong. That's seen quite dramatically in GPCR transducins, in which 10 genes occur in tandem pairs. Clearly these didn't come together by random rearrangement. Tandem pairs often separate rapidly in sequence because they must take on distinct roles to be supported by selection.
The transducins also illustrate the fallacy of parsimony. Gene families are a product of history, not statistics. That history is often but not always the most parsimonious -- Occam's razor cuts both ways. The third panel at left shows the correct history.
Gene families can also be exceedingly simple. Homogentisate 1,2-dioxygenase, HGD, is as simple as they get. It catalyzes an intermediate step in aromatic amino acid catabolism; without it, a toxic metabolite accumulates. This gene has apparently not been lost from any eukaryotic lineage nor fixed a duplicate or processed pseudogene in 50 billion years of branch length. Sequence conservation is high; there is never ambiguity with blast searches.
Yet HGD illustrates another pitfall in defining gene families. The most recent human genome assembly (NCBI Build 36.1) has a left-over 500 kbp piece of DNA that did not fit into the regular assembly (for unknown reasons). This piece on chr3_random:18,988-73,308 would give rise to an artefactual paralog of HGD with automated procedures, in effect doubling gene family size. Such problems are rare in near-complete genomes such as human but all too common in lower coverage assemblies.
Bad gene names complicate the task
Three sources of gene terminology error makes these histories harder to explain:
- The term 'gene copy' is sometimes used for what are really deeply diverged paralogs with dimly related functions; gene duplication processes can make highly inexact and inequivalent copies right at the event (so they are never copies), subsequent evolutionary trajectories (other than gene conversion) only dilute the comparison further.
- The Greek prefix 'iso' is used consistently throughout science to mean 'same'; hence 'isoform' is highly inappropriate for products of distinct diverged genes or alternative splicings of a single gene (the vast majority of which are transcriptional artefacts to begin with). This useage perpetuates long-gone days of starch gel electrophoresis, an experimental technique that could only distinguish enzymatic forms if surface charge happened to differ.
- The lack of awareness or refusal of authors to comply with agreed-upon international conventions for gene nomenclature (HUGO) causes many downstream problems. Gene names in the genomic era need to stay clear of subscripts, superscripts, mixed Roman and Greek letters, single or double letters, unreadable italics, lower case denoting mouse, easily confused i's and 1's (or o's and 0's), lab jokes, contrived acronyms, and naming by soon-forgotten experiment, tissue expression, end phenotype, or common disease symptom.
It's all been tried before and it doesn't work. There's a reason why Linnaean genus-species nomenclature took over for the species tree -- it's anchored in evolution. Gene trees too can have evolution-based nomenclature, say 3-4 letters to denote gene family, numbered paralogs after that (hopefully by sequence relatedness), and suffixed with a species code adequate for comparative genomics (as in SUMF2_homSap).
This creates a unique hierarchical identifier that works for Google, PubMed, and GenBank searches. It clusters members of gene families by name and extends to the many-genome era. With complete genomes, we are in a position to create stable nomenclature, imperfect perhaps but much improved. Gene terminology need not be immutable (Linnaean taxonomy certainly is not), but proposed improvements have a burden of published proof.
It's quite feasible to have global update cycles at browser centers that generate and maintain synonymy lookup tables at the same time. However 9 vertebrate gene families in 10 could be named forever stably today. For example, the official gene sets at UCSC and Ensembl assign genes random unmemorable alphanumeric strings such as uc003emt.1 (to serve as indexing fields in immense relational databases). We see already with opsins that strings for a gene family are entirely unrelated: uc003emt.1 uc003vnt.2 uc004fjz.2 uc001hza.1 uc003hzv.1.
Since geneSorter resource at UCSC *already* carries the all-vs-all blastp data needed for a quick gene tree as well as the requisite associated column for HUGO name, the uc and version number could be moved to the rear (.uc1, .uc2,...) and the 6 free characters be used for a gene tree-driven nomenclature that incorporates Hugo names to the extent these are available and unambiguous.
The nomenclatural problem has immense practical impacts in synteny. Relationships that should jump out from name along are instead obscured. For example, flawed gene names mask the extent of human paralogons on chr15, chr9, and chr19 -- gene names of close human paralogs often bear no resemblance (eg PRUNE and ATCAY). Comparative genomics in other mammals brings in still other names (more commonly no name) and uninformative transcript or RefSeq strings. It's not a sustainable practise.
Have vertebrate coding genes increased over time?
The number of coding human genes is still not known (Oct 2008) to within 1500-gene accuracy even seven years after sequencing. In fact, no such effort is even underway. Official compilations miss ordinary enzymes like COMT2 (transcribed only in special cells of inner ear) yet still include gravely decayed pseudogenes (that still have spliced transcripts) and non-genes (supported by transcript noise but no comparative genomics even to chimp). Beyond that, weak gene models may guess at initial methionines, lack stop codons and propose impossible alternate splices missing structurally essential core exons, active sites and any phylogenetic support.
Despite these limitations, it's worth compiling gene counts from original articles describing new genome assemblies and as subsequently amended. Being gene counts for contemporary species, they don't directly provide ancestral gene counts at divergence nodes. Yet since no trend whatsoever relates depth of divergence or simplicity of body plan with diminished gene count, ancestral counts may not be so different.
The scientific literature contains some incredulous assertions in the scientific literature about gene counts. If salmon truly experienced four rounds of whole genome duplications since cephalochordate, that implies (neglecting gene loss between rounds and gene gain in Branchiostoma) 350,400 coding genes. Yet the immense collection of salmon cDNA shows no indication whatsoever of any such growth in gene family size. On the contrary, salmon gene count seems very similar to every other vertebrate, as do all of the supposed 3R teleost fish. Their genomes show roughly 20,000 genes, nowhere near the 175,200 expected. How could mature gene-finding algorithms miss 155,200 coding genes given strong known paralogs?
Large-scale gene loss between rounds of duplication reduces the paradox somewhat, yet duplication followed shortly by loss of half the genes seem largely irrelevent to contemporary gene histories, like counting trees falling in the forest that got back on the stump.
Fish genomes have an astronomical numbers of gaps as of October 2008 assemblies. Unbridged gaps in particular imply a great many chromosomal misplacements. It is exceedingly premature to consider global syntenic relationships in a situation with 23,322 unbridged gaps because no known method can distinguish whole genome duplication from the steady accrual of ordinary segmental and re-translocated tandem duplications.
To the contrary, genome sequencing projects show if anything a decreasing trend in gene count in more recently emerging deuterostomes. For example, sea urchin genome has 23,300 coding genes whereas humans have but 20,176 in the 11 Sept 2008 tally of consensus CDS and even this number seems to be inflated over the 17,052 distinct locus count by assignment of multiple CCDS IDs numbers to single genes.
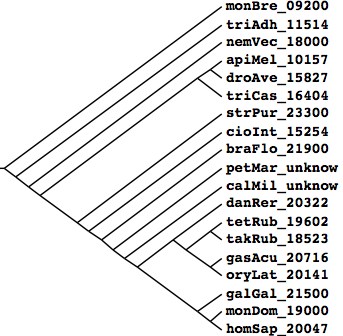
Tree generated at Phylodendron by the following Newick string and peer-reviewed literature compilation: (monBre_09200,(triAdh_11514,(nemVec_18000,(((apiMel_10157,droAve_15827),triCas_16404), (strPur_23300,(braFlo_21900,(petMar_unknow,(calMil_unknow,((danRer_20322,((tetRub_19602,takRub_18523),
(gasAcu_20716,oryLat_20141))),(galGal_21500,(monDom_19000,homSap_20047)))))))))));
Species Assembly Gaps Unbridged #Genes Organism homSap Mar 06 387 373 20,047 Homo sapiens (human) monDom Jan 06 72,803 5,336 19,000 Monodelphis domesticus (opossum) galGal May 06 78,478 17,335 21,500 Gallus gallus (chicken) anoCar Feb 07 43,237 ND ND Anolis carolinensis danRer Jul 07 49,727 11,857 20,322 Danio rerio (zebrafish) tetNig Feb 04 25,763 23,322 19,602 Tetraodon nigroviridis (pufferfish) takRub Oct 04 7,213 7,213 18,523 Takifugu rubripes (fugu) gasAcu Feb 06 16,945 1,913 20,716 Gasterosteus aculeatus (stickleback) oryLat Apr 06 134,426 8,131 20,141 Oryzias latipes (medaka) calMil Dec 06 ND ND ND Callorhinchus milii (eshark) petMar Mar 07 202,409 ND unpub Petromyzon marinus (lamprey) braFlo Mar 06 94,815 3,031 21,900 Branchiostoma floridae (amphioxus) cioInt Mar 05 22,521 ND 20,141 Ciona intestinals (tunicate) strPur Sep 06 80,391 ND 23,300 Strongylocentrotus purpuratus (urchin) monBre_09,200 Monosiga brevicollis triAdh_11,514 Trichoplax adhaerens nemVec_18,000 Nematostella vectensis apiMel_10,157 Apis melifera triCas_16,404 Tribolium castenatum droAve_15,827 Drosophila 12_species strPur_23,300 Strongylocentrotus purpuratus braFlo_21,900 Branchiostoma floridae petMar_unknow Petromyzon marinus calMil_unknow Callorhinchus milii danRer_20,322 Danio rerio tetRub_19,602 Tetraodon nigroviridis takRub_18,523 Takifugu rubripes gasAcu_20,716 Gasterosteus aculeatus oryLat_20,141 Oryzias latipes galGal_21,500 Gallus gallus monDom_19,000 Monodelphis domestica homSap_20,047 Homo sapiens