Phylogenetic Tree: Difference between revisions
Tomemerald (talk | contribs) |
m (→Web tools for drawing phylogenetic trees from Newick format: editing phyloPng url) |
||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 49: | Line 49: | ||
=== Web tools for drawing phylogenetic trees from Newick format === | === Web tools for drawing phylogenetic trees from Newick format === | ||
After grouping the species with nested parentheses (Newick format) that can include divergence dates or substitution rates, the tree can be drawn with various online tools such as [http://genome | After grouping the species with nested parentheses (Newick format) that can include divergence dates or substitution rates, the tree can be drawn with various online tools such as [http://genome.ucsc.edu/cgi-bin/phyloPng phyloPng], [http://iubio.bio.indiana.edu/treeapp/treeprint-sample1.html Phylodendron], or [http://cgi-www.daimi.au.dk/cgi-chili/phyfi/go PhyFi.] | ||
Here is a simple example of Newick format: (((((human,chimp),gorilla),orang),gibbon),rhesus); | Here is a simple example of Newick format: (((((human,chimp),gorilla),orang),gibbon),rhesus); | ||
| Line 119: | Line 119: | ||
=== The UCSC 100-way vertebrate genome phylogenetic tree in Newick format === | === The UCSC 100-way vertebrate genome phylogenetic tree in Newick format === | ||
Either of the two representations of the vertebrate genome phylogenetic tree, when pasted into the UCSC tree drawing utility [http://genome.ucsc.edu/cgi-bin/ | Either of the two representations of the vertebrate genome phylogenetic tree, when pasted into the UCSC tree drawing utility [http://genome.ucsc.edu/cgi-bin/phyloPng phyloPng], will reproduce species relatedness used at UCSC tracks and resources. Note coding genes are given for the 100-way in the order listed (see proteinFasta on gene details page) as both nucleotide and amino acid exonic format. | ||
The first representation shows various higher order tax as individual lines, for example great apes as the first line. (Line returns are not part of Newick format but are generally ignored by drawing tools.) This display makes it slightly easier to add new species. Thus, to add echidna (Tachyglossus aculeatus), the monotreme line can be hand-edited: ornAna), --> (tacAcu,ornAna), as this genome becomes available. | The first representation shows various higher order tax as individual lines, for example great apes as the first line. (Line returns are not part of Newick format but are generally ignored by drawing tools.) This display makes it slightly easier to add new species. Thus, to add echidna (Tachyglossus aculeatus), the monotreme line can be hand-edited: ornAna), --> (tacAcu,ornAna), as this genome becomes available. | ||
| Line 125: | Line 125: | ||
The second representation is linearized to spreadsheet format. This separates the genus species list from parenthetic nesting used to describe topology. Branching times (or evolutionary rates) can be taken from the literature and can be inserted (as shown above) to draw trees whose horizontal lines have quantitative significance. | The second representation is linearized to spreadsheet format. This separates the genus species list from parenthetic nesting used to describe topology. Branching times (or evolutionary rates) can be taken from the literature and can be inserted (as shown above) to draw trees whose horizontal lines have quantitative significance. | ||
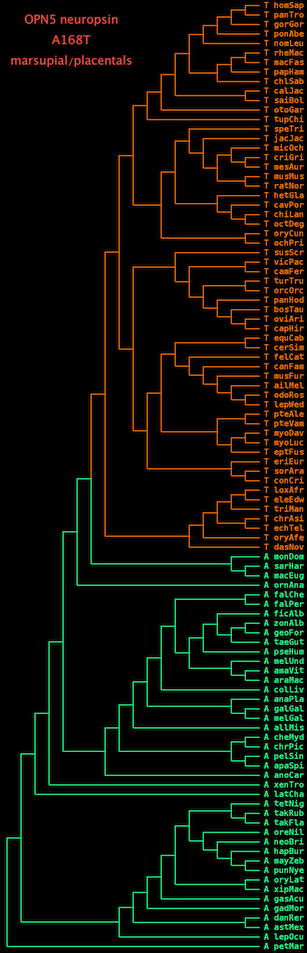
The graphic shows an advanced use of the linearized format. The 100-way and Blast searches were used to obtain 100 orthologs of the opsin gene ONP5 (neuropsin). Position 168 was then extracted from the protein alignment and its value placed in front of the appropriate species, followed by an underscore (_) which is interpeted as a space by | The graphic shows an advanced use of the linearized format. The 100-way and Blast searches were used to obtain 100 orthologs of the opsin gene ONP5 (neuropsin). Position 168 was then extracted from the protein alignment and its value placed in front of the appropriate species, followed by an underscore (_) which is interpeted as a space by phyloPng. | ||
The display was then colored to illustrate that a shift from alanine to threonine occurred at the divergence of placental mammals from marsupials. Alanine was ancestral in deuterostomes and continues to this day to be invariant in non-placentals. The threonine substitution for its part came under strong selection too and has been invariant over several billion years of summed placental branch length. Such substitutions are called phyloSNPs or [http://www.ncbi.nlm.nih.gov/pubmed/23733948 phylogenetically coherent events]. | The display was then colored to illustrate that a shift from alanine to threonine occurred at the divergence of placental mammals from marsupials. Alanine was ancestral in deuterostomes and continues to this day to be invariant in non-placentals. The threonine substitution for its part came under strong selection too and has been invariant over several billion years of summed placental branch length. Such substitutions are called phyloSNPs or [http://www.ncbi.nlm.nih.gov/pubmed/23733948 phylogenetically coherent events]. | ||
| Line 318: | Line 318: | ||
<span style="color: #666666;">Mar07 petMar Petromyzon marinus (lamprey)</span> | <span style="color: #666666;">Mar07 petMar Petromyzon marinus (lamprey)</span> | ||
=== | === Genus and species commonly used in comparative genomics === | ||
The list below provides correctly spelled genus and species for which complete <b>genes</b> are commonly available, either from whole genome sequencing, large-scale cDNA projects, or transcriptome sets. | |||
The first column of numbers | To align a set orthologous genes collected for a specific comparative project, replace the word 'gene' with its HUGO acronym (example PRNP). Then replace the '.' and the spaces with tabs and paste into spreadsheet columns. Then add accession numbers, taxon ID and sequence data to new columns. Replace the final ')' with a ')lineReturn' converts to standard fasta format. (Note the fasta header here is a database in its own right.) | ||
The first column of numbers then sort the rows into the same anthropocentric order of species as seen in the 100-species alignment at the UCSC human genome browser which is the same order as in the 100-way download page at proteinFasta link from the gene details page. | |||
The second column of numbers will sort rows into quasi-phylogenetic ordering (human taken arbitrarily as first). They're in that order now, but some [http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html important web alignment tools] do not have an option to retain input order, meaning that phylogenetic ordering needs to be restored after the alignment for purposes of comparative genomics. | The second column of numbers will sort rows into quasi-phylogenetic ordering (human taken arbitrarily as first). They're in that order now, but some [http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html important web alignment tools] do not have an option to retain input order, meaning that phylogenetic ordering needs to be restored after the alignment for purposes of comparative genomics. | ||
<pre> | <pre> | ||
Latest revision as of 18:17, 19 June 2017
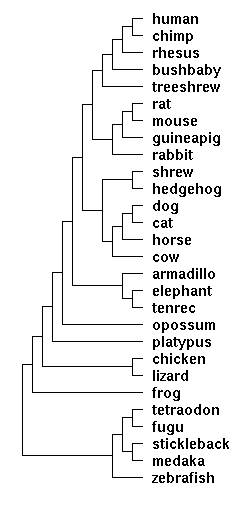
Vertebrate topology used at UCSC genome browser
The tree below shows the phylogenetic relationships of vertebrate species with assembled genomes. Lamprey, which recently became available, is not shown but would appear at the bottom as outgroup to all jawed vertebrates.
Adapted from: 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Research 17(12):1797-808 Dec 2007 Miller W, ...,Pringle TH, Lindblad-Toh K, Gibbs RA, Lander ES, Siepel A, Haussler D, Kent WJ.
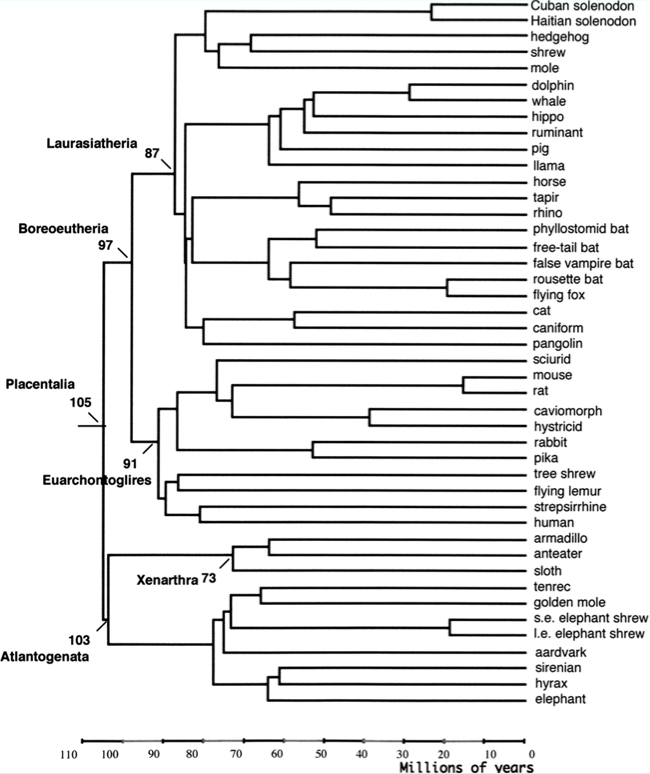
Placental mammal phylogenetic tree
Adapted from: Using genomic data to unravel the root of the placental mammal phylogeny. Genome Research Apr;17(4):413-21 2007 Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W.
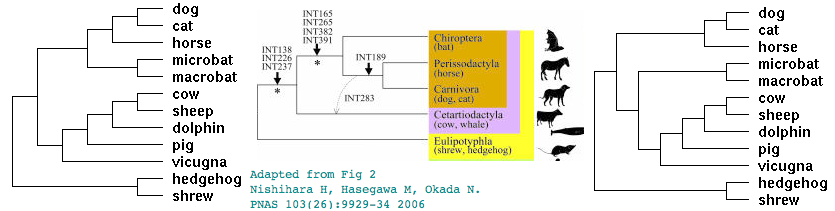
Alternative topologies for Laurasiatheres
The proper arrangement of species within Laurasiatheres is under active investigation. Two of many alternatives are shown along with L1MA9 retroposon data supporting the Pegasoferae arrangement. Pangolins, not shown and genome project apparently canceled, are now known to be the sister group to carnivores.
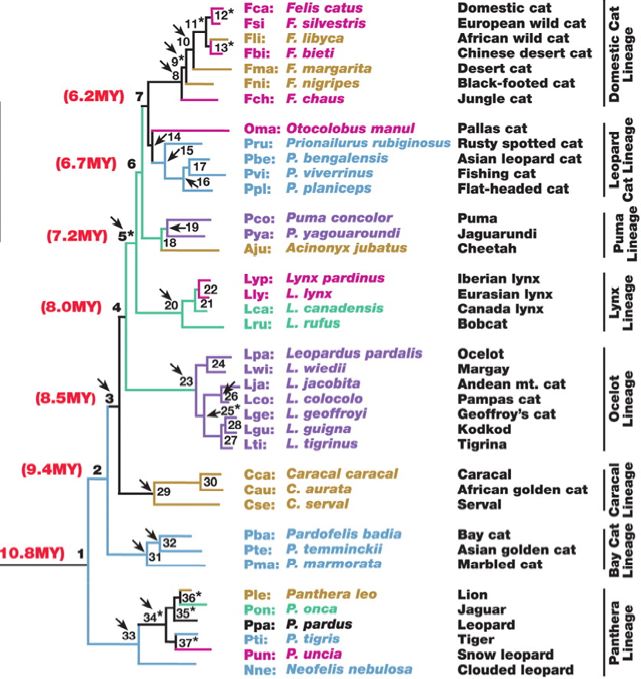
Felid phylogeny
Adapted from: Late Miocene Radiation of Modern Felidae: A Genetic Assessment Science Vol. 311 5757 73 - 77 Jan 2006 Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, O'Brien SJ
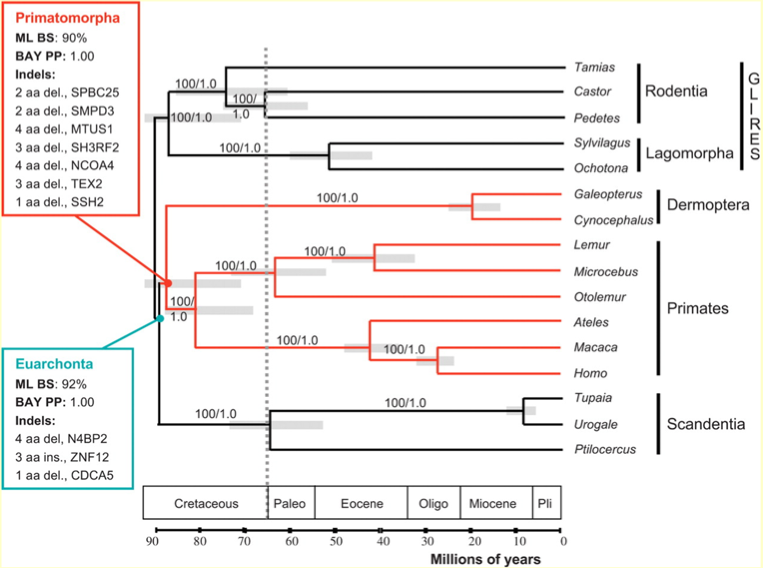
Euarchontoglires: rodents, rabbits, primates
Adapted from: Molecular and genomic data identify the closest living relative of primates. Science Nov 2;318(5851):792-4 2007 Janecka JE, Miller W, Pringle TH, Wiens F, Zitzmann A, Helgen KM, Springer MS, Murphy WJ.
Web tools for drawing phylogenetic trees from Newick format
After grouping the species with nested parentheses (Newick format) that can include divergence dates or substitution rates, the tree can be drawn with various online tools such as phyloPng, Phylodendron, or PhyFi.
Here is a simple example of Newick format: (((((human,chimp),gorilla),orang),gibbon),rhesus);
Two contrasting topologies for Laurasiatheres: (((((dog,cat),horse),(microbat,macrobat)),((((cow,sheep),dolphin),pig),vicugna)),(hedgehog,shrew)); ((((dog,cat),horse),((microbat,macrobat),((((cow,sheep),dolphin),pig),vicugna))),(hedgehog,shrew));
Placental mammals: (((((((((((((+_homSap:5,+_panTro:5):3,-_gorGor:8):3,+_ponPyg:11):14,-_nomLeu:25):10,+_macMul:35):20,+_calJac:55):10,-_tarSyr:65):2,(+_otoGar:58,-_micMur:58):9):10,-_cynVol:77):2,+_tupBel:79):2,((((+_musMus:15,+_ratNor:15):10,+_speTri:25):40,+_cavPor:65):13,+_oryCun:78):3):9,(((((+_canFam:15,+_felCat:15):10,+_equCab:25):20,(+_myoLuc:35,+_pteVam:35):10):20,(+_bosTau:50,+_susScr:50):15):21,(+_sorAra:75,+_eriEur:75):11):4):9,(((+_loxAfr:55, +_proCap:55):37,(-_eleRuf:89, (-_oryAfe:84,+_echTel:84):5):3):5,(+_dasNov:94,(+_choHof:65,-_cycDid:65):29):3):2);
Metazoans: ((((((((((((((((((((((((+_homSap:5,+_panTro:5):3,-_gorGor:8):3,+_ponPyg:11):14,-_nomLeu:25):10,+_macMul:35):20,+_calJac:55):10,-_tarSyr:65):2,(+_otoGar:58,-_micMur:58):9):10,-_cynVol:77):2,+_tupBel:79):2,((((+_musMus:15,+_ratNor:15):10,+_speTri:25):40,+_cavPor:65):13,+_oryCun:78):3):9,(((((+_canFam:15,+_felCat:15):10,+_equCab:25):20,(+_myoLuc:35,+_pteVam:35):10):20,(+_bosTau:50,+_susScr:50):15):21,(+_sorAra:75,+_eriEur:75):11):4):9,(((+_loxAfr:55, +_proCap:55):37,(-_eleRuf:89, (-_oryAfe:84,+_echTel:84):5):3):5,(+_dasNov:94,(+_choHof:65,-_cycDid:65):29):3):2):76, marsupials:175):55,monotremes:230):80,saura:310):90,amphibs:400):50,rayfinned:450):150,jawless:600):50,urochord:650):30,cephalo:680):20,(echino:660,hemi:660):40):100,protostome:800):150,((cnidarian:880,sponge:880):20,placozoa:900):50);
Lesser-known genSpp codes here are thaSir for Thamnophis sirtalis (garter snake) and sphPun for Sphenodon punctatus (tuatura). These species were approved for genome sequencing but to date not begun.
Few people can hand-edit Newick format to include more species or alter relationships. I've developed a linearization of Newick format that puts each species into its own spreadsheet row, separating species and metric data from the "grammar". This allows for easy editing and numerical spreadsheet operations such as to tally up branch lengths in comparative genomics projects. Tabs are ignored by the online tree tools, so the format works by simple paste-in.
((((((((((((((((( homSap : 5 , panTro : 5 ): 3 , gorGor : 8 ): 6 , ponPyg : 14 ): 3 , nomLeu : 17 ): 8 , macMul : 25 ): 20 , calJac : 45 ): 20 , tarSyr : 65 ): 12 ,( otoGar : 60 , micMur : 60 ): 17 ): 8 ,( cynVol : 82 , tupBel : 82 ): 3 ): 2 ,(((( musMus : 16 , ratNor : 16 ): 53 , cavPor : 69 ): 9 ,( dipOrd : 73 , speTri : 73 ): 5 ): 4 ,( ochPri : 80 , oryCun : 80 ): 2 ): 5 ): 8 ,(((((( canFam : 54 , felCat : 54 ): 8 , manPen : 62 ): 11 , equCab : 73 ): 7 ,( myoLuc : 69 , pteVam : 69 ): 11 ): 7 ,((( turTru : 53 , bosTau : 53 ): 8 , susScr : 61 ): 12 , vicPac : 73 ): 14 ): 4 ,( eriEur : 80 , sorAra : 80 ): 11 ): 4 ): 3 ,(( dasNov : 65 , choHof : 65 ): 27 ,(( loxAfr : 59 , proCap : 59 ): 16 , echTel : 75 ): 17 ): 6 ): 27 ,( monDom : 45 , macEug : 45 ): 80 ): 50 , ornAna : 175 ): 135 ,((( galGal : 218 , taeGut : 218 ): 57 , droNov : 275 ): 23 , allMis : 298 ): 12 ): 5 ,(( anoCar : 250 , thaSir : 250 ): 50 , sphPun : 300 ): 15 ): 5 , xenTro : 320 ): 3
The UCSC 100-way vertebrate genome phylogenetic tree in Newick format
Either of the two representations of the vertebrate genome phylogenetic tree, when pasted into the UCSC tree drawing utility phyloPng, will reproduce species relatedness used at UCSC tracks and resources. Note coding genes are given for the 100-way in the order listed (see proteinFasta on gene details page) as both nucleotide and amino acid exonic format.
The first representation shows various higher order tax as individual lines, for example great apes as the first line. (Line returns are not part of Newick format but are generally ignored by drawing tools.) This display makes it slightly easier to add new species. Thus, to add echidna (Tachyglossus aculeatus), the monotreme line can be hand-edited: ornAna), --> (tacAcu,ornAna), as this genome becomes available.
The second representation is linearized to spreadsheet format. This separates the genus species list from parenthetic nesting used to describe topology. Branching times (or evolutionary rates) can be taken from the literature and can be inserted (as shown above) to draw trees whose horizontal lines have quantitative significance.
The graphic shows an advanced use of the linearized format. The 100-way and Blast searches were used to obtain 100 orthologs of the opsin gene ONP5 (neuropsin). Position 168 was then extracted from the protein alignment and its value placed in front of the appropriate species, followed by an underscore (_) which is interpeted as a space by phyloPng.
The display was then colored to illustrate that a shift from alanine to threonine occurred at the divergence of placental mammals from marsupials. Alanine was ancestral in deuterostomes and continues to this day to be invariant in non-placentals. The threonine substitution for its part came under strong selection too and has been invariant over several billion years of summed placental branch length. Such substitutions are called phyloSNPs or phylogenetically coherent events.
((((((((((((((((((homSap,panTro),gorGor),ponAbe),nomLeu), (((rheMac,macFas),papHam),chlSab)), (calJac,saiBol)),otoGar),tupChi), (((speTri,(jacJac,((micOch,(criGri,mesAur)),(musMus,ratNor)))), (hetGla,(cavPor,(chiLan,octDeg)))), (oryCun,ochPri))), ((susScr,((vicPac,camFer),((turTru,orcOrc),(panHod,(bosTau,(oviAri,capHir)))))), ((((equCab,cerSim), (felCat,(canFam,(musFur,(ailMel,(odoRos,lepWed)))))), ((pteAle,pteVam),((myoDav,myoLuc),eptFus))), (eriEur,(sorAra,conCri))))), (((((loxAfr,eleEdw),triMan),(chrAsi,echTel)),oryAfe),dasNov)), (monDom,(sarHar,macEug))), ornAna), (((((((falChe,falPer),(((ficAlb,((zonAlb,geoFor),taeGut)),pseHum),(melUnd,(amaVit,araMac)))),colLiv),(anaPla,(galGal,melGal))), allMis), ((cheMyd,chrPic),(pelSin,apaSpi))), anoCar)), xenTro), latCha), (((((((tetNig,(takRub,takFla)),(oreNil,(neoBri,(hapBur,(mayZeb,punNye))))),(oryLat,xipMac)),gasAcu),gadMor),(danRer,astMex)), lepOcu)), petMar);
(((((((((((((((((( homSap , panTro ), gorGor ), ponAbe ), nomLeu ),((( rheMac , macFas ), papHam ), chlSab )),( calJac , saiBol )), otoGar ), tupChi ),((( speTri ,( jacJac ,(( micOch ,( criGri , mesAur )),( musMus , ratNor )))),( hetGla ,( cavPor ,( chiLan , octDeg )))),( oryCun , ochPri ))),(( susScr ,(( vicPac , camFer ),(( turTru , orcOrc ),( panHod ,( bosTau ,( oviAri , capHir )))))),(((( equCab , cerSim ),( felCat ,( canFam ,( musFur ,( ailMel ,( odoRos , lepWed )))))),(( pteAle , pteVam ),(( myoDav , myoLuc ), eptFus ))),( eriEur ,( sorAra , conCri ))))),((((( loxAfr , eleEdw ), triMan ),( chrAsi , echTel )), oryAfe ), dasNov )),( monDom ,( sarHar , macEug ))), ornAna ),((((((( falChe , falPer ),((( ficAlb ,(( zonAlb , geoFor ), taeGut )), pseHum ),( melUnd ,( amaVit , araMac )))), colLiv ),( anaPla ,( galGal , melGal ))), allMis ),(( cheMyd , chrPic ),( pelSin , apaSpi ))), anoCar )), xenTro ), latCha ),((((((( tetNig ,( takRub , takFla )),( oreNil ,( neoBri ,( hapBur ,( mayZeb , punNye ))))),( oryLat , xipMac )), gasAcu ), gadMor ),( danRer , astMex )), lepOcu )), petMar )
Available genome assemblies as of May 2008
The table is correct as of 01 May 08. The species are listed in quasi phylogenetic order (with human arbitrarily listed first and other subtree ordered by genome quality).
- Traces indicated in millions, eg Trc12 means 12 million traces but no wgs contigs or assembly available
- Wgs08 means wgs division of GenBank contains short assembled contigs searchable with tBlastn
- Mar06 etc means the March 2006 assembly is the most recent available at UCSC
Mar06 homSap Homo sapiens (human) Mar06 panTro Pan troglodytes (chimp) Trc04 gorGor Gorilla gorilla (gorilla) Jul07 ponPyg Pongo pygmaeus (orang_abelii) Trc19 nomLeu Nomascus leucogenys (gibbon) Jan06 macMul Macaca mulatta (rhesus) Trc12 papHam Papio hamadryas (baboon) Trc17 tarSyr Tarsius syrichta (tarsier) Jun07 calJac Callithrix jacchus (marmoset) Dec06 otoGar Otolemur garnettii (bushbaby) Wgs08 micMur Microcebus murinus (mouse_lemur) Trc00 cynVol Cynocephalus volans (flying_lemur) Dec06 tupBel Tupaia belangeri (treeshrew) Jul07 musMus Mus musculus (mouse) Nov04 ratNor Rattus norvegicus (rat) Wgs08 speTri Spermophilus tridecemlineatus (ground_squirrel) Trc07 dipOrd Dipodomys ordii (kangaroo_rat) Wgs08 cavPor Cavia porcellus (guinea_pig) May05 oryCun Oryctolagus cuniculus (rabbit) Wgs08 ochPri Ochotona princeps (pika) May05 canFam Canis familiaris (dog) Mar06 felCat Felis catus (cat) Jan07 equCab Equus caballus (horse) Wgs08 myoLuc Myotis lucifugus (microbat) Trc08 pteVam Pteropus vampyrus (macrobat) Aug06 bosTau Bos taurus (cow) Trc10 turTru Tursiops truncatus (dolphin) Trc06 susScr Sus scrofa (pig) Trc11 vicVic Vicugna vicugna (vicugna) Wgs08 sorAra Sorex araneus (shrew) Wgs08 eriEur Erinaceus europaeus (hedgehog) May05 loxAfr Loxodonta africana (elephant) Trc09 proCap Procavia capensis (hyrax) Jul05 echTel Echinops telfairi (tenrec) May05 dasNov Dasypus novemcinctus (armadillo) Trc09 choHof Choloepus hoffmanni (sloth) Jan06 monDom Monodelphis domestica (opossum) Trc10 macEug Macropus eugenii (wallaby) Mar07 ornAna Ornithorhynchus anatinus (platypus) May06 galGal Gallus gallus (chicken) Trc15 taeGut Taeniopygia guttata (finch) Feb07 anoCar Anolis carolinensis (lizard) Aug05 xenTro Xenopus tropicalis (frog) Jul07 danRer Danio rerio (zebrafish) Feb04 tetNig Tetraodon nigroviridis (pufferfish) Oct04 takRub Takifugu rubripes (fugu) Feb06 gasAcu Gasterosteus aculeatus (stickleback) Apr06 oryLat Oryzias latipes (medaka) Wgs08 calMil Callorhinchus milii (elephantfish) Mar07 petMar Petromyzon marinus (lamprey)
Genus and species commonly used in comparative genomics
The list below provides correctly spelled genus and species for which complete genes are commonly available, either from whole genome sequencing, large-scale cDNA projects, or transcriptome sets.
To align a set orthologous genes collected for a specific comparative project, replace the word 'gene' with its HUGO acronym (example PRNP). Then replace the '.' and the spaces with tabs and paste into spreadsheet columns. Then add accession numbers, taxon ID and sequence data to new columns. Replace the final ')' with a ')lineReturn' converts to standard fasta format. (Note the fasta header here is a database in its own right.)
The first column of numbers then sort the rows into the same anthropocentric order of species as seen in the 100-species alignment at the UCSC human genome browser which is the same order as in the 100-way download page at proteinFasta link from the gene details page.
The second column of numbers will sort rows into quasi-phylogenetic ordering (human taken arbitrarily as first). They're in that order now, but some important web alignment tools do not have an option to retain input order, meaning that phylogenetic ordering needs to be restored after the alignment for purposes of comparative genomics.
>10.10.gene_homSap Homo sapiens (human) >11.11.gene_panTro Pan troglodytes (chimp) >99.12.gene_gorGor Gorilla gorilla (gorilla) >99.13.gene_ponPyg Pongo pygmaeus (orang_sumatran) >99.14.gene_nomLeu Nomascus leucogenys (gibbon) >12.15.gene_macMul Macaca mulatta (rhesus) >12.15.gene_macFas Macaca fascicularis (crab-eating macaque) >12.15.gene_macNem Macaca nemestrina (pig-tailed macaque) >99.16.gene_papAnu Papio anubis (baboon) >99.17.gene_papHam Papio hamadryas (baboon) >99.18.gene_calJac Callithrix jacchus (marmoset) >99.19.gene_tarSyr Tarsius syrichta (tarsier) >13.20.gene_otoGar Otolemur garnettii (bushbaby) >99.21.gene_micMur Microcebus murinus (mouse_lemur) >99.22.gene_cynVol Cynocephalus volans (flying_lemur) >14.23.gene_tupBel Tupaia belangeri (tree_shrew) >15.24.gene_musMus Mus musculus (mouse) >16.25.gene_ratNor Rattus norvegicus (rat) >17.26.gene_cavPor Cavia porcellus (guinea_pig) >99.27.gene_speTri Spermophilus tridecemlineatus (squirrel) >99.28.gene_dipOrd Dipodomys ordii (kangaroo_rat) >18.29.gene_oryCun Oryctolagus cuniculus (rabbit) >99.30.gene_ochPri Ochotona princeps (pika) >21.31.gene_canFam Canis familiaris (dog) >22.32.gene_felCat Felis catus (cat) >23.36.gene_equCab Equus caballus (horse) >99.37.gene_myoLuc Myotis lucifugus (microbat) >99.38.gene_pteVam Pteropus vampyrus (macrobat) >99.39.gene_turTru Tursiops truncatus (dolphin) >24.33.gene_bosTau Bos taurus (cow) >99.34.gene_oviAri Ovis aries (sheep) >99.35.gene_susScr Sus scrofa (pig) >99.41.gene_vicVic Vicugna vicugna (vicugna) >19.42.gene_eriEur Erinaceus europaeus (hedgehog) >20.43.gene_sorAra Sorex araneus (shrew) >99.44.gene_borAnc Boreoeuthere ancestralis (ancestral) >25.45.gene_dasNov Dasypus novemcinctus (armadillo) >99.46.gene_choHof Choloepus hoffmanni (sloth) >26.47.gene_loxAfr Loxodonta africana (elephant) >99.48.gene_proCap Procavia capensis (hyrax) >99.49.gene_echTel Echinops telfairi (tenrec) >27.50.gene_monDom Monodelphis domestica (opossum) >99.51.gene_macEug Macropus eugenii (wallaby) >99.52.gene_triVul Trichosurus vulpecula (possum) >28.53.gene_ornAna Ornithorhynchus anatinus (platypus) >99.54.gene_tacAcu Tachyglossus aculeatus (echidna) >30.55.gene_galGal Gallus gallus (chicken) >99.56.gene_taeGut Taeniopygia guttata (finch) >29.57.gene_anoCar Anolis carolinensis (lizard) >31.58.gene_xenTro Xenopus tropicalis (frog) >99.59.gene_xenTro Xenopus laevis (frog) >99.60.gene_neoFor Neoceratodus forsteri (lungfish) >32.61.gene_danRer Danio rerio (zebrafish) >33.62.gene_tetNig Tetraodon nigroviridis (pufferfish) >34.63.gene_takRub Takifugu rubripes (fugu) >35.64.gene_gasAcu Gasterosteus aculeatus (stickleback) >36.65.gene_oryLap Oryzias latipes (medaka) >99.66.gene_ictPun Ictalurus punctatus (fish) >99.67.gene_oncMyk Oncorhynchus mykiss (trout) >99.68.gene_funHet Fundulus heteroclitis (flounder) >99.69.gene_calMil Callorhinchus milii (elephantfish) >99.70.gene_squAca Squalus acanthias (spiny dogfish) >99.71.gene_petMar Petromyzon marinus (lamprey) >99.72.gene_braFlo Branchiostoma floridae (amphioxus)