Opsin evolution: Peropsin phyloSNPs
Peropsin backgrounder
Peropsin, not being a glamorous visual photoreceptor, has a mere 11 PubMed matches since its discovery and initial characterization in 1997. The sole substantive google match attempts a diagnostic fingerprint for peropsins using 11 peptides (based solely on human and mouse representatives!).
The word peropsin was coined in 1997, with the first three letters a rearrangement of RPE (apical retinal pigment epithelium microvilli site of expression). The unmemorable gene name, RRH, arises from RPE-derived rhodopsin homologue and is used here only as synonym to PER (which better accompanies peropsin). The sites of expression are much broader than originally envisioned -- they include circadian expression levels in chicken pineal and fetal expression in mammals that precedes post-natal visual opsin expression, undercutting the notion of simple retinal photoisomerase. The RPE phagocytoses 10% of the mass of each photoreceptor outer segment on a diurnal schedule and isomerizes retinal from all-trans to 11-cis (which ciliary opsins cannot do on their own).
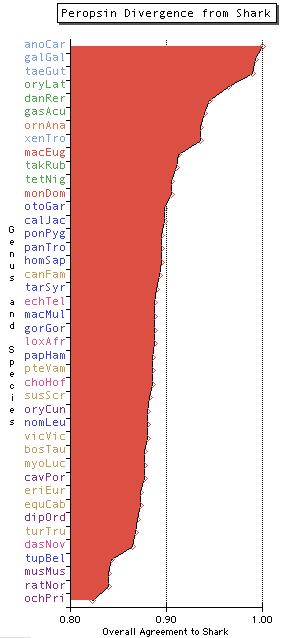
Peropsins are rather slowly evolving proteins (with some species like mouse anomalously faster): bird and shark remain 77% identical, this drops to 65% for the mouse comparison. The diagram at left provides a composite view of relative degree of divergence of bony vertebrates from their commmon outgroup chondrichthyes (which contributes a constant to each of the other species amounting to its own divergence back to ancestral node).
Since no single blast scoring parameter is fully satisfactory, bit score, log expectation, percent identity, and percent simility were separately normalized to their highest score (giving a 0-1 scale) and these 4 scales were averaged to provide the x-axis. This overall score averaged 89% with standard deviation 4% which will be interesting to compare with other opsins or the entire vertebrate proteome. Colors show the various major clades.
Although RGR mutations can lead to cause retinitis pigmentosa, peropsin to date does not, despite careful of screening 994 separate cases. One heterozyote for C98Y exhibited retinitis punctata albescens; the corresponding residue in RHO1 is a known disease allele. Knockout mice seem not yet to have been constructed. The protein in mammals is localized on the apical face of the retinal pigment epithelium, most prominently to microvilli that surround the photoreceptor outer segments.
Expression in chick retina and pineal gland is remarkable in that mRNA levels fluctuate on a circadian basis, highest late in the day (RGR is similar), where it may participate in regulation of the indoleamine hormone melatonin. The function is commonly stated to be auxillary replenishment photoisomerase, though that does not fit with developmental expression uncoupled to ciliary opsins, expression elsewhere in the brain, nor long evolutionary persistence. Peropsin may play a role in RPE physiology through its capacity to sense light directly or by monitoring concentrations of retinoids and related compounds.
Peropsin can be tracked back to amphioxus, sea urchin, and acornworm though it cannot be located in tunicate or lamprey genomes, much less protostomes or cnidarians. Amphioxus has three well-differentiated peropsin genes that have been reconstituted with 11-cis- and all-trans-retinal chromophores; these peropsins have photoisomerase activity but to what purpose is not clear (amphioxus lacks imaging eyes).
Peropsin signalling can be inferred from strong conservation of a classical DRY motif on the third intracellular loop; G-protein second-messenger signaling however is not understood. The carboxy terminus has various semi-conserved serines that could potentially by phosphorylated for purposes of signal termination, though interaction with a specific arrestin has to date not been documented. A conserved glycosylation site is evident in the extracellular amino terminus.
Peropsin represents a very old divergence deeply nested with the opsin gene family. The mystery with peropsin is its original gene function in early metazoa and its apparent loss in all other lineages than deuterostomes (where it experienced an unexplained resurgence in Branchiostoma). Its intron placements and blast clustering associate it with neuropsin and RGRopsin but only distantly so. Possibly the advent of rhabdomeric opsins rendered it superfluous; possibly it persists in cnidarians with imaging opsins not yet sequenced.
PhyloSNPs defined
PhyloSNPs are defined as single coding residue changes at a site (1) strictly conserved ancestrally over great branch lengths but (2) switching in an ancestral stem to a new residue that in turn is (3) strictly conserved in all descendent extant species. A certain amount of excursion from perfect conservation can be admitted (reduced alphabet, disproportionately attributable to sequencing error) and even low levels of homoplasy (where the ancestral value reoccurs in a later diverging a species or two. Peropsin (or RRH) has just a handful of them at its 337 positions, the number depending on quality cutoff. Most of these occured in the earliest mammals.
PhyloSNPs are thus similar to synapomorphies but are adapted to protein molecular evolution and its fixed alphabet of 20 distinct, unambiguously determinable charcters. Because phyloSNPs are fixed both before and after the stem event, they greatly enrich for defining characters of the stem clade (and its complement) -- in effect they answer the question, what makes a mammal a mammal. Synapomorphies (eg vertebral column) don't have this requirement on the clade complement -- ie sea urchins don't have notochords.
PhyloSNPs differ from conventional SNPs familiar from human variation studies in their phylogenetic depth. Differences in the human population or even human/chimp speak to a much shorter time scale with few or no intermediate divergence nodes (to extant or sequencable extinct species). Consequently these variations are not readily interpretable nor adaptive, whereas high quality phyloSNPs in stem placentals (at the current level of 37 mammalian genomes and 13 non-mammals) have accrued over a billion years of branch length during which the phyloSNP residue has been conserved in all species (either as the clade-specific change or ancestral value).
Consequently phyloSNPs must represent adaptive change at the residue in question. However phyloSNPs don't capture everything: features such as glycosylation sites and disulfides can evolve out of 'nothing' (ie variable sequence lacking a specific conserved alternative ancestral function). The opposite of an innovative phyloSNP is a decayed phyloSNP in which invariance is lost in one sister subclade but retained in the other. The opsin gene loss situation in mammals is an extreme version of decayed phyloSNPs.
The timing of phyloSNP establishment must be considered in relation to timing of gene duplications within the ambient gene family. If the phyloSNP precedes the duplication, then it may well descend into descendent clades of the gene tree (defining a phyletic phyloSNP). These phyloSNPs might appear to be independent events if the individual paralogs are considered in isolation. Alternately, phyloSNPs can arise at the same site independently of gene tree duplications as seen here in peropsin (defining a paraphyletic phyloSNP). Opsins are a moderately sized gene family with phyletic, paraphyletic and simple orthologous phyloSNPs (though so far peropsin only seems to have the latter two types).
The timing of phyloSNP establishment must be considered in relation to timing of gene duplications within the ambient gene family. If the phyloSNP precedes the duplication, then it may well descend into descendent clades of the gene tree. These phyloSNPs might appear to be independent events if the individual paralogs are considered in isolation. Opsins are a moderately sized gene family with both paralogous and orthologous phyloSNPs (though so far peropsin only seems to have the latter).
Interpretation of phyloSNPs is not so easy from bioinformatics alone. In opsins, a great deal of structural and experimental information is availble. For example, a peropsin douple phyloSNP in adacent N.S columns at positions 211-213 might appear to be an ancestral glycosylation site lost in a coordinated way in the post- marsupial stem but before atlantogenata diverged, perhaps suggesting a shift in subcellular location in placental mammals. However, the site in fact is on the cytoplasmic side and consequently cannot have been a glycosylation site to begin with.
Note the species are listed below in more or less phylogenetic order (up to an arbitrary choice of human at the top). More precisely, various admissible symmetry operations can result in some swapping of order (eg of marsupials or murid rodents). These reorderings are limited to superordinal clades -- no matter what combinations of tree reorderings (leaving human as fixed point), mouse always stays above dog (ie, Euarchontoglires stay above Laurasiatheres.)
PhyloSNPs are required to be invariant (stay a connected vertical chain) under the action of this finite abelian subgroup of the species tree's permutation group. In other words, they are defined on the quotient tree under this action (rather than on the tree itself). For example, if a phyloSNP continues into the marsupial macEug but not into monDom, it would not be a phyloSNP because swapping order breaks the vertical continuity. Thus, to the extent the assumed tree is valid, there is no arbitrariness in the phylogenetic ordering of the y axis (orthology). PhyloSNP is a strong condition.
In a cladesheet that considers a single phyloSNP, the x-axis can be used for the gene tree as shown here. That too has a distinct fixed topology and corresponding action of a separate abelian group of rearrangements. A phyletic phyloSNP must be well-defined under both group actions. In opsins, the counterion at position 113 has both phyletic and paraphyletic phyloSNPs with respect to the gene tree, with the latter still being valid phyloSNPs with respect to the species tree. In opsins, the counterion at position 113 has both phyletic and paraphyletic phyloSNPs with respect to the gene tree, with the latter still being valid phyloSNPs with respect to the species tree.
The ultimate double-cladesheet view is 3-dimensional. The y-axis can be taken as the species tree, the x-axis as linear sequence, and the z-axis as gene tree. The x-axis is now folded to reflect tertiary struture of the protein and bring non-adjacent but interacting residues (such as the counterion/Schiff base lysine and the disulfide), which for opsins is fairly conveniently represented by with a cartoon showing transmembrane segments as oriented columns. This object can be greatly reduced in complexity by cutting down only to residues that are some form of phyloSNP in at least one opsin paralog. Numbering must be based on a global alignment of the entire family; this introduces gaps that accommodate insertions in various famiy members and departs from numbering of individual opsin gene orthologs. These gaps will largely become superfluous in phyloSNP view, so a simple table is necessary to relate the numbering systems (ultimately to bovine rhodopsin to the extent feasible).
It's worth noting the tremendous distillation of sequencing effort that goes into defining a single phyloSNP. First 50 large vertebrate genomes must be carefully sequenced. Of that data, only one-fiftieth codes for protein. Of that, only one-hundredth is a phyloSNP. Of those perhaps one-tenth are high enough quality and on a stem of interest. Of those perhaps one-tenth are stem events on a gene tree (which for opsins involves comparison of 15 proteins). Of those perhaps two-fifths are interpretable. Overall, a good phyletic phyloSNP represents over a million-fold reduction of raw data. That's precious signal relative to the background level of near-neutral noise and the near-meaningless drift in protein evolution.
PhyloSNPs in vertebrate peropsins
PhyloSNPs emerge from aligning 48 peropsins from cartilaginous fish to human: they are columns towards the bottom of ancestral residues that continued past various divergence nodes in a conserved manner, only to change at a more recent node in its descendent clade (here typically mammals), in contrast to the ancestral value which continues to the present day in other descendent species.
PhyloSNPs can be classified in various ways, amounting to horizonal sorts of the cladesheet (a spreadsheet with topological ordering of rows) by the criteria below. When phyloSNPs become available in all opsin paralogs (indeed all GPCR receptors), it may be instructive to extend sorting across the entire gene family to identify structural hotspots and coldspots of change.
- -- derived amino acid (synapomorphy)
- -- phylogenetic depth: fish, land, mammal, theria, placental
- -- overall assessed phyloSNP quality: A best, B some noise, C some homoplasy
- -- physiochemical severity of change
- -- position within sequence
- -- coding exon number within gene
- -- relative order by phylogenetic depth
- -- relative order within linear sequence
- -- topology: membrane, cytoplasmic, or extracellular
- -- secondary structure: alpha helix, beta sheet, coil
Peropsin exhibits 12 phyloSNPs of varying degrees of quality and interest. The first four cannot yet be fully evaluated because no outgroup data is available from cartilaginous fish or lobe-finned fish -- these phyloSNPs may only have relevence in defining key events within ray-finned fish. This will be resolved by elephantfish and coelocanth genome projects.
In the second group, the high-quality glycine to serine phyloSNP occured in a transmembrane segment sometime after platypus but before marsupial divergence. While this is a commonly observed change (high PAM coefficient in the Dayhoff matrix), those statistics are averaged over thousands of mostly cytoplasmic proteins and so have little bearing on specific positions in specific integral membrane proteins.
In the third group, the alanine to proline A --> P change at position 59 in a cytoplasmic loop could potential result in a kink so more rigid conformation with potential conformational and functional implications. This residue position has been discussed before as a signature histidine uniquely characteristic of SWS2 opsins. Two independent phyloSNPs at the same position at vastly different evolutionary times shows the amino acid at position 59 -- proline, alanine, or histidine -- is adaptively exploitable in different ways yet always under strong selection. Peropsin is not closely related to SWS2 in the opsin gene tree, meaning these events have not descended down from a gene duplication.
The asperagine to basic histidine change at position 211 membrane boundary is temporally coupled with a serine to larger but similar threonine change at neighboring cytoplasmic position 213, possibly as coevolutionary change. (In opsins, because the 3D structure is available, we can also consider coevolutionary changes of residues that are adjacent not in the linear sequence but only in tertiary structure.)
PhyloSNPs in peropsin are readily located in the overall alignment of 230 opsins by web browser search with a few flanking residues. The conservation of residue position in the overall scheme of opsin structural and functional evolution can contribute to the interpretation of individual peropsin phyloSNPs (as we have seen in the DRY to GRY phyloSNP of RGR opsin).
When this is done for the A --> P change at position 59, a very interesting result emerges: proline is actually the deep ancestral value in all other classes of opsins including all invertebrate opsins (with the exception of the SWS2 mentioned above). Consequently the sequence of events in peropsin must have been: P --> A --> P with the first change predating vertebrates and the 'reversion' postdating marsupial divergences. Indeed, the peropsin story becomes very muddled in amphioxus and sea urchin homologs because of gapping and weakened alignment. Peropsin is undoubtedly an ancient GPCR persisting from early eumetazoa yet it cannot be located outside of deuterostomes -- perhaps this change was critical in recruiting it to photoreceptor functionality.
.......................*...... PERa_braBe P KFRSLR-SPTTMLLV PERa_braFl P KFRSLR-SPTTMLLV PERb_braBe A KWRQLCRKAPNLLVI PERb_braFl A KWRQLCRKAPNLLII PER1_strPu S RYGTFRKRSVNILLM PER2_strPu S RYRTFRKRSINLLLI
We've previously considered the case of the glutamate counterion at position 113 of cone/rod opsins but tyrosine ancestrally in most other opsins including peropsin. This locus thus exhibits a phyloSNP fixed prior to a series of gene family expansions in the stem of parapinopsin PPIN as well as later phyloSNPs localized within individual gene families. This suggests that considering paralogs helps even in evaluating orthologs to the extent that only a limited class of residue positions have phyloSNP potential, with that potential being used repeatedly at different junctures.
The phyloSNP at position 59 (human peropsin numbering) is quite interesting when considered across the entire gene family. The cladesheet is not fully populated -- only sequences from the opsin reference collection and the UCSC genome browser 28-way track are shown. (That data could be doubled by using genomes-in-progress stored at the NCBI trace archives.)
What is notable about this residue is not only the P --> A --> P phyloSNP in peropsin but also the P --> H --> gene loss seen in SWS2. This histidine is a departure from the expected proline that must have been present in the ancestral cone opsin protein that experienced considerable expansion between amphioxus and lamprey divergences. Details of that era are largely invisible to us for want of extant species or plausible sources for extinct species dna. Melanopsin 2 is also of interest for its leucine in early diverging species; this gene must be carefully separated from possibly similar extra melanopsins resulting from the fish-specific whole genome duplication.
The opsin family is somewhat restricted in terms of illustrating gene family aspects of phyloSNPs because of fairly massive gene loss (7 of 15) in mammals. We see here however that peropsin, neuropsin, and RGR (which are sometimes lumped as 'outsiders' to mainstream visual opsin development) have no deep connection at position 59.
A more systematic evaluation of this requires full scale curation of the full set of opsin genes in vertebrates. In essence, the cladesheet is 3-dimensional, with the species tree, residue position, and gene tree representing the 3 axes. The two trees have topological structure that is only indirectly indicated by choice of ordering.
Invertebrate peropsins
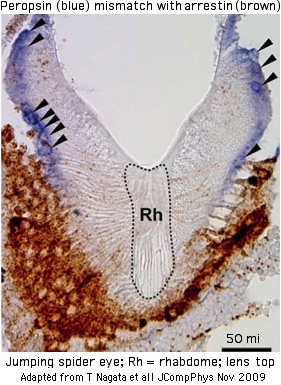
The first ecdysozoan peropsin (AB525082) has been identified in a Dec 2009 report on the eye of the jumping spider, Hasarius adansoni. The sequence clearly classifies as a peropsin by various criteria (though the NPxxY motif mentioned by the authors is irrelevent -- all GPCR have it). It bears an especially close relationship to amphioxus peropsins and is likely the ortholog.
Using the Hasarius peropsin as probe, a second chelicerate persopsin can barely be assembled from the Ixodes genome. Four exons can be located and their intronation determined. The amino terminus up to the first disulfide cysteine cannot yet be located. Otherwise the Ixodes peropsin has a conventional DRY motif, K296, and NPxxY motif KTASTINPFIYFLSNPCIR. Some of the introns match those of other peropsins. The Ixodes intron positions and phases can easily be transfered to Hasarius by homology.
Within peropsins, the spider protein groups with those of ancestral length in the N D P C region of TM2. In other words, it lacks the one residue insertion between N and D seen in a second group of 5 sea urchin and amphioxus peropsins. A third group of lohotrochozoan peropsins has slightly ambiguous associations. In any event, peropsins must have arisen in pre-Bilaterans even though no counterpart has yet been detected in cnidaria, ctenophores or earlier branching metazoa.
The spider peropsin is expressed in the retina but not in the imaging area of the eye. Furthermore it does not colocalize with spider arrestin. It seems that the signaling regions of this peropsin are intact but whether it signals and to what purpose remain obscure. The arrestin gene family may not be fully known in this species or cessation of signaling may not be important in an opsin able to internally recycle trans and cis retinal. If it does signal, then it should not be considered as a purely metabolic photoisomerase that merely replenishes cis-retinal for imaging opsins (which are presumably melanopsic rather than ciliary in this species).
The spider peropsin was localized only at the level of light microscopy so its ultrastructural associations are unclear. While surely localized in the cytoplasmic membrane, it may or may not be in a specialized substructure (ie microvillar extensions), columnated in a stack (for photon sensitivity), or backed by pigmentation (for directionality).
The only relevent study to date used electron microscopy on albino mouse peropsin, concluding "peropsin localizes to the apical face of the RPE, most prominently to the microvilli that surround the photoreceptor outer segments... The intimate association between RPE microvilli and photoreceptor outer segments is presumed to be important for adhesion of the retina to the RPE, transport of small molecules between the RPE and the photoreceptors, and orderly phagocytosis of photoreceptor outer segments by the RPE."
The RPE has melanin backing within the cell body so incidentally furnishes backing. It cannot be determined from these images whether peropsin is stacked (along the lines of rhabdomeric opsins), occurs in RPE microvillar patches, or is simply sporadically distributed as individual protein molecules. Peropsin seems not associated with the endoplasmic reticulum like RGR-opsin. One serious concern with both RGR and peropsin is expression (reported from genomewide arrays) in numerous other tissues besides RPE (and Muller cells) -- what would be the photobiological roles there?
The mouse 1999 peropsin study has seen little experimental followup other than this 2009 jumping spider study and a 2008 paper on RGR which, like peropsin, is also expressed in RPE. The authors concluded (without considering RPE peropsin) that RGR has a rather unexpected function (that peropsin therefore is not doing):
"Consider the sites of all-trans-RE storage inside RPE cells. One compartment is within lipid bilayers of the endoplasmic reticulum. Several retinoid-processing proteins are associated with the ER including LRAT, 11-cis-RDH, RGR, and RPE65. Another compartment in RPE cells for storage of all-trans-REs is lipid droplets (cytoplasmic structures containing a core of neutral lipids, including triglycerides, cholesterol and retinyl esters, surrounded by a phospholipid monolayer [which could not host a 7-transmembrane protein]). Lipid droplets are expandable three-dimensional structures of potentially far greater capacity for retinyl esters than ER membranes.
RPE65 does not co-localize with lipid droplets in RPE cells. Accordingly, lipid droplets may serve as a spill-over site for retinyl ester storage after ER membranes have been filled to capacity. Because of their water insolubility, all-trans-REs in lipid droplets must first be hydrolyzed by all-trans-REH, transported to the ER as all-trans-ROL, and secondarily re-esterified by LRAT into ER membranes before they can be used as substrate by the RPE65 isomerase. We propose that the translocation of all-trans-REs from lipid droplets to ER membranes is regulated by light through the RGR-opsin [indirectly via its Galpha signaling second messengers which lead to LRAT and REH inhibition]."
In other words, when there is sufficient light intensity that the photoreceptor cells are churning out all-trans retinal waste product and in need of cis-retinal replenishment, some of those photons are converting RGR-opsin, despite its much lower quantum efficiency, to its non-signaling cis-binding state, which reduces its inhibition of translocation from lipid droplets to ER, eventually allowing adequate replenishment of the imaging photoreceptors. Peropsin then may gate the last stages of this transfer, with both RGR and peropsin important in avoid futile ATP-wasting hydrolytic cycling of retinoids (and signaling compounds).
This scenario offers no role for peropsin and RGR outside of ciliary imaging support systems. Thus the presence of peropsin in jumping spider retina (which utilizes melanopsic imaging opsins not needing cycle replenishment) must be slightly different. It explains the absence of RGR outside of its current phylogenetic range (tunicate to mammal), though it must have a far more ancient pre-bilateran origin. Future studies of RGR must embrace the unprecedented shift of the DRY lock to GRY at boreoeuthere divergence -- mouse, dog, and human must have a vastly different RGR functionality than elephant, sloth, marsupial, platypus, birds, and reptiles.
In evo-devo terms, the RPE cell lineage gave rise to descendent photoreceptor cells. The ciliary imaging system, with its Rube Goldberg rapid replenishment system, is seemingly a late arrival in deuterostomes. The RPE may have been a standalone cell type specialized to storage and manipulation of retinoids and carotenoids that later came to service rudimentary (or auxillary) cell ciliary systems post-amphioxus, pre-tunicate (namely the parapinopsins of Ciona). This accords with the phylogenetic distribution of RPE65.
The alignment shows vertebrate peropsins have three specializations (indicated by * over the ClustalW polarity colored residues) -- a charged glutamate conserved through fish in CL1, a proline conserved only through placental mammals also in CL1, and an aspartate conserved through teleost fish, seemingly in TM2 (which calls for a salt bridge partner elsewhere.
Five amphioxus and sea urchin peropsins share an unusual extra residue at the boundary of CL1 with TM2. This may also occur in the high anomalous molluscan peropsin class (comprised of squid retinochrome and other molluscan opsins that best cluster to peropsins).
Note that the new ecdysozoan spider sequence is quite conventional. No currently available insect genome contains peropsin, include those still hosting ciliary opsins. These taxa have lost two of the three RPE65 homologs as well. Peropsin cannot be found in Ixodes despite its being, like spider, a chelicerate.
>PER_hasAda Hasarius adansoni (spider) AB525082 (not yet released at GenBank) peropsin PubMed 19960196 0 MDDNMSEIALADDMSTLSTQEPSENVYPYVFPLSTHTIVGTYLIIIGILGTLGNGLVLVTFLRFRVLVTPTTLLLVNLAVSDLGLILFGFPFSASSSLSAKWIFGE 1 2 GGCQWYAFMGFLFGSAHIGTLALLALDRYLIACRISL 1 2 RGKLTFKRYTQMITVVWTYAFFWAMPLLGWGR 2 1 YGLEPSVTTCTIDWQHNDSSYKSFLIVYFVL 1 2 GMVPFAIIAVSYIAIARRVGKKSKERPVVRDLWTNERSVTL 0 0 MAFILIVTFFVAWSPYAVLCLWTIFAEPNTVPPFLTLIPPLFAKSSTVVNPLIYFLSNPKLRTAILSTLSCCNEAPIQNIELPDSPERAANNDAI* 0
>PER_ixoSca Ixodes scapularis (tick) XM_002409761 XM_002435011 mRNAs ABJB010816023 contig frags 62% GMPCAIFAAQQARVRRFGKRAKHAPCARTLYTNEvglyf exon? 2 TGCQAYAFMGFLFGSAHIGTLTLLALDRYLATCRIGF 1 2 RSKPTFKRYFQLLLLVWLYGLFWAVMPLLGWAR 2 1 YGLEPSFQSCTIDWRHNDSSYKSFTLVYFVL 1 0 MVALIVVTFFFAWTPYAVLCLWAVFADTKSVPHLLAMVPPLFAKTASTINPFIYFLSNPCIRADVLQLLGCRARSSPHMAISSDAVEEERCCQDQA* 0
Alignment of 48 peropsins
..........................................................................................................1.........1.........1.........1.........1.........1.........1.........1.........1...1 ................1.........2.........3.........4.........5.........6.........7.........8.........9.........0.........1.........2.........3.........4.........5.........6.........7.........8...8 .......1234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234 .......eeeeeeeeeeeeeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMMMMccccccccccccMMMMMMMMMMMMMMMMMMMMMMMMMMeeeeeeeeeeeeeeMMMMMMMMMMMMMMMMMMMMcccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMMMMeeeeeeeeeeeeeeeeeeee exon...1111111111111111111111111111111111122222222222222222222222222222222222222222222222222222222222222233333333333333333333333333333333334444444444444444444444444444444444444444444444444444 marker........glyc....................................................................................diS.......................DRY................................................diS......... .......................y..............................xx.........x...............xx..........x...............y.........................y....................................................... opsins M........S..................A.Y...........N...L...v..K.LRTP.N.I..NLA..D......Gy.......LhG...FG..GC..........G......L.V.A.DRY..VC.P....R.........I...W......A..P..GW..Y.PD.....C.I....... Consen M.............e..S.Fsq.EHnIVA.YLI.AG.iSi.SN..VLgiF.k.keLRTpTNa!IiNLA.TDIGVs.IGYPMSAASDl.GsWkFG..GCQIYAgLNIFFGMaSIGLLTvVA.DRYLTiC.Pd.Gr.mt...Y..mIl.AW.N..FWa.mP..GWA.YAPDPTGATCTiNWR.ND. homSap MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMAGMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQVYAGLNIFFGMASIGLLTVVAVDRYLTICLPDVGRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR panTro ........................................................................................................................................................................................ gorGor ........G................................................................................................................................................L.............................. ponPyg ....................................I..............................................................................................I.................................................... nomLeu ........................................................................................................................................................................................ macMul ........................................L..........................................................................................I...........M........................................ papHam ........................................L..........................................................................................I...........M........................................ calJac .............................S..........L...............................................................................A..........I......S...IM................V....................... tarSyr ..K.D...................................L.........V................S...............................I............................R..I.........V.M............S...A...T..................A otoGar ........N...............................L..............................................H...........I............................R..I.......S...M.....V..........T......................A micMur I....................M.......R..I......H..V.M.....V......V...T...G..................A tupBel ..GSDA....NLG...S.A..........A..LT..V...L...V.....VTH...............F..................H.R....H....I..........................L.R.A.....GSS..AAM.....L......A...A...G..................A musMus ..SEASD...G.RS.G-....R...SVI.A...V..IT..L..VV.................V.....F..................H......H....I..........V.........M......SC............LSM....................................N..T ratNor ...DA.D...G.GS.G-...TKS..S.I.A...V..I...L.....................V.....F..................H......H...............V.........L......SC........G...LSMV...............V......................T speTri dipOrd .....V....G.R....................T..V...L..L........................L....................R.........I....................I.......H..I..G...R..VTM.......................................T cavPor ...HS........................A...L..L...L.......................M...L..................H........................................R..I.....SHS.V.M.......................................V oryCun ......S....F.H...................L......L..L........................F..................H...........I....................M.......H.........R..L.......V..........A......................T ochPri ...H......EA.V.A.............A...L......L..L........................F..................H...........I....................M.......Q..I......H..F.M................V......................K canFam ......D..T...................A...T..I...F..L.......................................................I....................M.......S..T..........SM.....L.................................A felCat ......D......................A...T. I....................M.......S.NS..........SM.....L.................................A bosTau ...........C...N.............A...T..V...L...........F..................................H...........I............................H..A.....A....SM.....T.................................V turTru ..................T..........A...T.....VL..............................................H...........I............................C.GA..........SM.....F..........V......................V susScr ..LMD...N................S...A...T......L...V.......H...............A..................H...........I............................R.EA..........SM.......................................V vicVic ...............Y......A......A...T......L...V.......H..............................................I....................M.......R..A..........SM............SV......G...............Q..V equCab .............................A..........L.........V.......S..............................R.........I....................L.....T.R..A......S..TSM.....V..........V...N..................A myoLuc ..K...S....F............................L..V.......T...................................H........... ..K...H...SM.......................................V pteVam ..........N......................I......L.........F................................................ ..........IM.......................................A sorAra ..E.RSD...GPTP.G.A.L.R...RL..A...V..V...L..TA.............R.....M...L..................H...R..H......................L........L.R..A..S....S.V..........F.......L.........M.........G..V eriEur ..E.....G...E................A...V. I............................R.HT..S.SA.S..AM.....L..........S...G...............E..G loxAfr ....S.D.........A.......................L..............................................H.R.....T...I............................H.HI.....S...VSM.........L...L..T......................A proCap ...............E.............A..... I..........S.........I.......H.N. echTel ..K..E.T..G.HS.E......A..R...S...T......L..F..................M........................H.............................A..........H..R.....S...V.M........FL...L.V....................E..A dasNov .....T.......D......................I...F......S.............T.....................................I..........T.........M.......R..T.....I....SM................A.....................NI choHof .I...L...V................TV....................................I............................H........I....SM.............L.VT......................G monDom .FK..SVKTLAPEK.GP....PI..K...A...T..V...V..V......V...A...A..T.................................D...I.................A..I.......Q..L.G...SYN.TLM..T..V..F.......V...G..................V macEug .FQ.DS---LEPEK.SY....P.......A...T..V...P.........V.......A..T.....................................I.................A..I.......Q..L ornAna .R..DSA.LLE.EHH.R.A....D.....A...T..IM..V..V......V.FE....A................G...........H......H....I..........S.................R.AI..K..RSN.TAM..A..M..F...S..LL......S...............A galGal .HW.DSA...E.DA.AH...T........A...T..V...F...V.....V.......A.........F......G...........H.......T...I..A.........................R..I......RN.AA...A....AV...S..TV...G..S........A......T taeGut .HW.DSS...E.DD.AH.A.T........A...T..V...F...V.....V.......A.........F......G...........H.......T...I..A.........................R..I......RS.AT...A....AV..SS..TA...............V......V anoCar .FL.DSA...E.DD.PH.A...A......A...T..V..LL...V.....V.......A.........F......G...........H.......T...I..A.................I.......K.HI.S.L.ATN.TT...A....A....S..VV...............V......T xenTro .AGTGTV.I..ASS.VH.....S......A...T..V...L.........V.......A.........F......G...........H.......V...I....................I.......R..I...ISGRH.TAM..A....AV..SV..VV..S...................A danRer .ESGL.NV.AETVYGEK.A.T........A...T..V..LS...V..LM.V.FR....A.........F.....AG...........H.......M...I..A.................I.......R..I.QKL..RS.TL..VA..L.AV..SS...V...G...............N..V takRub ..KVFSLVNISNEV.GK...T.W......G...I..V..LT...V..LM.V.F.....A..F......F.....AG..........IH......HT...I..A.................I...I...R..I..K..VQS.NL...A..L.AV..SS..VV...A...............Q.NV tetNig ..KVFSLVN.SNDV.GK.A.T.W......G...T..I..LT...V..LM.V.F.....A..F......F.....AG..........IH......HT...I..A.................I.......R..I..K..VQS.NL..AA..L.AV..SS..VV...A...............Q.NA gasAcu .DPEVNVTDDVTLYGGK.A.T.L......G...T..V..LF...V..LM.W.F.....A..F......F.....AG..........IH...........I..A.................I.......R..I.QK..MQS.NL...A..L.AV..SS..VV...................Q..A oryLat .DSAVNVSDAAAPYGGK.A.T.L......G...T..V..LS..VL..LM.V.FR....A..I......F..V..AG..........IH.......T...I..A.................I.......R..L.QK..MQS.NL...A..L.AV..SS...V...G...............Q..V calMil homSap MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMAGMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQVYAGLNIFFGMASIGLLTVVAVDRYLTICLPDVGRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR Consen M.............e..S.Fsq.EHnIVA.YLI.AG.iSi.SN..VLgiF.k.keLRT.TNa!IiNLA.TDIGVs.IGYPMSAASDl.GsWkFG..GCQIYAgLNIFFGMaSIGLLTvVA.DRYLTiC.Pd.Gr.mt...Y..mIl.AW.N..FWa.mP..GWA.YAPDPTGATCTiNWR.ND. opsins M........S..................A.Y...........N...L...v..K.LRT..N.I..NLA..D......Gy.......LHG...FG..GC..........G......L.V.A.DRY..VC.P....R.........I...W......A..P..GW..Y.PD.....C.I....... .......................y..............................xx.........x...............xx..........x...............y.........................y....................................................... .......1....1.........2.........2.........2.........2.........2.........2.........2.........2.........2.........2.........3........3..........3.........3.......3 .......8....9.........0.........1.........2.........3.........4.........5.........6.........7.........8.........9.........0........1..........2.........3.......3 .......5678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678901234567890123456789012345678 .......eeeeMMMMMMMMMMMMMMMMMMMMMMMMccccccccccccccccccccccccccccMMMMMMMMMMMMMMMMMMMMMMMMeeeeeeeeMMMMMMMMMMMMMMMMMMMMMMMMMcccccccccccccccccccccccccccccccccccccccc* exon...5555555555555555555555555555555555555555555555555555555566666666666666666666666666666666666666666666666666666666666677777777777777777777777777777777777777 ..........................................................................................................K...................................................... .................................x.y....................................................................................................y...x.................... opsins ...Sf........Fi.PL.V.F.CY.......K....................VT.M.iiMI..FL..W.PY..V.............P....I...FAK.S..YNP.IYV..NK.FR........C........................... Consen SFVSYTM.Vi.iNF..PL.VMfYCYY.V.........s.c....n.DWs.Q.DVTKMSvIMI.MFL.AWSPYSIVCLWasFGDPk.Ipp.mAIiAPLFAKSSTFYNPCIYV.ANKkFRrA..aM..Cqt.q.......lPM......l...... homSap SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTKMSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKKFRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* panTro .........................................................................................................................................................* gorGor ......V................................................. .....................................* ponPyg .........................................................................................................................................................* nomLeu .................................Y....................................................................................K.......WPN.....G.............T..K.* macMul ...................................A..........E..........................................................................................................* papHam ...................................A..........E...............................................................M..........................................* calJac .......A......................F......................................................N...A..................................L............V....I..........* tarSyr T......A...................A.Q.....VA.N.V..........V.... .....F..L....Y.A..A..S...N......T..KN* otoGar .......................L.........S...............................F.........A.A........I.......R..* micMur ...........V..V...M..........Q...R..ATN............V....................A.........N.E....S.....................I.........F.........A......F..G......P...T* tupBel .......A...V..V......S...VL.ARA.AG.AA.S.S.HRC....E.V.......L..L.....................QR...A...V.................L......K..C.........A.S...V...AS.PR...PA.V* musMus .......M..VV................SR.LRLYAA....AH.H...A..A..........L...L............C..N......S.....................A.H....K.........P.LAV.EP.T....MP.SS..PV..* ratNor .......M..VV................SQ.MRLSAA.N..TH.....AH.A.......M..L...L......V.....C..N......SL....................A......K..F..L...P..A..EP.T.A.G.PHS...PA..* speTri ...........V.................R.....AA.N..AY........V..........L...................N......S.....................A...R.....F.........A.....V.......S.R.....* dipOrd ...V..L......................E......................................L......A.....................* cavPor A.................A......L.I.RA.RR.VAG.RPPN.SG.....V.......V..L.....................RR.S.S.....................I.........F...Q.....AV..A......A..S.......* oryCun ....F..A......V..............Q...Q.RA.....Y........V..........F..........................A.....................A...R.....F.........A.....V..........P..I.* ochPri ....F..A..MV..V...........Y..Q......A....K.........V..........L.....................Q....S.....................A...RS....F......IP.AK....LS.R....S..S...T* canFam F...F......V.................R...C....N...Y........V..........F..........................S............................K.IF.........A..G.................N* felCat ....F.........N..............QT..C.D..N..GY......N.V.... ..K..F.....ENR.P................T...K* bosTau .......M.V...................Q.....G.NN...Y........V..........L..........................S.....................I.................T.A.....V.....P....T..KV* turTru .......M.V........IM.........R.....A..N...Y........V..........L..........................SV..L.................I...................A..ME.......P....T..KV* susScr .........V.V......I..........Q...S.A..N..AY........V..........L................................................I...................A..LE.T.....P........V* vicVic .......M.V........I..........R...CRA..N...Y........V..........L...L......................S.....................I...................A..M........P....T...L* equCab .......A.V...................R.M.R.P..N..QY..T.....V..........F..........................S...............................F........RA...........P..Q......* myoLuc ....F........................R...CRA..N...Y.....A..V..........F..........................S...............................F...........TTM.F.....P....T....* pteVam ....F.............A..........R...C....N...Y........V..........F..........................S...............................F......D..S.....V.....P....T....* sorAra .........V.V..VM.....A......IR.L.C...HSSPGH.DG...S.V.......V..LA..A.........F............S..V............................S..LT.RAQGA..AA.T....AAHS.Q....N* eriEur ..............M...A..L.......R.M.C..ARS.A.YVHG.....V..........L..........V...............S...M.................L.........F.........A....NT....IP.K-.D.R.N* loxAfr ..........V...V...A..........R...R..A.N.A.Y........L..........L....................S.....S...............................F.........AE...C....N.......A...* proCap ......V...MV..V...A........I.....C.AARN.A.P.C......L..........L...M.................T....S.........................................AV...N....T....SS.....* echTel .........VG...V...A..........QT..RQ.A.NGA.N.H......L.... .....F.LLQ..PQEARR.......N.....M....L* dasNov .........V....L...A..L.......R......K.N.S.YF.....N.V..........S.....................E....S..............................IF..L......A...M.....N..E........* choHof .........VV...V..VA........I.R...RRNS.N.S.Y........V..........L...L......................S............................TI.F..L......AV........N..E........* monDom ..........T...AM..G.......N.SQKM.QYSP.N.PDHI.....N.VA......V..L...L..................E...A...V.................A........IS..IR.....S..ISNA...N* macEug ..............VM..V.......N.S.KM.QY.R.S.P.HI.....N.V..........L...L......V...........E...A.....................A........IS..MR.E...S...SNA..LNLT* ornAna ..I........V..A...I.......N.SKAMRQYPA.RVL.N..I...E.V.......V..L...M...........S........S.AV..M............................S.VQ....REITI.DV...NR.RS..TL galGal .......S...V..V...........N.SRTM.QY.S.N.L..I.M.....V.......V..V...............S........S.A.....................I........I...VR...R.EITISNA...T..LSA.T. taeGut ..I....S...V..V...........N.SRTM.QYAS.N.L..I.I.....V.......V..I...............S........S.A.....................I........I...VR...R.EITINNA...S...SA.T.QNS* anoCar .......S...V..VI..S.......N.SKTM.YYMRNS.L.NI.I.....V.......V..I...L...........S........S.A...V.................I...R....I...IR...R.EITINNV...S...STI. xenTro .......S.V.V..V...M.......N.SRTM.GYGSRSSLGGI.A.....T......MV..V...............S.....R....A.....................I........I.S.VQ.KSR.EVTLDNHF..N...ST.TT* danRer ..........TV...I..S.......N.SATV.RFKA.N.LD.I.M.....M..........V...A.................Q...A........L.............I........IIG.IR...R.RVTINNQ...MA.SV..NP* takRub ..I....A..GV..ML..F.......N.SVTVNRCK..N.LDDI.IE..E.M......LV..I......................A..A......................I........IIG.IR...R.Q.TINTEI..TT..QTATQ* tetNig ..I....A..SV...L..F.......N.SVTV.QYKANN.LDNI.IE..E.M......IV..I......................T.SA......................IT.....Q.IIG.IR...R.QITINTDI..TA..QT.TQ* gasAcu ..I....A...V..VL..SA......N.SATV.RYKA.N.LD.A.I.....M......IV..I......................T..A......................I........IIG.VR...R.RITIN.QV..TT..Q..TQ* oryLat .......A...V..V...........N.SATV.RYKA.N.LD.I.I.....M......IV..V..........M...........T..A......................I........IIG.IR...R.RITISTQV..TI..Q..TQ* calMil L..........V..V...S...F...N.SKTMSRFIS.PSP.NI.L.....L.......V..V...L...............N..L...A.....................I......K.IM..IC..NR.EITINHT...TI.RV..TE* homSap SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTKMSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKKFRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* Consen SFVSYTM.Vi.iNF..PL.VMfYCYY.V.........s.c....n.DWs.Q.DVTKMSvIMI.MFL.AWSPYSIVCLWasFGDPk.Ipp.mAIiAPLFAKSSTFYNPCIYV.ANKkFRrA..aM..Cqt.q.......lPM......l..... opsins ...Sf........Fi.PL.V.F.CY.......K....................VT.M.iiMI..FL..W.PY..V.............P....I...FAK.S..YNP.IYV..NK.FR........C.......................... .................................x.y....................................................................................................y...x...................
Curated Set of 50 deuterostome peropsins
The sequences below represent all that are available as of March 2008. A few sequences such as chondrichthyes are incomplete. Note the main reference sequence collection contains additional deuterostome and even some lophotrochozoan peropsins.
>PER_homSap Homo sapiens (human) 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDV 1 2 GRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 >PER_panTro Pan troglodytes (chimp) 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDV 1 2 GRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 >PER_gorGor Gorilla gorilla (gorilla) 0 MLRNNLGNGSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDV 1 2 GRRMTTNTYIGLILGAWINGLLWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTVTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTK 0 0 2 1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 >PER_ponPyg Pongo pygmaeus (orang_abelii) 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GIISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDI 1 2 GRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHTTSDCTESLNRDWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 >PER_nomLeu Nomascus leucogenys (gibbon) 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISIISNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDV 1 2 GRRMTTNTYIGLILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKYHTTSDCTESLNRDWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRKAMLAMFKWPNHQTMPGTSILPMDVSQNPLTSGKI* 0 >PER_macMul Macaca mulatta (rhesus) 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISILSNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDI 1 2 GRRMTTNTYIGMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHATSDCTESLNREWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 >PER_papHam Papio hamadryas (baboon) 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISILSNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICLPDI 1 2 GRRMTTNTYIGMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMTVIAINFIVPLTVMFYCYYHVTLSIKHHATSDCTESLNREWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYMVANKK 2 1 FRRAMLAMFKCQTHQTMPVTSILPMDVSQNPLASGRI* 0 >PER_calJac Callithrix jacchus (marmoset) 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVASYLIMA 1 2 GMISILSNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAADRYLTICLPDI 1 2 GRRMTTSTYIIMILGAWINGLFWALMPIVGWASYAPDPTGATCTINWRKNDR 2 1 SFVSYTMAVIAINFIVPLTVMFYCYYHVTLFIKHHTTSDCTESLNRDWSDQIDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWASFGDPKNIPPAMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMLKCQTHQTMPVTSVLPMDISQNPLASGRI* 0 >PER_tarSyr Tarsius syrichta (tarsier) 0 MLKNDLGNSSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISILSNIIVLGIFVKYKELRTPTNAIIINLSVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTICRPDI 1 2 GRRMTTNTYVGMILGAWINGLFWASMPIAGWATYAPDPTGATCTINWRKNDA 2 1 TFVSYTMAVIAINFIVPLTVMFYCYYHATQSIKHHVASNCVESLNRDWSDQVDVTK 0 0 2 1 FRRAMFAMLKCQTYQAMPATSSLPMNVSQNPLTSGKN* 0 >PER_otoGar Otolemur garnettii (bushbaby) 0 MLRNNLGNNSDSKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISILSNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTICRPDI 1 2 GRRMTTNSYIGMILGAWVNGLFWALMPITGWASYAPDPTGATCTINWRKNDA 2 1 SFVSYTMTVIAVNFVVPLMVMFYCYYHVTQSIKRHTATNCTESLNRDWSDQVDVTK 0 0 MSVIMICMFLVAWSPYSIVCLWALFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMFAMFKCQTHQAMAVTSILPMDISQNPLASRRI* 0 >PER_micMur Microcebus murinus (mouse_lemur) 0 1 2 0 0 IYAGLNIFFGMASIGLLTVVAMDRYLTICRPDI 1 2 GRRMTTHTYVGMILGAWVNGLFWAVMPITGWAGYAPDPTGATCTINWRKNDA 2 1 0 0 MSVIMICMFLVAWSPYAIVCLWASFGNPEKIPPSMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAMFAMFKCQTHQAMPVTSIFPMGVSQNPLPSGRT* 0 >PER_tupBel Tupaia belangeri (treeshrew) 0 MLGSDAGNSSNLGNEDSSAFSQTEHNIVAAYLLTA 1 2 GVISILSNIVVLGIFVTHKELRTPTNAIIINLAFTDIGVSSIGYPMSAASDLHGRWKFGHAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTLCRPAV 1 2 GRRMGSSTYAAMILGAWLNGLFWAAMPIAGWAGYAPDPTGATCTINWRKNDA 2 1 SFVSYTMAVIAVNFVVPLTVMSYCYVLVARAIAGHAASSCSEHRCRDWSEQVDVTK 0 0 MSVLMILMFLVAWSPYSIVCLWASFGDPQRIPPAMAIVAPLFAKSSTFYNPCIYVLANKK 2 1 FRKAMCAMFKCQTHQAMSVTSVLPMASSPRPLAPARV* 0 >PER_musMus Mus musculus (mouse) 0 MLSEASDNSSGSRSE-GSVFSRTEHSVIAAYLIVA 1 2 GITSILSNVVVLGIFIKYKELRTPTNAVIINLAFTDIGVSSIGYPMSAASDLHGSWKFGHAGCQ 0 0 IYAGLNIFFGMVSIGLLTVVAMDRYLTISCPDV 1 2 GRRMTTNTYLSMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRNNDT 2 1 SFVSYTMMVIVVNFIVPLTVMFYCYYHVSRSLRLYAASDCTAHLHRDWADQADVTK 0 0 MSVIMILMFLLAWSPYSIVCLWACFGNPKKIPPSMAIIAPLFAKSSTFYNPCIYVAAHKK 2 1 FRKAMLAMFKCQPHLAVPEPSTLPMDMPQSSLAPVRI* 0 >PER_ratNor Rattus norvegicus (rat) 0 MLRDALDNSSGSGSE-GSVFTKSEHSIIAAYLIVA 1 2 GIISILSNIIVLGIFIKYKELRTPTNAVIINLAFTDIGVSSIGYPMSAASDLHGSWKFGHAGCQ 0 0 VYAGLNIFFGMVSIGLLTVVALDRYLTISCPDV 1 2 GRRMTGNTYLSMVLGAWINGLFWALMPIVGWASYAPDPTGATCTINWRKNDT 2 1 SFVSYTMMVIVVNFIVPLTVMFYCYYHVSQSMRLSAASNCTTHLNRDWAHQADVTK 0 0 MSVMMILMFLLAWSPYSVVCLWACFGNPKKIPPSLAIIAPLFAKSSTFYNPCIYVAANKK 2 1 FRKAMFAMLKCQPHQAMPEPSTLAMGVPHSPLAPARI* 0 >PER_speTri Spermophilus tridecemlineatus (ground_squirrel) 0 1 2 0 0 1 2 2 1 SFVSYTMTVIAVNFIVPLTVMFYCYYHVTRSIKHHAASNCTAYLNRDWSDQVDVTK 0 0 MSVIMILMFLVAWSPYSIVCLWASFGNPKKIPPSMAIIAPLFAKSSTFYNPCIYVAANKR 2 1 FRRAMFAMFKCQTHQAMPVTSVLPMDVSQSPRASGRI* 0 >PER_dipOrd Dipodomys ordii (kangaroo_rat) 0 MLRNNVGNSSGSRNEDGSVFSQTEHNIVATYLITA 1 2 GVISILSNLIVLGIFIKYKELRTPTNAIIINLALTDIGVSSIGYPMSAASDLYGRWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAIDRYLTICHPDI 1 2 GRGMTTRTYVTMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDT 2 1 0 0 MSVVMILMFLVAWSPYSIVCLWASFGDPKEIPPPMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMLKCQTHQAMPVTSILPMDVSQNPLASGRI* 0 >PER_cavPor Cavia porcellus (guinea_pig) 0 MLRHSLGNSSDSKNEDGSVFSQTEHNIVAAYLILA 1 2 GLISILSNIIVLGIFIKYKELRTPTNAIIMNLALTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTVVAVDRYLTICRPDI 1 2 GRRMTSHSYVGMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDV 2 1 AFVSYTMTVIAINFIVPLAVMFYCYLHITRAIRRHVAGDRPPNLSGDWSDQVDVTK 0 0 MSVVMILMFLVAWSPYSIVCLWASFGDPRRISPSMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAMFAMFQCQTHQAVPVASILPMDASQSPLASGRI* 0 >PER_oryCun Oryctolagus cuniculus (rabbit) 0 MLRNNLSNSSDFKHEDGSVFSQTEHNIVATYLILA 1 2 GMISILSNLIVLGIFIKYKELRTPTNAIIINLAFTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAMDRYLTICHPDV 1 2 GRRMTTRTYLGLILGAWVNGLFWALMPIAGWASYAPDPTGATCTINWRKNDT 2 1 SFVSFTMAVIAINFVVPLTVMFYCYYHVTQSIKQHRASDCTEYLNRDWSDQVDVTK 0 0 MSVIMIFMFLVAWSPYSIVCLWASFGDPKKIPPAMAIIAPLFAKSSTFYNPCIYVAANKR 2 1 FRRAMFAMFKCQTHQAMPVTSVLPMDVSQNPLPSGII* 0 >PER_ochPri Ochotona princeps (pika) 0 MLRHNLGNSSEAKVEAGSVFSQTEHNIVAAYLILA 1 2 GMISILSNLIVLGIFIKYKELRTPTNAIIINLAFTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAMDRYLTICQPDI 1 2 GRRMTTHTYFGMILGAWINGLFWALMPIVGWASYAPDPTGATCTINWRKNDK 2 1 SFVSFTMAVIMVNFVVPLTVMFYCYYYVTQSIKHHTASDCTKSLNRDWSDQVDVTK 0 0 MSVIMILMFLVAWSPYSIVCLWASFGDPQKIPPSMAIIAPLFAKSSTFYNPCIYVAANKR 2 1 SRRAMFAMFKCQIPQAKPVTSLSPRDVSQSPLSSGRT* 0 >PER_canFam Canis familiaris (dog) 0 MLRNNLDNSTDSKNEDGSVFSQTEHNIVAAYLITA 1 2 GIISIFSNLIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAMDRYLTICSPDT 1 2 GRRMTTNTYISMILGAWLNGLFWALMPIIGWASYAPDPTGATCTINWRKNDA 2 1 FFVSFTMTVIAVNFIVPLTVMFYCYYHVTRSIKCHTTSNCTEYLNRDWSDQVDVTK 0 0 MSVIMIFMFLVAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRKAIFAMFKCQTHQAMPGTSILPMDVSQNPLASGRN* 0 >PER_felCat Felis catus (cat) 0 MLRNNLDNSSDSKNEDGSVFSQTEHNIVAAYLITA 1 2 0 0 IYAGLNIFFGMASIGLLTVVAMDRYLTICSPNS 1 2 GRRMTTNTYISMILGAWLNGLFWALMPIIGWASYAPDPTGATCTINWRKNDA 2 1 SFVSFTMTVIAINFNVPLTVMFYCYYHVTQTIKCHDTSNCTGYLNRDWSNQVDVTK 0 0 2 1 FRKAMFAMFKCENRQPMPVTSILPMDVSQNPLTSGRK* 0 >PER_bosTau Bos taurus (cow) 0 MLRNNLGNSSDCKNENGSVFSQTEHNIVAAYLITA 1 2 GVISILSNIIVLGIFIKFKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTICHPDA 1 2 GRRMTANTYISMILGAWTNGLFWALMPIIGWASYAPDPTGATCTINWRKNDV 2 1 SFVSYTMMVVAINFIVPLTVMFYCYYHVTQSIKHHGTNNCTEYLNRDWSDQVDVTK 0 0 MSVIMILMFLVAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAMLAMFKCQTTQAMPVTSVLPMDVPQNPLTSGKV* 0 >PER_turTru Tursiops truncatus (dolphin) 0 MLRNNLGNSSDSKNEDGSTFSQTEHNIVAAYLITA 1 2 GMISVLSNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTICCPGA 1 2 GRRMTTNTYISMILGAWFNGLFWALMPIVGWASYAPDPTGATCTINWRKNDV 2 1 SFVSYTMMVVAINFIVPLIMMFYCYYHVTRSIKHHATSNCTEYLNRDWSDQVDVTK 0 0 MSVIMILMFLVAWSPYSIVCLWASFGDPKKIPPSVAILAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAMLAMFKCQTHQAMPMESILPMDVPQNPLTSGKV* 0 >PER_susScr Sus scrofa (pig) 0 MLMDLGNNSDSKNEDGSVFSQTEHSIVAAYLITA 1 2 GMISILSNIVVLGIFIKHKELRTPTNAIIINLAATDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTICRPEA 1 2 GRRMTTNTYISMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDV 2 1 SFVSYTMTVVAVNFIVPLIVMFYCYYHVTQSIKSHATSNCTAYLNRDWSDQVDVTK 0 0 MSVIMILMFLVAWSPYSIVCLWASFGDPKKIPPPMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAMLAMFKCQTHQAMPLESTLPMDVPQNPLASGRV* 0 >PER_vicVic Vicugna vicugna (vicugna) 0 MLRNNLGNSSDSKNEYGSVFSQAEHNIVAAYLITA 1 2 LRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAMDRYLTICRPDA 1 2 GRRMTTNTYISMILGAWINGLFWASVPIIGWAGYAPDPTGATCTINWRQNDV 2 1 SFVSYTMMVVAINFIVPLIVMFYCYYHVTRSIKCRATSNCTEYLNRDWSDQVDVTK 0 0 MSVIMILMFLLAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAMLAMFKCQTHQAMPMTSILPMDVPQNPLTSGRL* 0 >PER_equCab Equus caballus (horse) 0 MLRNNLGNSSDSKNEDGSVFSQTEHNIVAAYLIMA 1 2 GMISILSNIIVLGIFVKYKELRTSTNAIIINLAVTDIGVSSIGYPMSAASDLYGRWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVALDRYLTTCRPDA 1 2 GRRMTTSTYTSMILGAWVNGLFWALMPIVGWANYAPDPTGATCTINWRKNDA 2 1 SFVSYTMAVVAINFIVPLTVMFYCYYHVTRSMKRHPTSNCTQYLNTDWSDQVDVTK 0 0 MSVIMIFMFLVAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMFAMFKCQTHRAMPVTSILPMDVPQNQLASGRI* 0 >PER_myoLuc Myotis lucifugus (microbat) 0 MLKNNLSNSSDFKNEDGSVFSQTEHNIVATYLIMA 1 2 GMISILSNVIVLGIFITYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0 0 1 2 GRKMTTHTYISMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDV 2 1 SFVSFTMTVIAINFIVPLTVMFYCYYHVTRSIKCRATSNCTEYLNRDWADQVDVTK 0 0 MSVIMIFMFLVAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMFAMFKCQTHQTMTTMSFLPMDVPQNPLTSGRI* 0 >PER_pteVam Pteropus vampyrus (macrobat) 0 MLRNNLGNSSNSKNEDGSVFSQTEHNIVATYLIIA 1 2 GMISILSNIIVLGIFFKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 1 2 GRRMTTNTYIIMILGAWINGLFWALMPIIGWASYAPDPTGATCTINWRKNDA 2 1 SFVSFTMTVIAINFIVPLAVMFYCYYHVTRSIKCHTTSNCTEYLNRDWSDQVDVTK 0 0 MSVIMIFMFLVAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMFAMFKCQDHQSMPVTSVLPMDVPQNPLTSGRI* 0 >PER_sorAra Sorex araneus (shrew) 0 MLENRSDNSSGPTPEGGAVLSRTEHRLVAAYLIVA 1 2 GVISILSNTAVLGIFIKYKELRTRTNAIIMNLALTDIGVSSIGYPMSAASDLHGSWRFGHAGCQ 0 0 VYAGLNIFFGMASIGLLTLVAVDRYLTLCRPDA 1 2 GRSMTTNSYVGLILGAWINGFFWALMPILGWASYAPDPMGATCTINWRGNDV 2 1 SFVSYTMTVVAVNFVMPLTVMAYCYYHVIRSL-----HSSPGHLDGDWSSQVDVTK 0 0 MSVVMILAFLAAWSPYSIVCFWASFGDPKKIPPSMAVIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMSAMLTCRAQGAMPAASTLPMDAAHSPQASGRN* 0 >PER_eriEur Erinaceus europaeus (hedgehog) 0 MLENNLGNGSDSENEDGSVFSQTEHNIVAAYLIVA 1 2 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTICRPHT 1 2 GRSMSANSYIAMILGAWLNGLFWALMPISGWAGYAPDPTGATCTINWRENDG 2 1 SFVSYTMTVIAINFMVPLAVMLYCYYHVTRSMKCHTARSCAEYVHGDWSDQVDVTK 0 0 MSVIMILMFLVAWSPYSVVCLWASFGDPKKIPPSMAIMAPLFAKSSTFYNPCIYVLANKK 2 1 FRRAMFAMFKCQTHQAMPVTNTLPMDIPQK* 0LDSRRN* 0 >PER_loxAfr Loxodonta africana (elephant) 0 MLRNSLDNSSDSKNEDASVFSQTEHNIVATYLIMA 1 2 GMISILSNIIVLGIFIKYKELRTPTNAIIINLAVTDIGVSSIGYPMSAASDLHGRWKFGYTGCQ 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTICHPHI 1 2 GRRMTSNTYVSMILGAWINGLLWALLPITGWASYAPDPTGATCTINWRKNDA 2 1 SFVSYTMTVIVINFVVPLAVMFYCYYHVTRSIKRHTASNCAEYLNRDWSDQLDVTK 0 0 MSVIMILMFLVAWSPYSIVCLWASFGDSKKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMFAMFKCQTHQAEPVTCILPMNVSQNPLAAGRI* 0 >PER_proCap Procavia capensis (hyrax) 0 MLRNNLGNSSDSKNEEGSVFSQTEHNIVAAYLIMA 1 2 0 0 IYAGLNIFFGMSSIGLLTVVAIDRYLTICHPNV 1 2 2 1 SFVSYTVTVIMVNFVVPLAVMFYCYYHITLSIKCHAARNCAEPLCRDWSDQLDVTK 0 0 MSVIMILMFLMAWSPYSIVCLWASFGDPTKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLAMFKCQTHQAVPVTNILPMTVSQNSSASGRI* 0 >PER_echTel Echinops telfairi (tenrec) 0 MLKNNEGTSSGSHSEEGSVFSQAEHRIVASYLITA 1 2 GMISILSNFIVLGIFIKYKELRTPTNAMIINLAVTDIGVSSIGYPMSAASDLHGSWKFGYAGCQ 0 0 VYAGLNIFFGMASIGLLTAVAVDRYLTICHPDR 1 2 GRRMTSNTYVGMILGAWINGFLWALLPVIGWASYAPDPTGATCTINWRENDA 2 1 SFVSYTMTVVGINFVVPLAVMFYCYYHVTQTIKRQTASNGAENLHRDWSDQLDVTK 0 0 2 1 FRRAMFALLQCQPQEARRVTSILPMNVSQNPMASGRL* 0 >PER_dasNov Dasypus novemcinctus (armadillo) 0 MLRNNTGNSSDSKDEDGSVFSQTEHNIVATYLIMA 1 2 GIISIFSNIIVLSIFIKYKELRTPTNTIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 IYAGLNIFFGMTSIGLLTVVAMDRYLTICRPDT 1 2 GRRMTINTYISMILGAWINGLFWALMPIAGWASYAPDPTGATCTINWRKNNI 2 1 SFVSYTMTVVAINFLVPLAVMLYCYYHVTRSIKHHTKSNCSEYFNRDWSNQVDVTK 0 0 MSVIMISMFLVAWSPYSIVCLWASFGDPEKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAIFAMLKCQTHQAMPVMSILPMNVSENPLASGRI* 0 >PER_choHof Choloepus hoffmanni (sloth) 0 1 2 GIISILSNIVVLGIFIKYKELRTPTNTVIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTVVAVDRYLTICHPDV 1 2 GRRMTINTYISMILGAWINGLFWALLPVTGWASYAPDPTGATCTINWRKNDG 2 1 SFVSYTMTVVVINFVVPVAVMFYCYYHITRSIKRRNSSNCSEYLNRDWSDQVDVTK 0 0 MSVIMILMFLLAWSPYSIVCLWASFGDPKKIPPSMAIIAPLFAKSSTFYNPCIYVVANKK 2 1 FRTIMFAMLKCQTHQAVPVTSILPMNVSENPLASGRI* 0 >PER_macEug Macropus eugenii (wallaby) 0 MFQNDSLEPEKESYSVFSPTEHNIVAAYLITA 1 2 GVISIPSNIIVLGIFVKYKELRTATNTIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYAGCQ 0 0 IYAGLNIFFGMASIGLLTAVAIDRYLTICQPDL 1 2 2 1 SFVSYTMTVIAINFVMPLVVMFYCYYNVSLKMKQYTRSSCPEHINRDWSNQVDVTK 0 0 MSVIMILMFLLAWSPYSVVCLWASFGDPKEIPPAMAIIAPLFAKSSTFYNPCIYVAANKK 2 1 FRRAISAMMRCETHQSMPVSNALPLNLT* 0 >PER_monDom Monodelphis domestica (opossum) 0 MFKNNSVKTLAPEKEGPSVFSPIEHKIVAAYLITA 1 2 GVISIVSNVIVLGIFVKYKALRTATNTIIINLAVTDIGVSSIGYPMSAASDLYGSWKFGYDGCQ 0 0 IYAGLNIFFGMASIGLLTAVAIDRYLTICQPDL 1 2 GGRMTSYNYTLMILTAWVNGFFWALMPIVGWAGYAPDPTGATCTINWRKNDV 2 1 SFVSYTMTVITINFAMPLGVMFYCYYNVSQKMKQYSPSNCPDHINRDWSNQVAVTK 0 0 MSVVMILMFLLAWSPYSIVCLWASFGDPKEIPPAMAIVAPLFAKSSTFYNPCIYVAANKK 2 1 FRRAISAMIRCQTHQSMPISNALPMN* 0 >PER_ornAna Ornithorhynchus anatinus (platypus) 0 MRRNDSANLLESEHHDRSAFSQTDHNIVAAYLITA 1 2 GIMSIVSNVIVLGIFVKFEELRTATNAIIINLAVTDIGVSGIGYPMSAASDLHGSWKFGHAGCQ 0 0 IYAGLNIFFGMSSIGLLTVVAVDRYLTICRPAI 1 2 GRKMTRSNYTAMILAAWMNGFFWASMPLLGWASYASDPTGATCTINWRKNDA 2 1 SFISYTMTVIAVNFAVPLIVMFYCYYNVSKAMRQYPASRVLENLNIDWSEQVDVTK 0 0 MSVVMILMFLMAWSPYSIVCLWSSFGDPKKISPAVAIMAPLFAKSSTFYNPCIYVVANKK 2 1 FRRAMLSMVQCQTHREITITDVLPMNRSRSPLTL* 0 >PER_galGal Gallus gallus (chicken) 0 MHWNDSANSSESDAEAHSVFTQTEHNIVAAYLITA 1 2 GVISIFSNIVVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYTGCQ 0 0 IYAALNIFFGMASIGLLTVVAVDRYLTICRPDI 1 2 GRRMTTRNYAALILAAWINAVFWASMPTVGWAGYASDPTGATCTANWRKNDV 2 1 SFVSYTMSVIAVNFVVPLTVMFYCYYNVSRTMKQYTSSNCLESINMDWSDQVDVTK 0 0 MSVVMIVMFLVAWSPYSIVCLWSSFGDPKKISPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILAMVRCQTRQEITISNALPMTVSLSALTS* 0 >PER_taeGut Taeniopygia guttata (finch) 0 MHWNDSSNSSESDDEAHSAFTQTEHNIVAAYLITA 1 2 GVISIFSNIVVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYTGCQ 0 0 IYAALNIFFGMASIGLLTVVAVDRYLTICRPDI 1 2 GRRMTTRSYATLILAAWINAVFWSSMPTAGWASYAPDPTGATCTVNWRKNDA 2 1 SFISYTMSVIAVNFVVPLTVMFYCYYNVSRTMKQYASSNCLESINIDWSDQVDVTK 0 0 MSVVMIIMFLVAWSPYSIVCLWSSFGDPKKISPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILAMVRCQTRQEITINNALPMSVSQSALTSQNSSHLPA* 0 >PER_anoCar Anolis carolinensis (lizard) 0 MFLNDSANSSESDDEPHSAFSQAEHNIVAAYLITA 1 2 GVISLLSNIVVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYTGCQ 0 0 IYAALNIFFGMASIGLLTVVAIDRYLTICKPHI 1 2 GSRLTATNYTTLILAAWINALFWASMPVVGWASYAPDPTGATCTVNWRKNDT 2 1 SFVSYTMSVIAVNFVIPLSVMFYCYYNVSKTMKYYMRNSCLENINIDWSDQVDVTK 0 0 MSVVMIIMFLLAWSPYSIVCLWSSFGDPKKISPAMAIVAPLFAKSSTFYNPCIYVIANKR 2 1 FRRAILAMIRCQTRQEITINNVLPMSVSQSTIA* 0 >PER_xenTro Xenopus tropicalis (frog) 0 METLAEVSTLLPAGTGTVNISDASSEVHSVFSQSEHNIVAAYLITA 1 2 GVISILSNIIVLGIFVKYKELRTATNAIIINLAFTDIGVSGIGYPMSAASDLHGSWKFGYVGCQ 0 0 IYAGLNIFFGMASIGLLTVVAIDRYLTICRPDI 1 2 GRRISGRHYTAMILAAWINAVFWSVMPVVGWSSYAPDPTGATCTINWRKNDV 2 1 SFVSYTMSVVAVNFVVPLMVMFYCYYNVSRTMKGYGSRSSLGGINADWSDQTDVTK 0 0 MSMVMIVMFLVAWSPYSIVCLWSSFGDPRKIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAILSMVQCKSRQEVTLDNHFPMNVSQSTLTT* 0 >PER_danRer Danio rerio (zebrafish) 0 MESGLLNVSAETVYGEKSAFTQTEHNIVAAYLITA 1 2 GVISLSSNIVVLLMFVKFRELRTATNAIIINLAFTDIGVAGIGYPMSAASDLHGSWKFGYMGCQ 0 0 IYAALNIFFGMASIGLLTVVAIDRYLTICRPDI 1 2 GQKLTTRSYTLLIVAAWLNAVFWSSMPIVGWAGYAPDPTGATCTINWRNNDT 2 1 SFVSYTMTVITVNFIIPLSVMFYCYYNVSATVKRFKASNCLDSINMDWSDQMDVTK 0 0 MSVIMIVMFLAAWSPYSIVCLWASFGDPQKIPAPMAIIAPLLAKSSTFYNPCIYVIANKK 2 1 FRRAIIGMIRCQTRQRVTINNQLPMMASSVPLNP* 0 >PER_takRub Takifugu rubripes (fugu) 0 MKVFSLVNISNEVEGKSVFTQWEHNIVAGYLIIA 1 2 GVISLTSNIVVLLMFVKFKELRTATNFIIINLAFTDIGVAGIGYPMSAASDIHGSWKFGHTGCQ 0 0 IYAALNIFFGMASIGLLTVVAIDRYITICRPDI 1 2 GRKMTVQSYNLLILAAWLNAVFWSSMPVVGWAAYAPDPTGATCTINWRQNNA 2 1 SFISYTMAVIGVNFMLPLFVMFYCYYNVSVTVNRCKTSNCLDDINIEWSEQMDVTK 0 0 MSLVMIIMFLVAWSPYSIVCLWASFGDPKAIPAPMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAIIGMIRCQTRQQMTINTEIPMTTSQQTATQ* 0 >PER_tetNig Tetraodon nigroviridis (pufferfish) 0 MKSVNSSNDVEGKSAFTQWEHNIVAGYLITA 1 2 GIISLTSNIVVLLMFVKFKELRTATNFIIINLAFTDIGVAGIGYPMSAASDIHGSWKFGHTGCQ 0 0 IYAALNIFFGMASIGLLTVVAIDRYLTICRPDI 1 2 GRKMTVQSYNLLIAAAWLNAVFWSSMPVVGWAAYAPDPTGATCTINWRQNNV 2 1 SFISYTMAVISVNFILPLFVMFYCYYNVSVTVKQYKANNCLDNINIEWSEQMDVTK 0 0 MSIVMIIMFLVAWSPYSIVCLWASFGDPKTISAPMAIIAPLFAKSSTFYNPCIYVITNKK 2 1 FRQAIIGMIRCQTRQQITINTDIPMTASQQTLTQ* 0 >PER_gasAcu Gasterosteus aculeatus (stickleback) 0 MGIDPEVNVTDDVTLYGGKSAFTQLEHNIVAGYLITA 1 2 GVISLFSNIVVLLMFWKFKELRTATNFIIINLAFTDIGVAGIGYPMSAASDIHGSWKFGYAGCQ 0 0 IYAALNIFFGMASIGLLTVVAIDRYLTICRPDI 1 2 GQKMTMQSYNLLILAAWLNAVFWSSMPVVGWASYAPDPTGATCTINWRQNDV 2 1 SFISYTMAVIAVNFVLPLSAMFYCYYNVSATVKRYKASNCLDSANIDWSDQMDVTK 0 0 MSIVMIIMFLVAWSPYSIVCLWASFGDPKTIPAPMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAIIGMVRCQTRQRITINSQVPMTTSQQPLTQ* 0 >PER_oryLat Oryzias latipes (medaka) 0 DSAVNVSDAAAPYGGKSAFTQLEHNIVAGYLITA 1 2 GVISLSSNVLVLLMFVKFRELRTATNIIIINLAFTDVGVAGIGYPMSAASDIHGSWKFGYTGCQ 0 0 IYAALNIFFGMASIGLLTVVAIDRYLTICRPDL 1 2 GQKMTMQSYNLLILAAWLNAVFWSSMPIVGWAGYAPDPTGATCTINWRQNDA 2 1 SFVSYTMAVIAVNFVVPLTVMFYCYYNVSATVKRYKASNCLDSINIDWSDQMDVTK 0 0 MSIVMIVMFLVAWSPYSMVCLWASFGDPKTIPAPMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRRAIIGMIRCQTRQRITISTQVPMTISQQPLTQ* 0 >PER_calMil Callorhinchus milii (elephantfish) 0 1 2 0 0 1 2 2 1 LFVSYTMTVIAVNFVVPLSVMFFCYYNVSKTMSRFISSPSPENINLDWSDQLDVTK 0 0 MSVVMIVMFLLAWSPYSIVCLWASFGNPKLIPPAMAIIAPLFAKSSTFYNPCIYVIANKK 2 1 FRKAIMAMICCQNRQEITINHTLPMTISRVPLTE* 0 >PER1a_sacKol Saccoglossus kowalevskii (acornworm) provisional est FF635894 + trc .tg 0 MVTTDSLANSTDEPVPSILTLQQHYAASVTLLAL 1 2 AVIGTVLSSVNFRMLLSNPDYCSKAGNFFLSLAVTDLC 1 2 SVCIFETPFSAFSHHAGFWIFGDTACQ LYAFFGIFFGLVNIFMVTFISLDRYWATCSPVEV 1 2 MELKSKYYTRMTALGWMVALFWAAAPVFGWSRYFMEPSMASCSIDYMTNDF SYVTYITCLTLTCYVVPIVVMVYCYVKASKNIKYTGKVTEWAHENNATK 0 0 ISRLCVLQLVFCWSLYGFNCMWTVVADDVETLPKMLTVLAPILAKTTPILNSGLYFLHNKK FRGAAVDMFKAKEE* 0 >PER1b_sacKol Saccoglossus kowalevskii best hit: PERa_braFlo e = -49 Identities = 97/246 (39%) IIYYFFLLSTGLTIFGMSLSCVSSFAGRWLFGKFGCYFHGFAGMLFGLGSIGNLTVISIDRYIITCKRNL 1 2 WSYRHYYALLAVAWSNALFWSMMPLFGWSSYALEPEGTSCTIDWMNNDNQYISYVSCVTVTCFILPCAVMTYDYLAAYMKMVKAGYTLSEETEKPNND 0 0 MCIALVAAFLLSWFPSATVFLWAAFGNPGNIPLSFTGVADAFTKIPAVFNPVIYVALNPEFRKYFGKTIGCRRKRKKPIAVRLNGSEQNVENTI* 0